Abstract
Klebsiella pneumoniae is a notorious human pathogen involved in healthcare-associated infections. The worldwide expansion of infections induced by colistin-resistant and carbapenemase-producing Enterobacterales (CPE) isolates has been increasingly reported. This study aims to analyze the phenotypic and molecular profiles of 10 colistin-resistant (CR) isolates and 2 pairs of colistin-heteroresistant (ChR) (parental and the corresponding resistant mutants) isolates of K. pneumoniae CPE sourced from two hospitals. The phenotypes of strains in the selected collection had been previously characterized. Antimicrobial susceptibility testing was performed using a Vitek 2 Compact system (BioMérieux SA, Marcy l’Etoile, France), the disc diffusion method, and broth microdilution (BMD) for colistin. Whole-genome sequencing (WGS) did not uncover evidence of any mobile colistin resistance (mcr) genes, although the mgrB gene of seven isolates appeared to be disrupted by insertion sequences (ISKpn25 or ISKpn26). Possible deleterious missense mutations were found in phoP (L4F), phoQ (Q426L, L26Q, L224Q, Q317K), pmrB (R256G, P95L, T157P, V352E), and crrB (P151S) genes. The identified isolates belonged to the following clonal lineages: ST101 (n = 6), ST147 (n = 5), ST258 (n = 2), and ST307 (n = 1). All strains harbored IncF plasmids. OXA-48 producers carried IncL and IncR plasmids, while one blaNDM-1 genome was found to harbor IncC plasmids. Ceftazidime–avibactam remains a therapeutic option for KPC-2 and OXA-48 producers. Resistance to meropenem–vaborbactam has emerged in some blakPC-2-carrying isolates. Our study demonstrates that the results of WGS can provide essential evidence for the surveillance of antimicrobial resistance.
1. Introduction
Recently, it has been reported that carbapenemase-producing Enterobacterales (CPE) isolates are expanding at an alarming rate around the globe [1,2]. These Gram-negative bacilli are responsible for causing a variety of hospital-acquired and community-onset human infections, with Klebsiella pneumoniae being the principal pathogen associated with significant mortality [2,3,4].
Antimicrobial resistance of K. pneumoniae strains continues to be problematic [5]. Resistance to carbapenems is frequently associated with resistance to multiple other classes of antibiotics, which leads to limited possibilities for the treatment of infections induced by these multidrug-resistant (MDR) and extensively drug-resistant (XDR) pathogens [3,5,6]. Around the world, the geographical distribution of carbapenem resistance is reported to be heterogeneous [7].
The main carbapenem-hydrolyzing enzymes, K. pneumoniae carbapenemase (KPC), metallo-β-lactamases (MBL), and oxacillinase-48-like (OXA-48-like), are principally encoded on plasmids and linked with diverse mobile genetic elements [8].
The global propagation of carbapenemases is the result of a continuous interaction between the clonal dispersion of some efficient CPE lineages and the horizontal transfer of their resistance genes [9]. KPC-producing K. pneumoniae multilocus sequence type ST258/512 is a recognized “high-risk international clone” [2,10]. In Europe, ST258/512 and three additional clonal lineages, ST11, ST15, and ST101, along with their derivatives, are predominant [2,10], offering stability for carbapenemase genes [9]. The issues that remain unresolved are the distinct success of K. pneumoniae ST258 and the special relationship with epidemic resistance plasmids of incompatibility group F (IncF), which have diverse replicon types (FIA, FIB, and FII) [11,12]. The acquired antimicrobial resistance genes located on IncF can rapidly disseminate within species, while those situated on other types of plasmids, such as IncA/C, IncL/M, and IncN, can be transmitted between species [11,12]. The latter three types of plasmids are unusual in K. pneumoniae ST258 and have not been determined to contribute to international dominance [10]. IncL/M type plasmids are associated with blaOXA-48-like genes, and IncA/C and IncN types with blaNDM genes that encode New Delhi metallo-β-lactamases (NDMs) [9,11].
The situation has evolved with the emergence of K. pneumoniae strain ST23, which integrates both hypervirulence and resistance to carbapenems, with dramatic clinical consequences [2,4,13]. Biomarkers present on virulence plasmids have been shown to accurately differentiate hypervirulent from classical strains. The presence of the plasmid-associated gene loci peg-344, iro (salmochelin biosynthesis), iuc (aerobactin synthesis), rmpA (regulator of mucoid phenotype), and rmpA2 are highly predictive of strains as hypervirulent [4,13,14].
The worldwide escalation of colistin-resistant CPE isolates is of significant concern [15]. Colistin is a reserve antimicrobial agent with bactericidal action, principally as a consequence of interactions with the outer and inner membranes of Gram-negative bacilli [15,16]. Multiple chromosomal mutations constitute the main substrate of acquired resistance to colistin and promote alterations of the outer membrane lipopolysaccharide (LPS) with an abnormal positive charge, which decreases the affinity and action of colistin [17]. This strategy implies that the addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoethanolamine (PEtN) molecules induces the covalent transformation of lipid A of LPS [17]. A panel of various genes modulates these modifications, essentially based on the two-component regulatory systems (TCRSs) phoPQ and pmrAB and the feedback regulator mgrB gene and the colistin resistance regulation (crrAB) operon, respectively [17,18]. Transposition of insertion sequences (ISs) such as ISL3 (ISKpn25), IS5 (ISKpn26), ISKpn14, and IS903B from plasmids into the mgrB gene mediates the inactivation and disruption of this regulator gene, conferring colistin resistance [19,20,21]. Antibiotic heteroresistance is considered to be the presence of subpopulations with increased resistance within the majority cell population in the same culture [22]. However, a coherent definition and global guidelines for antibiotic heteroresistance detection are lacking, which complicates its diagnosis [22]. Of particular concern is the emergence of transferable colistin resistance via plasmids [5,15,16,18], with the discovery of a series of mobile colistin resistance (mcr) genes, mcr-1 to mcr-10, with diverse variants [23] that encode PEtN, which is incorporated into lipid A [15,23].
Despite continuous research activity, there is still an incomplete understanding of the molecular mechanisms of colistin resistance and heteroresistance [16,24], along with limited data regarding their dissemination and impact [5,18,25]. Whole-genome sequencing (WGS) has become an essential modern tool for interrogating both existing and emerging mechanisms of antibiotic resistance, and it has excellent potential in infection control surveillance [26].
Consequently, the aim of the study is to perform a detailed phenotypic and genomic characterization of 10 colistin-resistant (CR) isolates and 2 pairs of colistin-heteroresistant (ChR) (parental and the corresponding resistant mutants) isolates of K. pneumoniae CPE, collected from two Romanian hospitals in 2017 and 2021, toward delineating the variety of molecular substrates of colistin resistance and heteroresistance, identifying the genetic determinants of resistance to other classes of antibiotics, confirming the presence of circulating high-risk clones and dominant plasmids, and exploring potential therapeutic options, including novel β-lactam/β-lactamase inhibitor combinations.
2. Results
2.1. Phenotypic Antimicrobial Susceptibility Testing
All isolates presented resistance to multiple classes of antimicrobial agents, specifically meropenem, ertapenem, doripenem, aztreonam, cephalosporins, old generations of β-lactam/β-lactamase inhibitors, ciprofloxacin, and levofloxacin. The phenotypic results of antibacterial drug testing are given in Table 1.

Table 1.
Antimicrobial resistance profiles obtained by Vitek 2 Compact software and the disc diffusion method * for all strains.
Of the three aminoglycosides tested, gentamicin was the most active in vitro (n = 8 susceptible isolates), while total resistance was noted for tobramycin (Table 1). Most of the isolates (n = 12) were susceptible to tigecycline and ceftazidime–avibactam (Table 1). Imipenem–relebactam exhibited potent activity against all KPC producers, while meropenem–vaborbactam showed activity against only five out of the eight KPC producers (Table 1).
Minimum inhibitory concentrations (MICs) of colistin obtained by Vitek 2 Compact software are given in Table 1.
Colistin susceptibility test results obtained with other phenotypic methods and reference broth microdilution (BMD) are given in Table 2. MIC values of colistin obtained by BMD ranged from 0.25 to >64 mg/L (Table 2).

Table 2.
Colistin testing results obtained using different assays.
2.2. Whole-Genome Sequencing Data Analysis
2.2.1. Quality Metrics for the Assembled Draft Genomes
SPAdes assembly metrics for the sequenced genomes are presented in Table 3. They suggest that all the sequences are adequate for downstream analysis.

Table 3.
Quality control parameters for draft genomes.
2.2.2. Genetic Determinants of Antimicrobial Resistance
All genomes belonged to MDR strains, with several defined resistance genes against different categories of antibiotics, along with other antibiotic resistance determinants, such as insertions, point mutations, or substitutions that lead to the disruption of genes, thus leading to resistance to at least six antibiotic classes.
