Microbiological and Molecular Features Associated with Persistent and Relapsing Staphylococcus aureus Prosthetic Joint Infection
Abstract
:1. Introduction
2. Material and Methods
2.1. Setting and Patients
2.2. Antimicrobial Susceptibility Testing
2.3. Characterization of Hemolytic Activity
2.4. Biofilm Formation
2.5. Fibronectin Adhesion Assay
2.6. Cytotoxicity Assay on MG63 Osteoblasts
2.7. Adhesion, Invasion and Persistence Intracellular Assay
2.8. Whole Genome Sequencing
2.9. Statistical Analysis
3. Results
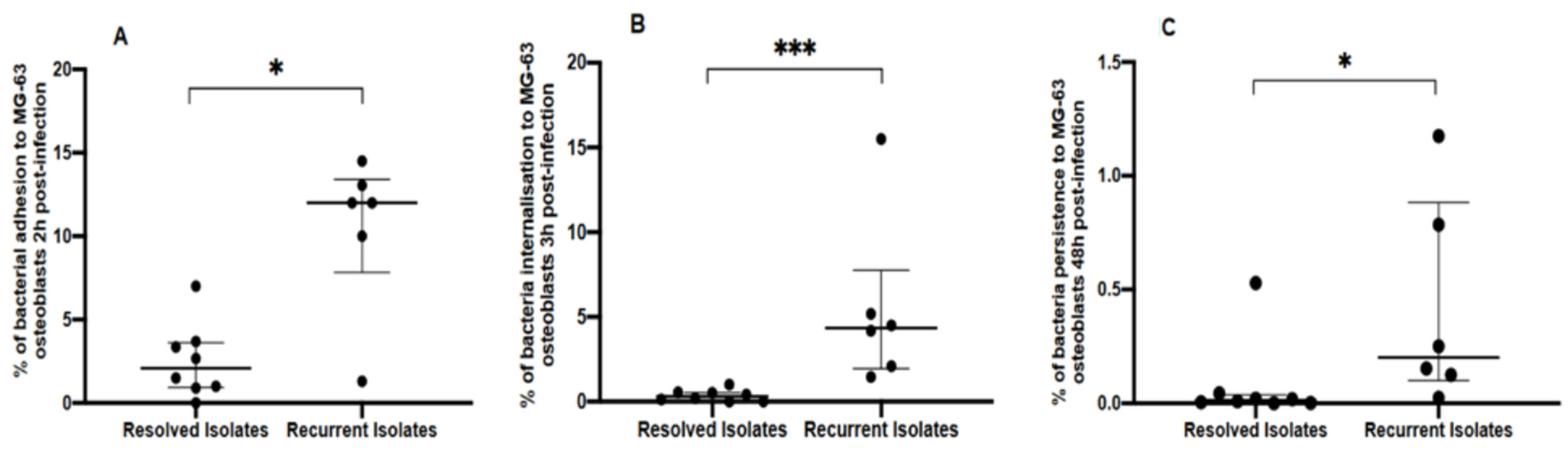
3.1. Genotypic and Phenotypic Comparisons of Isolates from Resolved and Recurrent PJIs
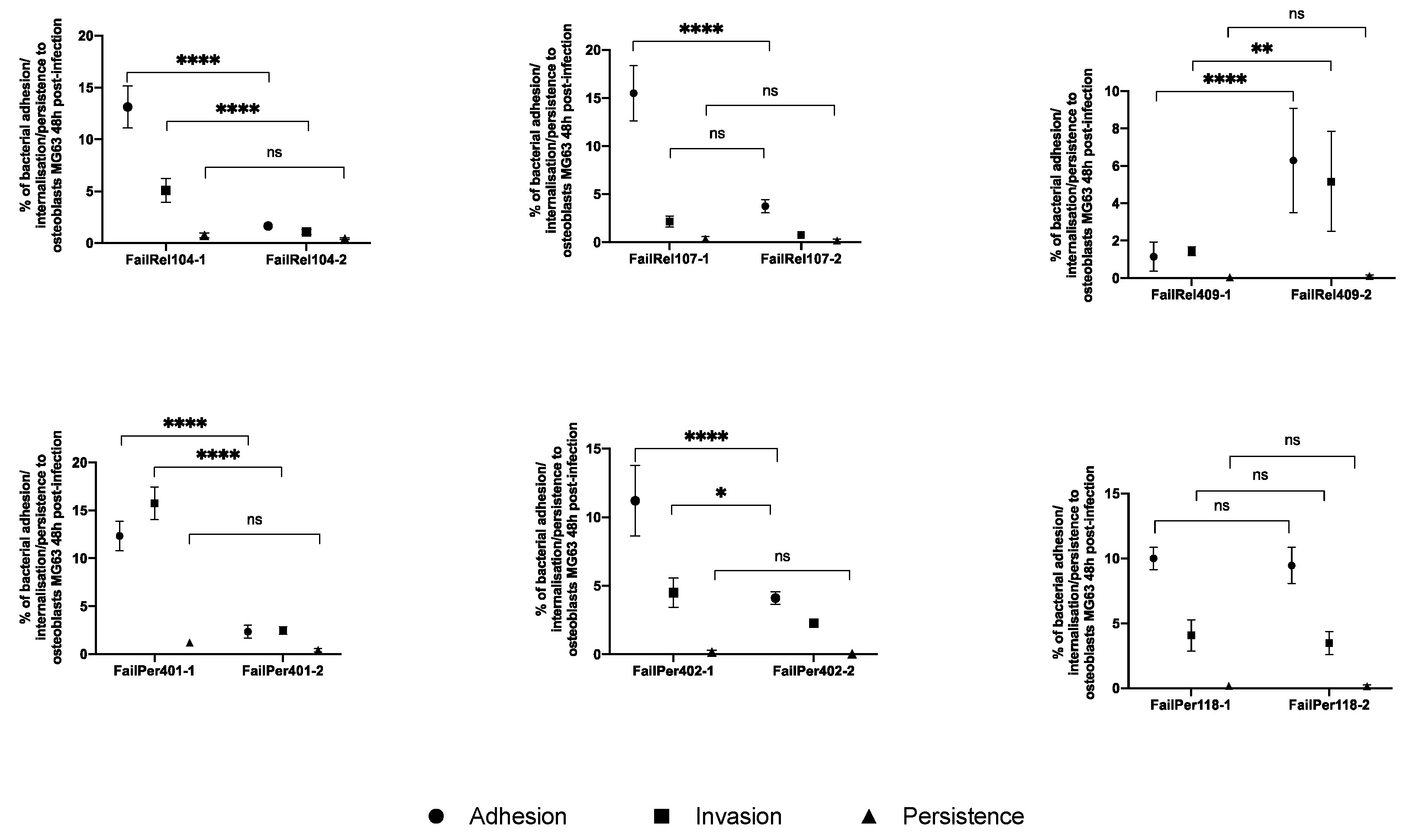
3.2. Phenotypic and Genotypic Changes during Persistent and Relapsing S. aureus PJIs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zimmerli, W.; Trampuz, A.; Ochsner, P.E. Prosthetic-joint infections. N. Engl. J. Med. 2004, 351, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Osmon, D.R.; Berbari, E.F.; Berendt, A.R.; Lew, D.; Zimmerli, W.; Steckelberg, J.M.; Rao, N.; Hanssen, A.; Wilson, W.R. Diagnosis and management of prosthetic joint infection: Clinical practice guidelines by the infectious diseases Society of America. Clin. Infect. Dis. 2013, 56, e1–e25. [Google Scholar] [CrossRef] [PubMed]
- El Helou, O.C.; Berbari, E.F.; Lahr, B.D.; Eckel-Passow, J.E.; Razonable, R.R.; Sia, I.G.; Virk, A.; Walker, R.C.; Steckelberg, J.M.; Wilson, W.R.; et al. Efficacy and safety of rifampin containing regimen for staphylococcal prosthetic joint infections treated with debridement and retention. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Lora-Tamayo, J.; Murillo, O.; Iribarren, J.A.; Soriano, A.; Sánchez-Somolinos, M.; Baraia-Etxaburu, J.M.; Rico, A.; Palomino, J.; Rodríguez-Pardo, D.; Horcajada, J.P.; et al. A large multicenter study of methicillin-susceptible and methicillin-resistant staphylococcus aureus prosthetic joint infections managed with implant retention. Clin. Infect. Dis. 2013, 56, 182–194. [Google Scholar] [CrossRef]
- Benito, N.; Franco, M.; Ribera, A.; Soriano, A.; Rodriguez-Pardo, D.; Sorlí, L.; Fresco, G.; Fernández-Sampedro, M.; Dolores del Toro, M.; Guío, L.; et al. Time trends in the aetiology of prosthetic joint infections: A multicentre cohort study. Clin. Microbiol. Infect. 2016, 22, 732.e1–732.e8. [Google Scholar] [CrossRef]
- Planet, P.J.; Narechania, A.; Chen, L.; Mathema, B.; Boundy, S.; Archer, G.; Kreiswirth, B. Architecture of a Species: Phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017, 25, 153–166. [Google Scholar] [CrossRef]
- Montanaro, L.; Speziale, P.; Campoccia, D.; Ravaioli, S.; Cangini, I.; Pietrocola, G.; Giannini, S.; Arciola, C.R. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 2011, 6, 1329–1349. [Google Scholar] [CrossRef]
- Alamanda, V.K.; Springer, B.D. Perioperative and Modifiable Risk Factors for Periprosthetic Joint Infections (PJI) and Recommended Guidelines. Curr. Rev. Musculoskelet. Med. 2018, 11, 325–331. [Google Scholar] [CrossRef]
- Costerton, W.; Veeh, R.; Shirtliff, M.; Pasmore, M.; Post, C.; Ehrlich, G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1466–1477. [Google Scholar] [CrossRef]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4, VMBF-0022–2015. [Google Scholar] [CrossRef]
- Valour, F.; Trouillet-Assant, S.; Riffard, N.; Tasse, J.; Flammier, S.; Rasigade, J.P.; Chidiac, C.; Vandenesch, F.; Ferry, T.; Laurent, F. Antimicrobial activity against intraosteoblastic Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Höök, M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998, 6, 484–488. [Google Scholar] [CrossRef]
- Post, V.; Wahl, P.; Uçkay, I.; Ochsner, P.; Zimmerli, W.; Corvec, S.; Loiez, C.; Richards, R.G.; Moriarty, T.F. Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal infections. Int. J. Med. Microbiol. 2014, 304, 565–576. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Heitmann, V.; Hussain, M.; Viemann, D.; Roth, J.; Von Eiff, C.; Peters, G.; Becker, K.; Löffler, B. Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. J. Infect. Dis. 2010, 202, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Lora-Tamayo, J.; Euba, G.; Cobo, J.; Horcajada, J.P.; Soriano, A.; Sandoval, E.; Pigrau, C.; Benito, N.; Falgueras, L.; Palomino, J.; et al. Short- versus long-duration levofloxacin plus rifampicin for acute staphylococcal prosthetic joint infection managed with implant retention: A randomised clinical trial. Int. J. Antimicrob. Agents 2016, 48, 310–316. [Google Scholar] [CrossRef]
- Kalinka, J.; Hachmeister, M.; Geraci, J.; Sordelli, D.; Hansen, U.; Niemann, S.; Oetermann, S.; Peters, G.; Löffler, B.; Tuchscherr, L. Staphylococcus aureus isolates from chronic osteomyelitis are characterized by high host cell invasion and intracellular adaptation, but still induce inflammation. Int. J. Med. Microbiol. 2014, 304, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Trouillet-Assant, S.; Lelièvre, L.; Martins-Simões, P.; Gonzaga, L.; Tasse, J.; Valour, F.; Rasigade, J.-P.; Vandenesch, F.; Guedes, R.L.M.; de Vasconcelos, A.T.R.; et al. Adaptive processes of Staphylococcus aureus isolates during the progression from acute to chronic bone and joint infections in patients. Cell Microbiol. 2016, 18, 1405–1414. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Gallego, I.; Viedma, E.; Esteban, J.; Mancheño-Losa, M.; García-Cañete, J.; Blanco-García, A.; Rico, A.; García-Perea, A.; Garbajosa, P.R.; Escudero-Sánchez, R.; et al. Genotypic and phenotypic characteristics of Staphylococcus aureus prosthetic joint infections: Insight on pathogenesis and prognosis of a multicentre prospective cohort. Open Forum Infect. Dis. 2020, 7, ofaa344. [Google Scholar] [CrossRef]