(A) Molecular mechanism of resistance to various antibiotic classes, except colistin
The highest heterogeneity of resistance-conferring elements was noted for β-lactams (n = 13), aminoglycosides (n = 11), and fluoroquinolones (n = 8) (Table 4). Isolates blaNDM-1-positive BC7_BM and BC1_TM presented the highest numbers of genetic determinants (n = 28 and n = 22, respectively), while the strain BC2_BM OXA-48 producer had the lowest (n = 12) (Table 4).

Table 4.
Antimicrobial resistance determinants (except colistin) detected in all sequenced genomes.
In agreement with the phenotypic results, the genotypic analysis confirmed the presence of carbapenem-hydrolyzing enzymes responsible for carbapenem resistance.
For cephalosporin resistance, extended-spectrum β-lactamases (ESBLs) were the molecular elements most frequently detected, represented by blaCTX-M-15 (n = 12), followed by blaSHV-12 (n = 2). All blaOXA-48 genes were accompanied by blaCTX-M-15 genes. The NDM-1 producer isolate BC1_TM co-presented blaCTX-M-15 and blaSHV-12 genes. In the genome of isolate BC7_BM, blaNDM-1, blaCTX-M-15, and blaCMY-16 genes were found (Table 4).
Genes encoding the aminoglycoside-modifying enzyme (AME) were the main elements conferring resistance to aminoglycosides in all strains, with the aac(6′)-Ib gene being the most commonly encountered (n = 12 strains) (Table 4). All isolates harbored two to six AME genes, and only isolate BC7_BM co-expressed the rmtC gene, a type of 16S rRNA methyltransferase (RMT) (Table 4). Amikacin nonsusceptible isolates encoded the following genes: aac(6′)-Ib (n = 12), aac(6′)-Ib-cr5 (n = 2), aph(3′)-VI (n = 2), and rmtC (n = 1) (Table 4). Gentamicin nonsusceptible strains possessed the aac(3)-IIe gene (n = 3), followed by the aac(3)-IId gene (n = 1) (Table 4). The aac(6′)-Ib gene (n = 6) and the rmtC gene (n = 1) were responsible for tobramycin resistance.
The oqxAB efflux pump genes and various chromosomal mutations in the quinolone resistance-determining region (QRDR) of gyrA and parC genes contributed to fluoroquinolone resistance in all the analyzed strains. Six strains were observed with the genotype profile of gyrA D87N and parC S80I mutations (Table 4). All the isolates carried a gyrA S83 mutation, with gyrA S83Y identified in six strains and gyrA S83I in eight. Additionally, plasmid-mediated quinolone resistance (PMQR) genes were noted: aac(6′)-Ib-cr5 (n = 2) and qnrS1 (n = 1) (Table 4).
A number of alterations in the principal outer membrane of porin-encoding genes ompK36 and ompK37, which potentially confer resistance to cephalosporins and carbapenems, identified by ResFinder, are summarized in Supplementary Table S1.
Similarly, using BioEdit for sequence alignment, the glycine–aspartic acid duplication (GD) (position 134–135 duplicated in 136–137) in ompK36 was visualized in five strains (BC2_BM, BC4_BM, BC5_TM, BC9_TM, and BC10_TM). The insertion of ISKpn26 (IS5) upstream of ompK36 (position -48 versus the start of ompK36 gene), which leads to ompK36 inactivation, was visualized in WGS data and further confirmed by Sanger sequencing (data not shown) in the BC3_TM and BC8_BM isolates. The BC1_TM strain possessed an N186K mutation, while BC7_BM carried several single nucleotide polymorphisms (SNPs), along with deletions, insertions, and substitutions. Similarly, the isolates BC3_TM, BC7_BM, and BC8_BM presented SNPs and substitutions in ompK37, while an insertion in ompK35 that produced a frame shift (FS) at amino acid 166 was noted in isolate BC6_BM. ClustalW (BioEdit) alignment of ompK35, ompK36, and ompK37 genes is shown in Supplementary Figure S1.
(B) Molecular determinants of colistin resistance
The genomic results indicate the absence of plasmidial mcr genes.
At least one colistin-resistance chromosomal determinant was noted in the mgrB gene or in the two-component regulatory systems phoPQ, pmrAB, and crrAB (Table 5).

Table 5.
Cumulative colistin resistance determinants.
For the three CR strains (BC1_TM, BC6_BM, and BC9_TM) with the highest colistin MIC value by BMD, 32 mg/L, two distinct classes of insertion sequences, ISL3 (ISKpn25) and IS5 (ISKpn26), were identified to disrupt the mgrB gene in three different nucleotide positions, rendering it nonfunctional (Table 5). Four other CR strains had an M27K SNP in the mgrB gene. Additionally, other SNPs identified in CR isolates with the potential to change colistin sensitivity were in phoP (L4F), phoQ (L26Q, Q426L), and pmrB genes (P95L, T157P, R256G, V352E) (Table 5).
As in the case of BC6_BM, the two ChR strains and their corresponding resistant mutants harbored ISL3 (ISKpn25) in the same nucleotide position and an R256G SNP in the pmrB gene, but with a supplementary presumptive intolerant L224Q SNP in the phoQ gene. Notably, only the resistant mutants BC12_TM_B_m and BC14_TM_C_m presented deleterious Q317K SNPs in the phoQ gene and P151S in the crrB gene (Table 5).
In contrast, no changes to pmrA and crrA genes resulting in functional amino acid substitutions were identified in any isolates; only changes encoding neutral amino acid substitutions were identified (Table 5).
The Protein Variation Effect Analyzer (PROVEAN) scores for amino acid changes with a potentially harmful influence on the biological function of the analogous proteins are given in Table 6. The sorting intolerant from tolerant (SIFT) algorithm also confirmed the tolerant/intolerant classification.

Table 6.
PROVEAN prediction for all possible deleterious amino acid substitutions detected in PhoP, PhoQ, PmrB, and CrrB proteins.
(C) Association between phenotypic antimicrobial susceptibility test results and genotypic prediction
In the case of strains BC3_TM and BC8_BM, despite being characterized by Vitek 2 Compact as being resistant to gentamicin, no corresponding molecular resistance element was noted (Table 1 and Table 4). Similarly, the instrument indicated that BC1_TM, BC2_BM, BC4_BM, and BC10_TM were resistant to chloramphenicol, but no genetic resistance markers were detected (Table 1 and Table 4). In these two situations, the disc diffusion method indicated susceptibility to the same antimicrobial agents in all cases. Strain BC9_TM was phenotypically susceptible to amikacin and trimethoprim–sulfamethoxazole, but resistance genes aac(6′)-Ib-cr5 and dfrA14 were detected. Two isolates, BC7_BM and BC9_TM, were categorized as susceptible to tigecycline, but tet(A) and tet(D) genes were identified. Two other strains (BC4_BM and BC10_TM) that showed intermediate susceptibility to tigecycline did not carry tetracycline resistance markers. Conversely, isolate BC14_TM_C_m was interpreted as resistant to chloramphenicol based on both testing methods, but no resistance genes were found upon genetic analysis (Table 1 and Table 4). For the remaining antimicrobial compounds, except colistin, no disagreement between phenotypic profiles and molecular elements was noted (Table 1 and Table 4).
Interestingly, in the case of BC3_TM, the colistin susceptibility profile based on Vitek 2 Compact revealed a discrepancy between the CMI results obtained concomitantly by AST-N222 and AST-XN05 cards, though molecular markers of colistin resistance were detected (Table 1 and Table 5). In the case of BC2_BM, disagreement was noted between the Vitek 2 Compact results and the molecular analysis of colistin. All BMD colistin results correlated strongly with genotypic findings, except in the case of two ChR isolates.
The concordance between molecular and phenotypic susceptibility testing results for Vitek 2 Compact and for disc diffusion was 96.42% (297/308 × 100) and 98.21% (330/336 × 100), respectively.
2.2.3. Molecular Serotyping, Plasmid Replicon Identification, and Sequence Type Determination
Molecular serotyping determined that six strains were KL17;O1v1, five were KL112;O2v2, two were KL106;O2v2, and one isolate belonged to serotype KL102;O2v2 (Table 7). No gene characteristics of hypervirulence were identified by the Virulence Factor Database (VFDB) tool.

Table 7.
Serotype, MLST, cgMLST, and plasmid replicons of all isolates.
The identified replicons covered mainly Col, IncFIA, IncFIB, IncFII, IncC, IncL, IncR, and IncX3 type plasmids, with a total of 12 types (Table 7). Virulence-encoding plasmid replicon types IncFIB(K) and IncFII(K) were identified in 12 strains. More than half of the isolates were positive for ColRNAI. IncFIB(pQil) was observed, along with other replicon types, in six strains (Table 7). The genome of all the OXA-48 producers carried IncL and IncR plasmids, while one blaNDM-1 genome was associated with the IncC plasmid (Table 7). The correlation between ST258 clones and the IncF-carrying plasmid with FII(K) replicons was ascertained. The IncX3 plasmid was positive in two KPC-2-producing isolates. In the genome of strains BC6_BM, BC11_TM_B_hR, BC12_TM_B_m, BC13_TM_C_hR, and BC14_TM_C_m, two replicons from the same incompatibility class were identified, IncFIB(K) and IncFIB(pQil), suggesting the presence of multi-replicon plasmids along with other replicons.