- Stulik, L.; Malafa, S.; Hudcova, J.; Rouha, H.; Henics, B.Z.; Craven, D.E.; Sonnevend, A.M.; Nagy, E. α-hemolysin activity of methicillin-susceptible Staphylococcus aureus predicts ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 2014, 190, 1139–1148. [Google Scholar] [CrossRef]
- Traber, K.E.; Lee, E.; Benon, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar] [CrossRef]
- Salgado-Pabón, W.; Herrera, A.; Vu, B.G.; Stach, C.S.; Merriman, J.A.; Spaulding, A.R.; Schlievert, P.M. Staphylococcus aureus β-toxin production is common in strains with the β-toxin gene inactivated by bacteriophage. J. Infect. Dis. 2014, 210, 784–792. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Seidl, K.; Bayer, A.S.; Fowler, V.G.; McKinnell, J.A.; Hady, W.A.; Sakoulas, G.; Yeaman, M.R.; Xiong, Y.Q. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob. Agents Chemother. 2011, 55, 575–582. [Google Scholar] [CrossRef]
- Richards, R.L.; Haigh, R.D.; Pascoe, B.; Sheppard, S.K.; Price, F.; Jenkins, D.; Rajakumar, K.; Morrissey, J.A. Persistent Staphylococcus aureus isolates from two independent cases of bacteremia display increased bacterial fitness and novel immune evasion phenotypes. Infect. Immun. 2015, 83, 3311–3324. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontéen, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Bradley, P.; Gordon, N.C.; Walker, T.M.; Dunn, L.; Heys, S.; Huang, B.; Earle, S.; Pankhurst, L.J.; Anson, L.; de Cesare, M.; et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun. 2015, 6, 10063. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with sanger sequencing for spa typing of methicillin-resistant staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Petkau, A.; Mabon, P.; Sieffert, C.; Knox, N.C.; Cabral, J.; Iskander, M.; Iskander, M.; Weedmark, K.; Zaheer, R.; Katz, L.S.; et al. SNVPhyl: A single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb. Genom. 2017, 3, e000116. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef]
- Wildeman, P.; Tevell, S.; Eriksson, C.; Lagos, A.C.; Söderquist, B.; Stenmark, B. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci. Rep. 2020, 10, 5938. [Google Scholar] [CrossRef]
- Giulieri, S.G.; Baines, S.L.; Guerillot, R.; Seemann, T.; da Silva, A.G.; Schultz, M.; Massey, R.C.; Holmes, N.E.; Stinear, T.P.; Howden, B.P.; et al. Genomic exploration of within-host microevolution reveals a distinctive molecular signature of persistent staphylococcus aureus bacteraemia. BioRxiv 2018, 10, 273904. [Google Scholar] [CrossRef]
- Lozano, C.; Aspiroz, C.; Sáenz, Y.; Ruiz-García, M.; Royo-García, G.; Gómez-Sanz, E.; Ruiz-Larrea, F.; Zarazaga, M.; Torres, C. Genetic environment and location of the lnu(A) and lnu(B) genes in methicillin-resistant Staphylococcus aureus and other staphylococci of animal and human origin. J. Antimicrob. Chemother. 2012, 67, 2804–2808. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Brothers, K.M.; Maher, P.L.; Phillips, N.J.; Simonetti, D.; William Pasculle, A.; Richardson, A.R.; Cooper, V.S.; Urish, K.L. Staphylococcus aureus genotype variation among and within periprosthetic joint infections. J. Orthop. Res. 2021, 40, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, E.M.; Thaden, J.T.; Sharma-Kuinkel, B.; Park, L.P.; Rude, T.H.; Ruffin, F.; Hos, N.J.; Seifert, H.; Rieg, S.; Kern, W.V.; et al. Polymorphisms in fibronectin binding proteins A and B among staphylococcus aureus bloodstream isolates are not associated with arthroplasty infection. PLoS ONE 2015, 10, e0141436. [Google Scholar] [CrossRef] [PubMed]
- Löffler, B.; Tuchscherr, L.; Niemann, S.; Peters, G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014, 304, 170–176. [Google Scholar] [CrossRef]
- Loughman, A.; Sweeney, T.; Keane, F.M.; Pietrocola, G.; Speziale, P.; Foster, T.J. Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 2008, 8, 74. [Google Scholar] [CrossRef]
- Lewandowski, T.; Huang, J.; Fan, F.; Rogers, S.; Gentry, D.; Holland, R.; DeMarsh, P.; Aubart, K.; Zalacain, M. Staphylococcus aureus formyl-methionyl transferase mutants demonstrate reduced virulence factor production and pathogenicity. Antimicrob. Agents Chemother. 2013, 57, 2929–2936. [Google Scholar] [CrossRef]
- Zhu, X.; Yan, X.; Carter, L.G.; Liu, H.; Graham, S.; Coote, P.J.; Naismith, J. A model for 3-methyladenine recognition by 3-methyladenine DNA glycosylase i (TAG) from Staphylococcus aureus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 610–615. [Google Scholar] [CrossRef]
- Loss, G.; Simões, P.M.; Valour, F.; Cortês, M.F.; Gonzaga, L.; Bergot, M.; Trouillet-Assant, S.; Josse, J.; Diot, A.; Ricci, E.; et al. Staphylococcus aureus Small Colony Variants (SCVs): News from a Chronic Prosthetic Joint Infection. Front. Cell Infect. Microbiol. 2019, 9, 363. [Google Scholar] [CrossRef]

| Strain Code | Sex, Age (years) | Infection location | Type of Infection 2 | Duration of Antibiotherapy (days) 3 | Outcome 4 | Antimicrobial Resistance | Hemolytic Activity | Biofilm | ST 5 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| α-Hemolysin (hla) | β-Hemolysin (hlb) | δ-Hemolysin (agr) | |||||||||
| FailRel104.1 | F, 63 | Knee | EPI | 60 | - | PEN | Positive | Negative | Strong | Weak | 15 |