In the mapping of raw reads of the two blaNDM-1 sequenced strains to 2014–2015 Târgu Mureș blaNDM-1 plasmid sequences, no similarity was found between reference plasmids and blaNDM-1 strain BC1_TM. However, the historical plasmid pNDM_18ES had 96% coverage with 99.89% identity by the reads of BC7_BM strain, and plasmids 1TM and 6TM had 76% and 73% coverage, respectively, with 99.9% identity.
Four sequence types (STs) were assigned by multilocus sequence typing (MLST): ST101 (n = 6), ST147 (n = 5), ST258 (n = 2), and ST307 (n = 1) (Table 7). The colistin-resistant mutants presented the same ST as those of their parental strains, namely, ST147.
Nine complex types were identified by core genome MLST (cgMLST). Five of them (CT5839, CT5840, CT5853, CT5854, and CT5848) were considered new founders by SeqSphere and were included in the K. pneumoniae database of the cgMLST.org Nomenclature Server.
Overall, ST101 isolates presented the highest number of distinct plasmid replicon types (n = 7 types). KL17 and KL112 serotypes were exclusively associated with ST101 and ST147, respectively.
The minimum spanning tree of the 14 CR and ChR strains revealed three clusters of closely related strains (Figure 1):
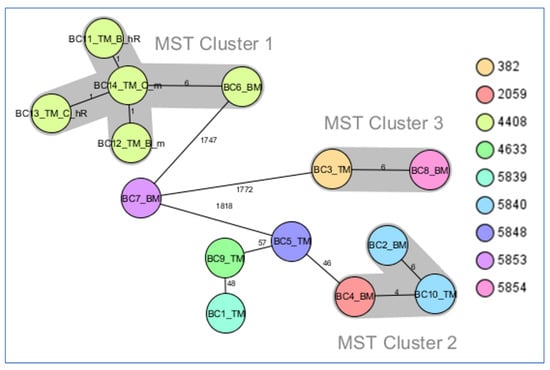
Figure 1.
Minimum spanning tree for K. pneumoniae strains investigated in this study generated from cgMLST data. Ridom SeqSphere+ MST for 14 samples based on 2358 columns, pairwise ignoring missing values, logarithmic scale. Distance based on columns from K. pneumoniae sensu lato cgMLST (2358). MST cluster distance threshold: 15. Nodes colored by complex type.
- MST cluster 1, comprising blaKPC-2 producers: two pairs of ChR with their corresponding resistant mutants (from Târgu Mureș) and BC6_BM (from Baia Mare), collected in the period 2019–2020.
- MST cluster 2, consisting of three blaOXA-48 strains, BC2_BM, BC4_BM, and BC10_TM (two from Baia Mare and one from Târgu Mureș), collected in the period 2018–2019.
- MST cluster 3, comprising two blaKPC-2 strains, BC3_TM and BC8_BM (from Târgu Mureș and Baia Mare, respectively), collected in the period 2017–2018.
3. Discussion
In this study, we analyzed the phenotypic features, resistome, virulome, and plasmid content of 10 CR and 2 pairs of ChR (parental and the corresponding resistant mutants) isolates of K. pneumoniae CPE, collected from two Romanian hospitals between 2017 and 2021. This information is of critical epidemiological importance in the context of the continuous global expansion of high-risk MDR K. pneumoniae clones [1,2,9,10,27]. Additionally, the European Antimicrobial Resistance Surveillance Network (EARS-Net) received reports of an increasing trend of invasive infections due to K. pneumoniae CPE isolates at Romanian hospitals, from 32.3% in 2019 to 48.3% in 2020 [5,28]. The varying degrees of resistance observed between European countries may be partially explained by differences in antimicrobial consumption, with the lowest rates of both resistance and use in Northern and Central Europe [10,28].
To the best of our knowledge, this is the first Romanian study that focuses on evaluating molecular determinants of resistance in CR and ChR K. pneumoniae CPE isolates. The results may also contribute to mitigating the spread of antimicrobial resistance in Romanian hospitals.
In line with previous reports [2,3,5], our phenotypic findings reflect the remarkable rates of resistance to several classes of antimicrobial agents; only a few pathogens remain susceptible to gentamicin and trimethoprim–sulfamethoxazole, but most exhibit susceptibility to tigecycline. Unsurprisingly, the two NDM-1-positive isolates were susceptible only to tigecycline. New compounds with targeted action against NDM producers have been developed, but none have yet been approved for clinical use [29]. Additionally, among the new combinations of β-lactam/β-lactamase inhibitors, ceftazidime–avibactam and imipenem/cilastatin–relebactam were found to be reliable treatment options for all of our KPC producers, while ceftazidime–avibactam exhibited supplementary in vitro activity against all blaOXA-48-carrying isolates. In contrast, the resistance to meropenem–vaborbactam in three out of the eight KPC producers represents an unwelcome occurrence in the context of patients who have not previously been exposed to this promising novel antimicrobial compound.
The resistome analysis confirms the existence of an extensive repertoire of antibiotic resistance genes, along with insertions or point mutations, and all our isolates possessed determinants of resistance to β-lactams, amikacin, tobramycin, fosfomycin, fluoroquinolones, colistin, and phenicol. Similarly, several previous reports described a great variety of resistance genes in K. pneumoniae isolates with the MDR or XDR phenotype [27,30,31,32] and pointed out the potential flexibility of these pathogens to accumulate and exchange antimicrobial resistance [27]. Furthermore, the concurrent carriage of multiple β-lactamase genes with overlapping hydrolytic activity was confirmed in each of our analyzed strains. The implications of this have not yet been entirely elucidated, but it might have provided an evolutionary advantage for K. pneumoniae by offering an additional reliable basis for resistance [27].
In the current evaluation, concordance of more than 96% between phenotypic and genomic predictions of antimicrobial resistance was observed. Similarly, Ruppe et al., obtained more than 96% concordance in a study that included 187 Enterobacterales strains tested phenotypically using the disc diffusion method [33]. However, despite WGS being considered a robust surveillance tool [26,32], there is still insufficient published evidence to support its use as a highly accurate instrument to predict antimicrobial susceptibility phenotypes from genomic traits, and it is necessary to establish a consensus regarding which database to interrogate for the detection of antimicrobial resistance genes [33,34].
Of note, some discrepancies were identified in this study. Two isolates, BC3_TM and BC8_BM, were categorized as resistant to gentamicin by Vitek 2 Compact and susceptible by disc diffusion, but no AME or RMT genes were detected. However, inadequate correspondence between genotypes and inference of the presence of substrates with resistance to aminoglycosides have been extensively reported based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints in association with expert rules [35]. Instead, consistent with Ruppe et al. [33], the BC9_TM strain was found to possess the aac(6′)-Ib-cr5 gene but without phenotypic expression of amikacin resistance. Vaziri et al. underscored the fundamental role of the qnrB gene in the K. pneumoniae genome, which, in association with the aac(6′)-Ib-cr5 gene, contributes to increased resistance to aminoglycosides, fluoroquinolones, and cephalosporins [36], a conclusion that was not supported by our findings. A recent nationwide epidemiological survey conducted in Greece confirmed a divergence between the resistance phenotype and the AME genotype for aminoglycosides, probably as a consequence of various competing resistance mechanisms associated with the distinct catalytic activities of AME genes [37]. In addition, molecular detection of a resistance gene does not necessarily indicate expression and activity [38].
The catA1 acetyltransferase and the nonenzymatic cmlA5 genes, encoding resistance to chloramphenicol, were detected in most of our strains. However, discrepant results for chloramphenicol were noted in the case of four strains (BC1_TM, BC2_BM, BC4_BM, and BC10_TM), showing resistance based on Vitek 2 Compact and susceptibility based on the Kirby–Bauer method, while no resistance genes were detected by WGS. The contribution of another mechanism responsible for chloramphenicol resistance (high expression of efflux systems) should not be underestimated [32], given that all of our isolates harbor the oqxAB efflux pump.