| FailRel104.2 | - | Relapse | † | Negative | Negative | Strong | Weak | 15 | |||
| FailRel107.1 | F, 86 | Knee | AHI | 66 | - | PEN, CL, GEN, CIP, LEV | Positive | Positive | Negative | Weak | 398 |
| FailRel107.2 | - | Relapse | PEN, CL, GEN CIP, LEV | Positive | Positive | Negative | Weak | 398 | |||
| FailRel409.1 | F, 87 | Hip | EPI | 37 | - | PEN, CIP | Positive | Negative | Weak | Weak | 188 |
| FailRel409.2 | - | Relapse | PEN, CIP | Positive | Negative | Weak | Moderate | 188 | |||
| FailPer401.1 | F, 65 | Knee | EPI | 144 | - | PEN, GEN | Positive | Negative | Strong | Weak | 45 |
| FailPer401.2 | - | Persistence | PEN | Positive | Negative | Strong | Weak | 45 | |||
| FailPer402.1 | M, 81 | Knee | AHI | 78 | - | PEN | Positive | Negative | Weak | Weak | 5 |
| FailPer402.2 | - | Persistence | PEN | Positive | Negative | Weak | Weak | 5 | |||
| FailPer118.1 | F, 78 | Hip | EPI | 25 | - | PEN, MET, ER, CIP, LEV | Positive | Negative | Strong | Moderate | 125 |
| FailPer118.2 | - | Persistence | PEN, MET, ER, CIP, LEV | Positive | Negative | Strong | Moderate | 125 | |||
| Cure116 | F, 62 | Knee | EPI | 79 | Resolved | PEN | Positive | Negative | Weak | Weak | 5 |
| Cure804 | F, 84 | Knee | AHI | 89 | Resolved | ‡ | Positive | Negative | Weak | Weak | 10 |
| Cure806 | M, 37 | Knee | EPI | 81 | Resolved | PEN, GEN | Positive | Negative | Strong | Weak | 45 |
| Cure111 | M, 69 | Knee | EPI | 82 | Resolved | PEN | Positive | Negative | Strong | Weak | 30 |
| Cure112 | F, 83 | Knee | AHI | 90 | Resolved | PEN | Positive | Negative | Negative | Weak | 6 |
| Cure208 | M, 53 | Hip | EPI | 93 | Resolved | PEN, ER, CL | Positive | Negative | Negative | Strong | 509 |
| Cure807 | F, 63 | Knee | EPI | 57 | Resolved | PEN | Positive | Negative | Strong | Weak | 45 |
| Cure808 | F, 72 | Knee | EPI | 100 | Resolved | PEN | Positive | Negative | Strong | Weak | 852 |
| Antibiotic Type | Phenotypic Resistance N (%) | Genotypic Resistance N (%) | ||
|---|---|---|---|---|
| Betalactams | Penicillin Methicillin | 13 (92.9) 1 (7.1) | blaZ mecA | 13 (92.9) 1 (7.1) |
| Aminoglycosides | Gentamicin | 3 (21.4) | aac(6′)-aph(2″) aphA ant | 2 (14.3) 0 (0.0) 1 (7.1) |
| Fluoroquinolones | Ciprofloxacin Levofloxacin | 3 (21.4) 2 (14.3) | norA gryA mutation (S8AL) grlA mutation (S80F) | 14 (100) 2 (14.3) 3 (21.4) |
| Macrolides, lincosamides, streptogramin B | Erythromycin Clindamycin | 2 (14.3)2 (14.3) | erm(A) erm(C) msr(A) | 1 (7.1) 0 (0.0) 1 (7.1) |
| Trimethoprim | Cotrimoxazole | 0 (0.0) | dfrA dfrB mutation (2 H31N, 1 F99Y) dfrC | 0 (0.0) 3 (21.4) 0 (0.0) |
| Fosfomycin | Fosfomycin | 0 (0.0) | fosB | 5 (35.7) |
| Mupirocin | Mupirocin | 0 (0.0) | mupA | 0 (0.0) |
| Fusidic acid | Fusidic acid | 0 (0.0) | fusA mutation | 0 (0.0) |
| Rifampin | Rifampin | 0 (0.0) | rpoB mutation | 0 (0.0) |
| Isolates | Nucleotide Position | Initial Isolate | Relapsing/ Persistent Isolate | Annotation | Type of Mutation | Description (Gene) |
|---|---|---|---|---|---|---|
| FailRel104 | 82110 | A | C | G57G | SYN | lipoprotein |
| 126756 | Δ8 bp | 83–90/741 nt | DEL | GntR family regulatory protein | ||
| 241537–243250 | 4 MBS, 3 NSYN, 5 SYN | staphylocoagulase precursor | ||||
| 428158–431375 | 6 MBS, 11 NSYN, 4 SYN | exotoxin | ||||
| 437491–437497 | 3 NSYN | exotoxin5 | ||||
| 442703, 442706 | SYN, NSYN | exotoxin | ||||
| 446421–448928 | 5 NSYN, 6 SYN, 1 MBS | lipoprotein | ||||
| 479985, 479991 | 2 SYN | glutamate synthase, small subunit | ||||
| 560689 | Δ3 bp | 343–345/1401 nt | DEL | cysteinyl-tRNA synthetase | ||
| 598737 | T | A | A364A | SYN | serine-aspartate repeat-containing protein D (sdrD) | |
| 604977 | C | T | D947D | SYN | bone sialoprotein-binding protein | |
| 842802, 842812 | MBS, SYN | clumping factor A (clfA) | ||||
| 946908–947417 | 3 SYN, 1 DEL, 1 MBS | transport system extracellular binding lipoprotein | ||||
| 1059255 | +TGTTGGTTTCGACGGTGT | 1279/3753 nt | INS | bifunctional autolysin precursor | ||
| 1388164, 1388182 | MBS, SYN | topoisomerase IV subunit A | ||||
| 1883013 | C | T | A234T | NSYN | serine protease | |
| 2088541–2088547 | 3SYN | autoinducer sensor protein | ||||
| 2275393–2275829 | 2 DEL, 4 NSYN, 1 SYN, 1 MBS | hyaluronate lyase precursor 2 | ||||
| 2464027 | Δ4 bp | 126–129/780 nt | DEL | extracellular solute-binding lipoprotein | ||
| 2538425–2538596 | 6 SYN, 1 NSYN, 4MBS | lipoprotein | ||||
| 2557255–2558646 | 2 SYN, 2MBS | fibronectin-binding protein B precursor (fnbB) | ||||
| 2560605–2562933 | 5 NSYN, 7 SYN, 2 MBS, 2 DEL | fibronectin-binding protein A precursor (fnbA) | ||||
| FailRel107 | 915183 | A | G | G139D | NSYN | lpxtg-motif cell wall anchor domain |
| 1642042 | T | C | Y18Y | SYN | integrase | |
| 1699959 | A | C | T582T | SYN | penicillin-binding Protein dimerisation domain family | |
| FailRel409 | 81598–83027 | 6 NSYN, 6 SYN, 3 MBS | lipoprotein | |||
| 98820 | G | A | G298G | SYN | immunoglobulin G binding protein A precursor | |
| 243016 | C | T | N532N | SYN | staphylocoagulase precursor | |
| 431559, 431562 | 2 NSYN | exotoxin | ||||
| 598737 | A | T | A364A | SYN | serine-aspartate repeat-containing protein D (sdrD) | |
| 843649 | G | T | V867V | SYN | clumping factor A (clfA) | |
| 1059255 | +TGTTGGTTTCGACGGTGT | 1279/3753 nt | INS | bifunctional autolysin precursor | ||
| 1148233 | +68 bp | 931/939 nt | INS | ribonuclease HIII | ||
| 1622493, 1622511 | DEL, NSYN | DNA primase | ||||