Previous studies in Romania have reported on the worrisome emergence of the rmtC gene in NDM-1-positive isolates belonging to the order Enterobacterales [39,40]. This observation is supported by the results of the present study, in which isolate BC7_BM was found to co-carry the rmtC gene along with blaNDM-1, blaCTX-M-15, and blaCMY-16 resistance genes and express pan-aminoglycoside resistance. Similarly, the co-expression of NDM-1 and rmtC genes, along with at least one ESBL-encoding gene in K. pneumoniae isolates, has previously been documented in Kenya, Turkey, and Saudi Arabia [41,42,43]. Since the first description of the RMT gene aminoglycoside resistance methylase (armA) in K. pneumoniae in 2003 [44], eight other plasmid-mediated variants (rmtA to rmtH) and N1-A1408 methyltransferase (MTase) (npmA) have emerged in Gram-negative pathogens in various parts of the world [45]. In contrast to our findings, a study performed in Switzerland between 2017 and 2020, which analyzed 103 carbapenem- and aminoglycoside-resistant Enterobacterales strains, did not find any K. pneumoniae isolates harboring the rmtC gene; the most frequently identified was the armA gene [46]. Among the RMT genes, armA, rmtB, and rmtC genes are distributed worldwide and confer high-level, broad-range aminoglycoside resistance, including against plazomicin, a newly approved aminoglycoside compound that can evade virtually all clinically relevant AMEs, including acetyltransferase (aac), phosphotransferase (aph), and adenylyltransferase (aad or ant) [45]. The association between RMT genes and blaNDM, blaKPC, and mcr genes is a multifaceted topic because of the possible involvement in the expansive dissemination of extensively pandrug-resistant organisms [45].
Among our three blaKPC-2-carrying isolates resistant to meropenem–vaborbactam (BC3_TM, BC5_TM, and BC8_BM), detailed molecular analysis verified their affiliation with the international high-risk lineages ST258 and ST101, with evidence of either GD134-135 duplication in the ompK36 gene or insertion of ISKpn26 (IS5) upstream of ompK36 in association with SNPs and substitutions in the ompK37 porin gene. Furthermore, even though BC6_BM showed susceptibility to meropenem–vaborbactam, an FS mutation at amino acid 166 in ompK35 was recorded. These mutations are responsible for alterations in permeability that contribute to reduced susceptibility to meropenem–vaborbactam, as previously reported [47,48,49]. Meropenem–vaborbactam penetrates the outer membrane of K. pneumoniae mainly through ompK35 and ompK36 porins, but vaborbactam prefers the latter, which has a narrower inner channel [50]. However, the single presence of a nonfunctional ompK35 porin, without mutation in ompK36, is associated with MIC values of ≤0.06 mg/L meropenem–vaborbactam [47], and a higher degree of meropenem–vaborbactam inactivation was demonstrated only upon ompK36 porin loss of function, either in isolation or concurrently with ompK35 [50].
In agreement with other reports from diverse geographical regions [30,31,32], chromosomal mutations associated with colistin resistance were revealed in all examined strains in the current study, and no plasmid-borne mcr genes were identified.
MgrB, a small negative regulatory transmembrane protein, represses phoPQ signaling, but the expression of this TCRS regulator is upregulated in the presence of a modified mgrB gene, and it remodels LPS through the addition of cationic L-Ara4N, which prevents colistin molecules attaching to the LPS membrane [17,18,51]. Mutation of mgrB gene was the basis for resistance most frequently encountered in our study (11 out of the total strains) and was induced by either ISKpn25 (n = 6), ISKpn26 (n = 1), or a missense mutation M27K (n = 4), predicted as deleterious by bioinformatics tools. Several recent studies have highlighted the pivotal role of ISs in contributing to the emergence of colistin resistance by disrupting the mgrB gene [19,20,21,30,31,32]. Our findings provide evidence that plasmid IncFIB(pQil) encodes ISkpn25, while the ISKpn26 element is associated with IncFIA(HI1) and IncR plasmids. Fordham et al. demonstrated similar connections between these IS elements and their companion plasmid families, with the exception of the relationship between ISKpn26 and IncFII(pHN7A8) plasmids [19]. The M27K mutation in the mgrB gene was also reported to mediate colistin resistance in a previous study [52]. In our analysis, only strain BC5_TM presented the M27K mutation as the sole potential marker of resistance to colistin. In contrast, Liu et al. concluded that the expression of MgrB protein was not affected by the M27K mutation, even though the strain exhibited an MIC of 32 mg/L colistin [53]. Inactivation of mgrB has also been shown to promote virulence in K. pneumoniae isolates by suppressing the initiation of host defense reactions and limiting the action of multiple antimicrobial peptides [54].
In comparison with prior studies, our analysis indicates the occurrence of five previously unreported point mutations in phoP and phoQ genes, which potentially mediate colistin resistance and heteroresistance. We observed an alteration in the phoP gene induced by a novel deleterious L4F substitution in the BC2_BM isolate, while several new intolerant SNPs (L26Q, L224Q, Q426L, and Q317K), as anticipated by bioinformatics tools, were detected in phoQ in both CR and ChR strains. Meanwhile, other researchers have mentioned the deleterious L26Q substitution in the phoP gene [55]. Furthermore, some mutations closely related to those above were detected in the phoQ gene (such as L30Q, L26P, L96R, and L257P substitutions) [31,56,57,58] and the phoP gene (V3F) [58] and have previously been described as mediating colistin resistance.
Within the complex chromosomal cascade mechanism that confers colistin resistance, phoP can also stimulate the production of PmrA protein, which belongs to the pmrAB TCRS, either directly or indirectly through the adaptor PmrD protein, leading to the addition of cationic pEtN and L-Ara4N moieties to LPS [18,51,59]. In our investigation, only pmrB, as part of the pmrAB TCRS, was subject to diverse mutations (R256G, T157P, P95L, and the newly reported V352E). The R256G substitution has been widely reported to contribute to colistin resistance [30,31,32,53], but this deleterious SNP has also been found in polymyxin-susceptible strains [30,58], suggesting that this alteration alone might not be sufficient to increase MIC values for colistin [58]. Interestingly, all seven isolates detected with this substitution presented at least an additional colistin-resistant element and phenotypically expressed resistance, except the two parental ChR strains, with MIC ≤0.5 mg/L colistin by BMD. In addition, T157P substitution has been reported to disrupt the α-helix secondary structure of mutated PmrB protein, with consecutive activation of PmrA [60], and this SNP was mentioned by several authors [32,52,56,60]. In agreement with the analysis of KPC-3-producing Colombia strains by Jajol et al. [60], our KPC-2-producing BC8_BM strain, with the same T157P mutation, belongs to the same ST258 clone. The two PmrB mutations, R256G and T157P, are among the most frequently reported as being associated with colistin resistance [61], and the rare P95L alteration in PmrB was confirmed to confer resistance by complementation assay [62].
Notably, our BC2_BM and BC3_TM isolates, with colistin-susceptible results by Vitek 2 Compact AST N222 cards but an MIC value of ≥8 mg/L colistin by BMD, did not present any mgrB gene disruption by ISs. Consequently, in the first case, a combination of mutations in mgrB (M27K), phoP (L4F), and phoQ (Q426L) genes was noted, while in the second case, an L26Q substitution in the phoQ gene was associated with R256G mutation in the pmrB gene. However, several reports have mentioned that Vitek 2 Compact is inappropriate for colistin susceptibility testing [63,64,65], especially for strains showing an MIC > 1 mg/L by BMD [63]. The manufacturer of Vitek 2 Compact has also recommended using an alternative method prior to reporting colistin results obtained with several types of testing cards in this automated system [66], at least until this issue can be addressed through technology upgrades to be implemented in the future.
Moreover, the crrAB TCRS can activate Pmr A [51,59], and gain-of-function mutations in crrB alone can activate the expression of genes leading to colistin resistance without any contribution from the pmrAB TCRS [59]. Interestingly, our findings revealed a rare P151S substitution in the crrB gene only in the resistant mutant BC14_TM_C_m isolate, which, in contrast to its susceptible parental strain BC13_TM_C_hR, expressed an MIC value of >64 mg/L colistin. The P151S substitution detected in the putative histidine kinase domain has been validated to induce elevated resistance to colistin [67]. Similarly, Jajol et al. reported a mutation with a subtle difference, P151L in crrB, as conferring colistin resistance [52], while Pitt et al. confirmed a P158R substitution in the same gene [30].
Remarkably, another significant aspect of our study is the molecular basis of the two pairs of ChR (parental and the corresponding resistant mutants) isolates, which are reported for the first time in Romania [65]. These two pairs of strains co-harbored an mgrB alteration caused by an ISKpn25 element insertion at the same position (with the deletion of nucleotides 1–5) and an R256G substitution in pmrB. An identical molecular profile was observed with the BC6_BM strain, which, unlike the two parental ChR strains, had an MIC value of 32 mg/L colistin indicated by BMD. In particular, the two pairs of isolates were supplementarily accompanied by a new L224Q substitution in phoQ, and the two mutants presented either an additional amino acid change P151S in crrB or Q317K in phoQ. Despite the presence of a disrupted mgrB gene in the two parental ChR strains, a possible explanation for the results of colistin susceptibility according to BMD might be the potential suppressor effect of the L224Q mutation in phoQ. In addition, BMD has been considered an unreliable method for the detection of colistin heteroresistance [25,68,69]. Pitt el al., reported that K. pneumoniae isolates exclusively carrying an ISKpn26-like element exhibit an MIC of ≥64 mg/L colistin, while the introduction of a mutated phoP (P47L or A95S) or phoQ (N253T or V446G) gene into an IS mgrB-altered strain led to a decrease in the MIC [30]. These mutations have been shown to disturb the pathways involved in phoQ, phoP, and pmrD expression [30]. However, the complex and challenging mechanisms underlying colistin resistance have not been entirely decoded, and accurate detection accompanied by functional analysis is required in order to clarify their influence on resistance [17,30,51,59]. Moreover, there are still insufficient data on the genetic basis of colistin heteroresistance, although there is evidence that mutations in phoPQ, pmrAB, and mgrB regulatory systems or in lpxM and yciM alleles are involved [61,69,70,71,72] and that the mcr-1 gene is not connected with this phenomenon based on PCR results [68,71].
The strains included in our collection were concentrated in either established (ST258, ST101, and ST147) or emerging (ST307) international clonal lineages [11,27,73,74]. The circulation of bacterial clones ST258, ST101, and ST307 in Romania has been previously documented in both clinical and wastewater specimens [39,40,75]. The present analysis reaffirms the prominent relationship between ST258 and blaKPC genes [27], whereas our strains assigned to ST101 carried blaOXA-48, blaKPC-2, and blaNDM-1 genes, consistent with Palmieri et al. [76]. K. pneumoniae genotype ST101 has been associated with resistance to carbapenem, colistin, aminoglycosides, fluoroquinolones, and fosfomycin, with the potential to become a “perfect storm” clone, considering the similarity with the genetic profiles of hypervirulent strains [77]. In addition, a recent Romanian study demonstrated that it is able to persist in wastewater samples after chlorine treatment [78]. All of our ST101 isolates expressed capsular KL17 and somatic O1v1 antigens, and these findings are also supported by other studies [77,79]. K. pneumoniae ST147 and ST307 have been associated with pandrug resistance and have been reported from endemic regions as well as in global nosocomial outbreaks, with proven links to KPC, NDM, OXA-48-like, and Verona integron-encoded MBL (VIM) [73]. In agreement with Peirano et al. [73], our six strains, assigned to ST147 and ST307, were found to carry the ESBL blaCTX-M-15 gene along with gyrA S83I and parC S80I mutations. Contrary to a recent report on an outbreak in northeast Germany induced by K. pneumoniae ST307 co-harboring blaNDM-1 and blaOXA-48 genes and colistin resistance [74], the blaOXA-48 gene was not detected in our ST307 isolate, nor were any virulence genes or elements conferring susceptibility to chloramphenicol detected.
It is noteworthy that the two parental strains, BC11_TM_B_hR and BC13_TM_C_hR, showed the same phenotypic and genomic profiles belonged to the successful ST147 clone, carried the same mgrB, phoQ, pmrB, and crrB alterations, and were recovered from the same intensive care unit (ICU) approximately 3 weeks apart from different patients, all of which are indicative of silent clonal expansion and intraward dissemination. There is also concern regarding the close relationship of these two ChR isolates from Târgu Mureș with the BC6_BM strain isolated in Baia Mare, suggesting the interregional propagation of ST147. Furthermore, the OXA-48-producer cluster ST101 revealed both interward and interregional spread, while the KPC-2-producer cluster ST258 showed interregional transmission. The constituent isolates of each cluster possessed similar or almost similar plasmids, implying plasmid-mediated propagation of blaOXA-48 and blaKPC-2 genes.
The interdependence between KPC-2-producing isolates with FII(K) and IncX3 plasmids and between OXA-48 producers with IncL and IncR plasmids concurs with previous observations made by Becker et al. in Germany [80]. In contrast to a prior study conducted at the same medical institution in Târgu Mureș on strains collected between 2012 and 2013 that demonstrated the presence of the IncR plasmid replicon in five K. pneumoniae NDM-1-positive isolates [81], our two NDM-1 producers did not harbor this replicon type. Moreover, a significant match between the BC7_BM strain and the reference plasmids suggests the persistence and evolution of a blaNDM plasmid in this geographical area, which was previously named pKOX_NDM1-like by Phan et al. [82]. Additionally, the concurrent carriage of IncFIB(K) and IncFIB(pQil) replicons belonging to the same incompatibility class is in agreement with the results of Villa et al., demonstrating the great versatility of IncF plasmids [83]. These extrachromosomal DNA molecules often exhibit a multi-replicon status [12,83], which enables the acquisition of plasmids harboring incompatible replicons when replication is promoted by a compatible replicon [83]. However, the second-generation raw reads used in our research did not offer the same possibility to construct the entire sequence of a plasmid as in the case of long-read sequencing [32,38].
The present investigation has some limitations. The retrospective nature of the study did not allow us to provide a reliable picture of all significant clinical and therapeutic aspects or previous hospitalizations, which was beyond the scope of this manuscript. However, the data on previous colistin therapy in patients diagnosed with ChR strains may be found elsewhere [65]. BMD was performed only for colistin as the research was focused on detecting the molecular basis of resistance and heteroresistance to this antimicrobial agent. Complementation experiments with wild-type alleles to validate novel mutations potentially conferring colistin resistance and heteroresistance were not performed because of logistical constraints and should be included in future investigations.
Future Directions
Future studies should be oriented toward the reliable detection of both existing and novel mutations, combined with functional analysis, to establish their real contribution to colistin resistance and heteroresistance. Additional research should be conducted to expand our knowledge of the complex mechanisms underlying colistin resistance and heteroresistance; for the last phenomenon to establish clinical relevance, we should formulate a harmonized international definition and develop a standardized detection methodology. Furthermore, investigations using long-read sequencing technology will offer an adequate resolution for exploring plasmid structures.
4. Materials and Methods
4.1. Bacterial Strains, Setting, and Design of the Study
A total of 10 unique clinical CR K. pneumoniae CPE strains were analyzed: 5 selected from patients admitted to Dr. Constantin Opriș County Emergency Hospital, Baia Mare, Romania, between January 2017 and April 2021 and 5 from patients at Târgu Mureș County Emergency Clinical Hospital, Romania, between January 2017 and April 2019. Additionally, 2 ChR K. pneumoniae strains obtained from Târgu Mureș in March 2019 and their corresponding colistin-resistant mutants were included in the study. The collection comprising the 10 CR and 2 ChR isolates has been phenotypically characterized in an earlier study [65].
The first medical center is a public 920-bed general acute care nonteaching hospital in the northwest region of Romania, and the second is a large 1089-bed teaching hospital located in Transylvania, in the central region; they are located approximately 200 km apart.
CR K. pneumoniae CPE isolates were randomly selected based on (i) MIC values greater than 2 mg/L colistin (categorized as resistance) [84], as determined using the BMD method; (ii) diverse specimens collected from various hospital wards; (iii) carbapenemase types identified phenotypically (a set of n = 2 KPC, n = 2 OXA-48-like, and n = 1 MBL for each medical institution); and (iv) date of collection.
Pathogens were mostly isolated from patients in the ICU (n = 6) and were obtained from different anatomical sites, as summarized in Table 8.

Table 8.
Demographic data of patients and general details of CR and ChR K. pneumoniae CPE isolates.
4.2. Demographic Data of Patients
Data from electronic medical records available in the 2 laboratories are presented in Table 8.
4.3. Phenotypic Bacterial Identification and Antimicrobial Susceptibility Testing
All strains were identified at the species level using standard techniques and a Vitek 2 Compact system (BioMérieux SA, Marcy l’Etoile France). AST-XN05 and AST-N233 pair testing cards, starting from the same inoculum, were used with the Vitek 2 Compact system, and AST-N222 cards were added for particular strains. Vitek 2 Compact version 9.02 software was used. The Kirby–Bauer disc diffusion method was used in all cases. Meropenem–vaborbactam (30 µg), imipenem–relebactam (35 µg), ceftazidime–avibactam (14 µg), and doripenem (10 µg) were tested exclusively by disc diffusion.
All K. pneumoniae CPE strains were additionally assessed for colistin resistance by BMD and using the following 5 phenotypic methods, as described elsewhere [65]: Micronaut MIC-Strip (Merlin Diagnostika GmbH, Bornheim-Hersel, Germany), Etest gradient diffusion strip on Mueller Hinton E agar (MHE) (BioMérieux SA, Marcy l’Etoile, France), ChromID Colistin R agar (COLR) assay (BioMérieux SA, Marcy l’Etoile, France), Rapid Polymyxin NP test (ELITechGroup, Signes, France), and colistin broth disc elution. Moreover, the 2 ChR strains were further confirmed using the population analysis profiling (PAP) assay, as also described in [65].
The EUCAST breakpoint [84] was applied for the interpretation of all antibiotic susceptibility test results, except for tigecycline, for which US Food and Drug Administration (FDA) criteria were adopted [85].
The modified carbapenem inactivation method (mCIM) [86,87] and the combination disc test (KPC, MBL, and OXA-48 Confirm Kit, Rosco Diagnostica, Denmark) were applied to phenotypically categorize the carbapenemase producers, as described elsewhere [65].