| 2088515 | 3 bp→CTC | 610–612/1293 nt | MBS | autoinducer sensor protein | ||
| 2496948–2497602 | 7 SYN, 1 NSYN, 7MBS | protein flp | ||||
| 2538455, 2539355 | SYN, MBS, NSYN | exported protein (lipoprotein) | ||||
| 2560599–2562279 | 5 SYN, 3 NSYN | fibronectin-binding protein A precursor (fnbA) | ||||
| 2567893–2567901 | 2 SYN, 2 NSYN | MerR family regulatory protein | ||||
| 2700008, 2700011 | 2 SYN | fibrinogen and keratin-10 binding surface anchored protein | ||||
| FailPer401 | 19218 | +G | 874/1968 nt | INS | DHHA1 domain protein | |
| 103426 | T | A | I503F | NSYN | N-acetyl-ornithine/N-acetyl-lysine deacetylase family protein | |
| 199157 | Δ131 bp | 730-860/960 nt | DEL | cation diffusion facilitator transporter family protein | ||
| 1197044 | A | C | K309Q | NSYN | methionyl-tRNA formyltransferase (fmt) | |
| 1598596 | Δ8 bp | 695-702/1020 nt | DEL | maltose operon transcriptional repressor-like protein | ||
| 2042765 | Δ107 bp | 1167-1273/1539 nt | DEL | sodium/proline symporter (putP) | ||
| 2215085 | A | T | N169I | NSYN | ATP synthase F1, gamma subunit (atpG) | |
| 2397306 | +T | 193/348 nt | INS | transcriptional regulator family protein | ||
| FailPer402 | No mutations were found in the strains belonging to the persistent PJI 402. | |||||
| FailPer118 | 1696986 | (AATG)3→4 | 11/561 nt | DEL | DNA-3-methyladenine glycosidase | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Gallego, I.; Meléndez-Carmona, M.Á.; Lora-Tamayo, J.; Garrido-Allepuz, C.; Chaves, F.; Sebastián, V.; Viedma, E. Microbiological and Molecular Features Associated with Persistent and Relapsing Staphylococcus aureus Prosthetic Joint Infection. Antibiotics 2022, 11, 1119. https://doi.org/10.3390/antibiotics11081119
Muñoz-Gallego I, Meléndez-Carmona MÁ, Lora-Tamayo J, Garrido-Allepuz C, Chaves F, Sebastián V, Viedma E. Microbiological and Molecular Features Associated with Persistent and Relapsing Staphylococcus aureus Prosthetic Joint Infection. Antibiotics. 2022; 11(8):1119. https://doi.org/10.3390/antibiotics11081119
Chicago/Turabian StyleMuñoz-Gallego, Irene, María Ángeles Meléndez-Carmona, Jaime Lora-Tamayo, Carlos Garrido-Allepuz, Fernando Chaves, Virginia Sebastián, and Esther Viedma. 2022. "Microbiological and Molecular Features Associated with Persistent and Relapsing Staphylococcus aureus Prosthetic Joint Infection" Antibiotics 11, no. 8: 1119. https://doi.org/10.3390/antibiotics11081119
APA StyleMuñoz-Gallego, I., Meléndez-Carmona, M. Á., Lora-Tamayo, J., Garrido-Allepuz, C., Chaves, F., Sebastián, V., & Viedma, E. (2022). Microbiological and Molecular Features Associated with Persistent and Relapsing Staphylococcus aureus Prosthetic Joint Infection. Antibiotics, 11(8), 1119. https://doi.org/10.3390/antibiotics11081119