Concordance between phenotypic and genotypic susceptibility test results was calculated individually for Vitek 2 Compact (14 isolates × 22 antimicrobial agents = 308 combinations) and the disc diffusion method (14 isolates × 24 agents = 336 combinations) as the proportion of concordant results in the total combinations tested by each method. In this study, results of susceptibility or susceptibility to increased exposure were considered to be discrepant if a resistance molecular marker was noted or resistance results were not linked with genetic elements.
Routine and extended quality control were performed according to EUCAST [84] with the following reference strains: Escherichia coli ATCC 25922, E. coli NCTC 13846, E. coli ATCC 35218, K. pneumoniae ATCC 700603, and K. pneumoniae BAA 2814. Other details regarding quality control for all methods used can be found elsewhere [65].
The isolates were frozen at −70 °C and subcultured twice on solid medium before additional testing.
4.4. Genotypic Characterization Using WGS
4.4.1. Sequencing and Assembly of Draft Genomes
The 14 genomes of CR, ChR, and corresponding resistant mutants of CPE strains were sequenced using the Ion Torrent PGM platform (Thermo Fisher Scientific, Waltham, MA, USA) according to the 400 bp protocol for library preparation, which includes enzymatic shearing, Ion OneTouch2 emulsion PCR, enrichment, and Hi-Q View sequencing kits (Thermo Fisher Scientific). The sequences obtained were subjected to de novo assembly into contigs using the Assembler SPAdes plugin version 5.12 (21, 33, 55, 77, and 99 k-mers) [88] installed on the Ion Torrent Server. For epidemiological interconnections and worldwide distribution, all the raw reads in the study were deposited in the publicly available European Nucleotide Archive (ENA) database under project PRJEB53146 (ERR9860230–ERR9860243) (https://www.ebi.ac.uk/ena/browser/home, accessed on 11 August 2022).
WGS data were analyzed using both commercial and free online bioinformatics tools.
4.4.2. Resistome Analysis
For antibiotic resistance characterization, we used AMRFinderPlus version 1.1 [89] through Ridom SeqSphere+ commercial software (Ridom GmbH). The analysis of antimicrobial resistance determinants was complemented with ResFinder version 4.1 [90,91], which is available on the public Center of Genomic Epidemiology (CGE) server (http://www.genomicepidemiology.org/services/, accessed on 20 April 2022) (identity 85%, minimum length 60%), the Basic Local Alignment Search Tool (BLAST) search against reference sequences from the National Center for Biotechnology Information (NCBI) nucleotide repository, and BioEdit software version 7.0.5.3. [92].
Plasmid-mediated mcr genes, the presence and integrity of TCRS genes (phoPQ, pmrAB, and crrAB), and the regulatory transmembrane protein-coding mgrB gene were investigated to identify colistin resistance determinants. ISfinder was used to depict the insertion sequence (IS) types that led to gene disruption [93]. In order to predict whether amino acid substitutions identified in PhoPQ, PmrAB, and CcrAB had an impact on the biological function of the proteins, 2 bioinformatics tools were applied: the Protein Variation Effect Analyzer (PROVEAN) version 1.1 [94], with a default score threshold set at -2.5 for binary classification, and the sorting intolerant from tolerant (SIFT) algorithm [95], with default parameters. The corresponding genes from colistin-susceptible K. pneumoniae NC_009648.1 were used as references.
4.4.3. Molecular Typing of Isolates and Plasmid Profiling
In order to depict the allelic profile of the genomes, MLST, ST [96], and cgMLST complex type (CT) genes (2358 target genes) were used with Ridom SeqSphere+ software [97]. For cgMLST, a default threshold of 15 allele differences was selected, and a minimum spanning tree with the option pairwise ignoring missing values was generated.
Capsular serotyping was conducted using the Kaptive web interface (https://kaptive-web.erc.monash.edu/, accessed on 16 June 2022) [98]. The Virulence Factor Database (VFDB) [14], included in SeqSphere+, was interrogated for the detection of genes characteristic of hypervirulent strains.
Plasmid replicons were detected using the web-based PlasmidFinder version 2.1 [99] on the CGE server (with default parameters). In an article published in 2018, Phan et al. identified blaNDM plasmids circulating during the period 2014–2015 in the town of Târgu Mureș [82]. Since these plasmid sequences are publicly available, Burrows–Wheeler alignment (BWA) was adopted for the mapping of raw reads from the 2 blaNDM-1-positive strains (BC1_TM and BC7_BM) to 3 reference plasmid sequences from 2014–2015 for comparison, namely, K. pneumoniae strain 1TM plasmid pNDM_1TM (MF042353.1), K. pneumoniae strain 6TM plasmid pNDM_6TM (MF042354.1), and K. pneumoniae strain 18ES plasmid pNDM_18ES (MF042350.1).
5. Conclusions
This study highlights the significant challenges involved in the phenotypic and molecular diagnoses of colistin resistance and heteroresistance. Each of our isolates presented at least one mutation in the mgrB, phoPQ, pmrAB, or crrAB gene, predicted to confer colistin resistance. We report on a novel and rare potential suppressor mutation in phoQ, L224Q, with possible involvement in heteroresistance to colistin, which in the presence of an altered mgrB, modified by IS, leads to low MIC values for colistin, as determined using BMD and other phenotypic methods aside from the Rapid Polymyxin NP test and PAP assays. Evidence for the silent intrahospital dissemination of heteroresistant mutant ST147 clones is thus provided.
This research underlines the importance of WGS in identifying a catalog of molecular markers of various classes of antibiotics coupled with descriptions of circulating plasmids and clones, data that can be useful when investigating the dynamics of the dispersion of K. pneumoniae MDR isolates, in addition to being an innovative and promising strategy for continuous surveillance of antimicrobial resistance in order to limit worldwide transmission.
Ceftazidime-avibactam, imipenem/cilastatin-relebactam, meropenem-vaborbactam, tigecycline, gentamicin, and trimethoprim-sulfamethoxazole are still potential in vitro agents with activity against some of the studied pathogens.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics11091171/s1: Figure S1. ClustalW alignment of ompK35, ompK36, and ompK37 genes (BioEdit); Table S1. Mutations in ompK36 and ompK37 genes detected by ResFinder for all strains.
Author Contributions
Conceptualization, A.F., M.O., E.S., C.-R.U. and M.D.; methodology, A.F., M.O., E.S. and C.-R.U.; software, M.O.; validation, A.F., M.O., E.S. and C.-R.U.; formal analysis, M.O. and A.F.; investigation, A.F., M.O. and C.-R.U.; resources, A.F., M.O., E.S., C.-R.U. and M.D.; data curation, M.O.; writing—original draft preparation, A.F. and M.O; writing—review and editing, A.F., M.O., E.S., C.-R.U. and M.D.; visualization, A.F.; supervision, M.O., E.S., C.-R.U. and M.D.; project administration, A.F.; funding acquisition, A.F., M.O., C.-R.U. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received partial internal funding from George Emil Palade University ofMedicine, Pharmacy, Science, and Technology, Târgu Mureș, Romania, code 25/P3 AA/08.07.2020, ct.2401/21/31.10.2018 BB. The DNA sequencing was carried out within the framework of the “Installations and Special Objectives of National Interest” research and development program granted by the Romanian Ministry of Research, Innovation, and Digitization to Cantacuzino National Military Medical Institute for Research and Development through HG no. 78/2014.
Institutional Review Board Statement
Ethical approval for this study was granted by the ethics committees of Dr. Constantin Opriș County Emergency Hospital, Baia Mare, Romania (reference number 14598/04.06.2019); Târgu Mureș County Emergency Clinical Hospital (reference number Ad. 14925/27.05.2019); and George Emil Palade University of Medicine, Pharmacy, Science, and Technology, Târgu Mureș, Romania (reference numbers 405/11.10.2019, 1024/13.07.2020, and 1217/18.12.2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article, and all the raw reads are available in the ENA database.
Acknowledgments
We would like to express our sincere gratitude to Dunia Vago, Marketing Director of the company AMS 2000 Trading Impex S.R.L., Romania for sponsoring the reference strain K. pneumoniae ATCC 2814 used for the experiments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Logan, L.K.; Weinstein, R.A. The Epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 15 (Suppl. S1), S28–S36. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Carbapenem-Resistant Enterobacteriaceae; Second Update; Rapid Risk Assessment; ECDC: Stockholm, Sweden, 2019; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/carbapenem-resistant-enterobacteriaceae-risk-assessment-rev-2.pdf (accessed on 10 August 2022).
- Rodriguez-Bano, J.; Gutierrez-Gutierrez, B.; Machuca, I.; Pascuala, A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018, 31, e00079-17. [Google Scholar] [CrossRef]
- Lan, P.; Jiang, Y.; Zhou, J.; Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 25, 26–34. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019; ECDC: Stockholm, Sweden, 2020; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2019.pdf (accessed on 10 August 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin. Infect. Dis. 2019, 69 (Suppl. S7), S521–S528. [Google Scholar] [CrossRef]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacte-riaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control. Emergence of Hypervirulent Klebsiella pneumoniae ST23 Carrying Carbapenemase Genes in EU/EEA Countries; Rapid Risk Assessment; ECDC: Stockholm, Sweden, 2021; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Emergence-of-hypervirulent-Klebsiella-pneumoniae-ST23-carrying-carbapenemase-genes.pdf (accessed on 10 August 2022).
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis––10 years. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Expert Consensus Protocol on Colistin Resistance Detection and Characterisation for the Survey of Carbapenem- and/or Colistin-Resistant Enterobacteriaceae; Version 1.0; ECDC: Stockholm, Sweden, 2019; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/expert-consensus-protocol-colistin-resistance.pdf (accessed on 10 August 2022).
- World Health Organization. The Detection and Reporting of Colistin Resistance, 2nd ed.; Global Antimicrobial Resistance and use Surveillance System (GLASS); WHO: Geneva, Switzerland, 2021; Available online: https://apps.who.int/iris/bitstream/handle/10665/343654/9789240019041-eng.pdf?sequence=1 (accessed on 10 August 2022).
- El-Sayed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Fordham, S.M.E.; Mantzouratou, A.; Sheridan, E. Prevalence of insertion sequence elements in plasmids relating to mgrB gene disruption causing colistin resistance in Klebsiella pneumoniae. MicrobiologyOpen 2022, 11, e1262. [Google Scholar] [CrossRef]
- Hamel, M.; Chatzipanagiotou, S.; Hadjadj, L.; Petinaki, E.; Papagianni, S.; Charalampaki, N.; Tsiplakou, S.; Papaioannou, V.; Skarmoutsou, N.; Spiliopoulou, I.; et al. Inactivation of mgrB gene regulator and resistance to colistin is becoming endemic in carbapenem-resistant Klebsiella pneumoniae in Greece: A nationwide study from 2014 to 2017. Int. J. Antimicrob. Agents 2020, 55, 105930. [Google Scholar] [CrossRef]
- Yang, T.; Wang, S.; Lin, J.; Griffith, B.T.; Lian, S.; Hong, Z.; Lin, L.; Lu, P.; Tseng, S. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2020, 55, 105894. [Google Scholar] [CrossRef]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Stojowska-Swedrzynska, K.; Lupkowska, A.; Kuczynska-Wisnik, D.; Laskowska, E. Antibiotic heteroresistance in Klebsiella pneumoniae. Int. J. Mol. Sci. 2021, 23, 449. [Google Scholar] [CrossRef]
- Band, V.I.; Satola, S.W.; Smith, R.D.; Hufnagel, D.A.; Bower, C.; Conley, A.B.; Rishishwar, L.; Dale, S.E.; Hardy, D.J.; Vargas, R.L.; et al. Colistin heteroresistance is largely undetected among Carbapenem-Resistant Enterobacterales in the United States. mBio 2021, 12, e02881-20. [Google Scholar] [CrossRef]
- Koser, C.U.; Ellington, M.J.; Peacock, S.J. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014, 30, 401–407. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Centre for Disease Prevention and Control, Surveillance Atlas of Infectious Diseases. 2021. Available online: https://www.ecdc.europa.eu/en/surveillance-atlas-infectious-diseases (accessed on 10 August 2022).
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Pitt, M.E.; Elliott, A.G.; Cao, M.D.; Ganesamoorthy, D.; Karaiskos, I.; Giamarellou, H.; Abboud, C.S.; Blaskovich, M.A.T.; Cooper, M.A.; Coin, L.J.M. Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb. Genom. 2018, 4, e000158. [Google Scholar] [CrossRef]
- Esposito, E.P.; Cervoni, M.; Bernardo, M.; Crivaro, V.; Cuccurullo, S.; Imperi, F.; Zarrilli, R. Molecular epidemiology and virulence profiles of colistin-resistant Klebsiella pneumoniae blood isolates from the Hospital Agency “Ospedale dei Colli,” Naples, Italy. Front. Microbiol. 2018, 9, 1463. [Google Scholar] [CrossRef]
- Lomonaco, S.; Crawford, M.A.; Lascols, C.; Timme, R.E.; Anderson, K.; Hodge, D.R.; Fisher, D.J.; Pillai, S.P.; Morse, S.A.; Khan, E.; et al. Resistome of carbapenem- and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS ONE 2018, 13, e0198526. [Google Scholar] [CrossRef]
- Ruppe, E.; Cherkaoui, A.; Charretier, Y.; Girard, M.; Schicklin, S.; Lazarevic, V.; Schrenzel, J. From genotype to antibiotic susceptibility phenotype in the order Enterobacterales: A clinical perspective. Clin. Microbiol. Infect. 2020, 26, 643.e1–643.e7. [Google Scholar] [CrossRef]
- Ellington, M.J.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.T.G.; Hopkins, K.L.; et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef]
- Mancini, S.; Marchesi, M.; Imkamp, F.; Wagner, K.; Keller, P.M.; Quiblier, C.; Bodendoerfer, E.; Courvalin, P.; Bottger, E.C. Population-based inference of aminoglycoside resistance mechanisms in Escherichia coli. eBioMedicine 2019, 46, 184–192. [Google Scholar] [CrossRef]
- Vaziri, S.; Afsharian, M.; Mansouri, F.; Azizi, M.; Nouri, F.; Madadi-Goli, N.; Afshar, Z.M.; Zamanian, M.H.; Alvandi, A.; Ahmadi, K. Frequency of qnr and aac(6′)Ib-cr genes among ESBL-producing Klebsiella pneumoniae strains isolated from burn patients in Kermanshah, Iran. Jundishapur J. Microbiol. 2020, 13, e100348. [Google Scholar] [CrossRef]
- Galani, I.; Nafplioti, K.; Adamou, P.; Karaiskos, I.; Giamarellou, H.; Souli, M. Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect. Dis. 2019, 19, 167. [Google Scholar] [CrossRef]
- Shelenkov, A.; Mikhaylova, Y.; Yanushevich, Y.; Samoilov, A.; Petrova, L.; Fomina, V.; Gusarov, V.; Zamyatin, M.; Shagin, D.; Akimkin, V. Molecular typing, characterization of antimicrobial resistance, virulence profiling and analysis of whole-genome sequence of clinical Klebsiella pneumoniae isolates. Antibiotics 2020, 9, 261. [Google Scholar] [CrossRef]
- Szekely, E.; Damjanova, I.; Janvari, L.; Vas, K.E.; Molnar, S.; Bilca, D.V.; Lorinczi, L.K.; Toth, A. First description of blaNDM−1, blaOXA−48, blaOXA−181 producing Enterobacteriaceae strains in Romania, Int. J. Med. Microbiol. 2013, 303, 697–700. [Google Scholar] [CrossRef]
- Dortet, L.; Flonta, M.; Boudehen, Y.M.; Creton, E.; Bernabeu, S.; Vogel, A.; Naas, T. Dissemination of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in Romania. Antimicrob. Agents Chemother. 2015, 59, 7100–7103. [Google Scholar] [CrossRef]
- Poirel, L.; Revathi, G.; Bernabeu, S.; Nordmann, P. Detection of NDM-1-Producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 2011, 55, 934–936. [Google Scholar] [CrossRef]
- Gokmen, T.G.; Nagiyev, T.; Meral, M.; Onlen, C.; Heydari, F.; Koksal, F. NDM-1 and rmtC-producing Klebsiella pneumoniae isolates in Turkey. Jundishapur J. Microbiol. 2016, 9, e33990. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; El-Mahdy, T.S.; Radwan, H.H.; Poirel, L. Cooccurrence of NDM-1, ESBL, rmtC, aac(6)-Ib, and qnrB in clonally related Klebsiella pneumoniae isolates together with coexistence of CMY-4 and aac(6)-Ib in Enterobacter cloacae isolates from Saudi Arabia. BioMed Res. Int. 2019, 2019, 6736897. [Google Scholar] [CrossRef]
- Galimand, M.; Courvalin, P.; Lambert, T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 2003, 47, 2565–2571. [Google Scholar] [CrossRef]
- Wachino, J.I.; Doi, Y.; Arakawa, Y. Aminoglycoside resistance: Updates with a focus on acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 2020, 34, 887–902. [Google Scholar] [CrossRef]
- Fournier, C.; Poirel, L.; Despont, S.; Kessler, J.; Nordmann, P. Increasing trends of association of 16S rRNA methylases and carbapenemases in Enterobacterales clinical isolates from Switzerland, 2017–2020. Microorganisms 2022, 10, 615. [Google Scholar] [CrossRef]
- Sun, D.; Rubio-Aparicio, D.; Nelson, K.; Dudley, M.N.; Lomovskaya, O. Meropenem-vaborbactam resistance selection, resistance prevention, and molecular mechanisms in mutants of KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2017, 61, e01694-17. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Meropenem/vaborbactam: A review in complicated urinary tract infections. Drugs 2018, 78, 1259–1270. [Google Scholar] [CrossRef]
- Gaibani, P.; Lombardo, D.; Bussini, L.; Bovo, F.; Munari, B.; Giannella, M.; Bartoletti, M.; Viale, P.; Lazzarotto, T.; Ambretti, S. Epidemiology of meropenem/vaborbactam resistance in KPC-producing Klebsiella pneumoniae causing bloodstream infections in Northern Italy, 2018. Antibiotics 2021, 10, 536. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Sun, D.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Griffith, D.C.; Dudley, M.N. Vaborbactam: Spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 61, e01443-17. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Jayol, A.; Nordmann, P.; Andre, C.; Poirel, P.; Dubois, V. Evaluation of three broth microdilution systems to determine colistin susceptibility of Gram-negative bacilli. J. Antimicrob. Chemother. 2018, 73, 1272–1278. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Y.; Zhu, Y.; Jia, P.; Li, X.; Jia, X.; Yu, W.; Cui, Y.; Yang, R.; Xia, W.; et al. Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerg. Microbes. Infect. 2022, 11, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Kidd, T.J.; Mills, G.; Sa-Pessoa, J.; Dumigan, A.; Frank, C.G.; Insua, J.L.; Ingram, R.; Hobley, L.; Bengoechea, J.A. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 2017, 9, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Zaman, T.U.; Albladi, M.; Siddique, M.I.; Aljohani, S.M.; Balkhy, H.H. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect. Drug Resist. 2018, 11, 1183–1187. [Google Scholar] [CrossRef]
- Azam, M.; Gaind, R.; Yadav, G.; Sharma, A.; Upmanyu, K.; Jain, M.; Singh, R. Colistin resistance among multiple sequence types of Klebsiella pneumoniae is associated with diverse resistance mechanisms: A report from India. Front. Microbiol. 2021, 12, 609840. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Lin, T.L.; Pan, Y.J.; Wang, Y.P.; Lin, Y.; Wang, J.T. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 2015, 59, 2909–2913. [Google Scholar] [CrossRef]
- Castanheira, M.; Doyle, T.B.; Davis, A.P.; Deshpande, L.M.; Mendes, R.E. Disruption of mgrB and Alterations on pmrB Are Most Common Resistance Mechanisms among Colistin-Resistance among Klebsiella pneumoniae from a Global Surveillance Program. ASM Microbe 2018. Available online: https://www.jmilabs.com/data/posters/ASM-Microbe-2018-Klebsiella-pneumoniae-colistin-resistance.pdf (accessed on 10 August 2022).
- Binsker, U.; Kasbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef] [PubMed]
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.V.; Yilmaz, M.; Nordmann, P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766. [Google Scholar] [CrossRef] [PubMed]
- Elias, R.; Duarte, A.; Perdigao, J. A Molecular Perspective on colistin and Klebsiella pneumoniae: Mode of action, resistance genetics, and phenotypic susceptibility. Diagnostics 2021, 11, 1165. [Google Scholar] [CrossRef]
- Pitt, M.E.; Cao, M.D.; Butler, M.S.; Ramu, S.; Ganesamoorthy, D.; Blaskovich, M.A.T.; Coin, L.J.M.; Cooper, M.A. Octapeptin C4 and polymyxin resistance occur via distinct pathways in an epidemic XDR Klebsiella pneumoniae ST258 isolate. J. Antimicrob. Chemother. 2019, 74, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Lellouche, J.; Schwartz, D.; Elmalech, N.; Ben Dalak, M.A.; Temkin, E.; Paul, M.; Geffen, Y.; Yahav, D.; Eliakim-Raz, N.; DuranteMangoni, E.; et al. Combining VITEK 2 with colistin agar dilution screening assist timely reporting of colistin susceptibility. Clin. Microbiol. Infect. 2019, 25, 711–716. [Google Scholar] [CrossRef]
- Pfennigwerth, N.; Kaminski, A.; Korte-Berwanger, M.; Pfeifer, Y.; Simon, M.; Werner, G.; Jantsch, J.; Marlinghaus, L.; Gatermann, S.G. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin. Microbiol. Infect. 2019, 25, 1385–1389. [Google Scholar] [CrossRef]
- Foldes, A.; Szekely, E.; Voidazan, S.T.; Dobreanu, M. Comparison of six phenotypic assays with reference methods for assessing colistin resistance in clinical isolates of carbapenemase-producing Enterobacterales: Challenges and opportunities. Antibiotics 2022, 11, 377. [Google Scholar] [CrossRef]
- BioMerieux. Urgent Product Correction Notice. 2017. Available online: https://www.bfarm.de/SharedDocs/Kundeninfos/EN/08/2017/04963-17_kundeninfo_en.pdf?__blob=publicationFile&v=1 (accessed on 10 August 2022).
- Cheng, Y.H.; Lin, T.L.; Lin, Y.T.; Wang, J.T. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2016, 60, 3709–3716. [Google Scholar] [CrossRef]
- Bardet, L.; Rolain, J.M. Development of new tools to detect colistin-resistance among Enterobacteriaceae strains. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 3095249. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Brink, A.; Poirel, L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 2015, 59, 2780–2784. [Google Scholar] [CrossRef] [PubMed]
- Bardet, L.; Baron, S.; Leangapichart, T.; Okdah, L.; Diene, S.M.; Rolain, J.M. Deciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, France. Antimicrob. Agents Chemother. 2017, 61, e00356-17. [Google Scholar] [CrossRef] [PubMed]
- Cheong, H.S.; Kim, S.Y.; Wi, Y.M.; Peck, K.R.; Ko, K.S. Colistin heteroresistance in Klebsiella pneumoniae isolates and diverse mutations of PmrAB and PhoPQ in resistant subpopulations. J. Clin. Med. 2019, 8, 1444. [Google Scholar] [CrossRef]
- Halaby, T.; Kucukkose, E.; Janssen, A.B.; Rogers, M.R.C.; Doorduijn, D.J.; Van Der Zanden, A.G.M.; Al Naiemi, N.; Vandenbroucke-Grauls, C.M.J.E.; Van Schaik, W. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob. Agents Chemother. 2016, 60, 6837–6843. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Outbreak of Carbapenemase-Producing (NDM-1 and OXA-48) and Colistin-Resistant Klebsiella Pneumoniae ST307, North-East Germany, 2019; ECDC: Stockholm, Sweden, 2019; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Klebsiella-pneumoniae-resistance-Germany-risk-assessment.pdf (accessed on 10 August 2022).
- Surleac, M.; Czobor Barbu, I.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, M.; D’Andrea, M.M.; Pelegrin, A.C.; Mirande, C.; Brkic, S.; Cirkovic, I.; Goossens, H.; Rossolini, G.M.; van Belkum, A. Genomic epidemiology of carbapenem- and colistin-resistant Klebsiella pneumoniae isolates from Serbia: Predominance of ST101 strains carrying a novel OXA-48 plasmid. Front. Microbiol. 2020, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Roe, C.C.; Vazquez, A.J.; Esposito, E.P.; Zarrilli, R.; Sahl, J.W. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front. Microbiol. 2019, 10, 542. [Google Scholar] [CrossRef]
- Popa, L.I.; Gheorghe, I.; Czobor Barbu, I.; Surleac, M.; Paraschiv, S.; Marutescu, L.; Popa, M.; Gradisteanu Pîrcalabioru, G.; Talapan, D.; Niţă, M.; et al. Multidrug resistant Klebsiella pneumoniae ST101 clone survival chain from inpatients to hospital effluent after chlorine treatment. Front. Microbiol. 2021, 11, 610296. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.H. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Becker, L.; Kaase, M.; Pfeifer, Y.; Fuchs, S.; Reuss, A.; von Laer, A.; Sin, M.A.; Korte-Berwanger, M.; Gatermann, S.; Werner, G. Genome-based analysis of carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008-2014. Antimicrob. Resist. Infect. Control 2018, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Molnar, S.; Vas, K.E.; Székely, E. Carbapenemase producing Enterobacterales in Romania: Investigating the origins. Revista Romana de Medicina de Laborator 2020, 28, 341–348. [Google Scholar] [CrossRef]
- Phan, H.T.T.; Stoesser, N.; Maciuca, I.E.; Toma, F.; Szekely, E.; Flonta, M.; Hubbard, A.T.M.; Pankhurst, L.; Do, T.; Peto, T.E.A.; et al. Illumina short-read and MinION long-read WGS to characterize the molecular epidemiology of an NDM-1 Serratia marcescens outbreak in Romania. J. Antimicrob. Chemother. 2018, 73, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; Garcia-Fernandez, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST), Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 12.0. 2022. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf (accessed on 10 August 2022).
- Tygacil. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/021821s026s031lbl.pdf (accessed on 10 August 2022).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; M100-S27; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2017. [Google Scholar]
- van der Zwaluw, K.; de Haan, A.; Pluister, G.N.; Bootsma, H.J.; de Neeling, A.J.; Schouls, L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP Test to assess phenotypic carbapenemase activity in Gram-negative rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulilov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemotherapy. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Zankari, E.; Allesae, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.A.D.; Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [PubMed]
- Junemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296, Erratum in: Nat. Biotechnol. 2013, 31, 1148. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).