Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia
Abstract
1. Introduction
2. Results
2.1. Antibiotic Resistance and Antibiograms of Enterococcal Isolates
2.2. Multidrug Resistance (MDR) among the Enterococcal Isolates Recovered from the Poultry Farm Environment
3. Discussion
4. Materials and Methods
4.1. Study Design and Source of Sampling
4.2. Sample Collection, Preparation, and Culture
4.3. Identification and Antibiotic Susceptibility Testing
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Karna, A.; Baral, R.; Khanal, B. Characterization of Clinical Isolates of Enterococci with Special Reference to Glycopeptide Susceptibility at a Tertiary Care Center of Eastern Nepal. Int. J. Microbiol. 2019, 2019, 7936156. [Google Scholar] [CrossRef] [PubMed]
- Said, M.S.; Tirthani, E.; Lesho, E. Enterococcus Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.F.; Tiago, I.; VerÃ-ssimo, A.; Boaventura, R.A.R.; Nunes, O.C.; Manaia, C.M. Antibiotic Resistance of Enterococci and Related Bacteria in an Urban Wastewater Treatment Plant: Antibiotic Resistance of Enterococci in Wastewater. FEMS Microbiol. Ecol. 2006, 55, 322–329. [Google Scholar] [CrossRef]
- Iweriebor, B.; Gaqavu, S.; Obi, L.; Nwodo, U.; Okoh, A. Antibiotic Susceptibilities of Enterococcus Species Isolated from Hospital and Domestic Wastewater Effluents in Alice, Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Kim, Y.B.; Seo, K.W.; Ha, J.S.; Noh, E.B.; Lee, Y.J. Characteristics of Linezolid-Resistant Enterococcus Faecalis Isolates from Broiler Breeder Farms. Poult. Sci. 2020, 99, 6055–6061. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Webster, T.J. Bacteria Antibiotic Resistance: New Challenges and Opportunities for Implant-Associated Orthopedic Infections: Bacteria Antibiotic Resistance. J. Orthop. Res. 2017, 36, 22–32. [Google Scholar] [CrossRef]
- Bourgeois-Nicolaos, N.; Nguyen, T.T.; Defrance, G.; Massias, L.; Alavoine, L.; Lefort, A.; Noel, V.; Senneville, E.; Doucet-Populaire, F.; Mentré, F.; et al. The Emergence of Linezolid Resistance among Enterococci in Intestinal Microbiota of Treated Patients Is Unrelated to Individual Pharmacokinetic Characteristics. Antimicrob. Agents Chemother. 2014, 58, 2681–2687. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sing, A. (Ed.) Zoonoses—Infections Affecting Humans and Animals: Focus on Public Health Aspects; Springer: Dordrecht, The Netherlands, 2015; ISBN 978-94-017-9456-5. [Google Scholar]
- Basak, S.; Singh, P.; Rajurkar, M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J. Pathog. 2016, 2016, 4065603. [Google Scholar] [CrossRef] [PubMed]
- Moussa, A.A.; Md Nordin, A.F.; Hamat, R.A.; Jasni, A.S. High Level Aminoglycoside Resistance And Distribution Of The Resistance Genes In Enterococcus Faecalis And Enterococcus Faecium From Teaching Hospital In Malaysia. Infect. Drug Resist. 2019, 12, 3269–3274. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Espinosa-Gongora, C.; Guardabassi, L. Human Health Risks Associated with Antimicrobial-Resistant Enterococci and Staphylococcus Aureus on Poultry Meat. Clin. Microbiol. Infect. 2016, 22, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Usui, M.; Ozawa, S.; Onozato, H.; Kuge, R.; Obata, Y.; Uemae, T.; Ngoc, P.T.; Heriyanto, A.; Chalemchaikit, T.; Makita, K.; et al. Antimicrobial Susceptibility of Indicator Bacteria Isolated from Chickens in Southeast Asian Countries (Vietnam, Indonesia and Thailand). J. Vet. Med. Sci. 2014, 76, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.C.; Conly, J.M.; Andremont, A.; McEwen, S.A.; Aidara-Kane, A. World Health Organization Ranking of Antimicrobials According to Their Importance in Human Medicine: A Critical Step for Developing Risk Management Strategies to Control Antimicrobial Resistance from Food Animal Production. Clin. Infect. Dis. 2016, 63, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Treskova, M.; Kuhlmann, A.; Freise, F.; Kreienbrock, L.; Brogden, S. Occurrence of Antimicrobial Resistance in the Environment in Germany, Austria, and Switzerland: A Narrative Review of Existing Evidence. Microorganisms 2022, 10, 728. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th rev.; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-151552-8. [Google Scholar]
- Bi, R.; Qin, T.; Fan, W.; Ma, P.; Gu, B. The Emerging Problem of Linezolid-Resistant Enterococci. J. Glob. Antimicrob. Resist. 2018, 13, 11–19. [Google Scholar] [CrossRef]
- Ament, P.W. Linezolid: Its Role in the Treatment of Gram-Positive. Drug-Resist. Bact. Infect. 2002, 65, 8. [Google Scholar]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Conwell, M.; Daniels, V.; Naughton, P.J.; Dooley, J.S.G. Interspecies Transfer of Vancomycin, Erythromycin and Tetracycline Resistance among Enterococcus Species Recovered from Agrarian Sources. BMC Microbiol. 2017, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in Food and Feedingstuffs: From Regulation to Analytical Methods, Bacterial Resistance, and Environmental and Health Implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef]
- Blanch, A.R.; Caplin, J.L.; Iversen, A.; Kuhn, I.; Manero, A.; Taylor, H.D.; Vilanova, X. Comparison of Enterococcal Populations Related to Urban and Hospital Wastewater in Various Climatic and Geographic European Regions. J. Appl. Microbiol. 2003, 94, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.B.; Zakaria, M.P.; Latif, P.A.; Saari, N. Occurrence of Veterinary Antibiotics and Progesterone in Broiler Manure and Agricultural Soil in Malaysia. Sci. Total Environ. 2014, 488–489, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Joint External Evaluation of IHR Core Capacities of Malaysia: Mission Report 2020. Available online: https://apps.who.int/iris/bitstream/handle/10665/336716/9789240015296-eng.pdf?sequence=1&isAllowed=y (accessed on 10 March 2022).
- Diab, M.; Salem, D.; El-Shenawy, A.; El-Far, A.; Abdelghany, A.; Awad, A.R.; El Defrawy, I.; Shemis, M. Detection of High Level Aminoglycoside Resistance Genes among Clinical Isolates of Enterococcus Species. Egypt. J. Med. Hum. Genet. 2019, 20, 28. [Google Scholar] [CrossRef]
- Rosselli Del Turco, E.; Bartoletti, M.; Dahl, A.; Cervera, C.; Pericàs, J.M. How Do I Manage a Patient with Enterococcal Bacteraemia? Clin. Microbiol. Infect. 2021, 27, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance in Enterococci. Expert Rev. Anti Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Ligozzi, M.; Bernini, C.; Bonora, M.G.; de Fatima, M.; Zuliani, J.; Fontana, R. Evaluation of the VITEK 2 System for Identification and Antimicrobial Susceptibility Testing of Medically Relevant Gram-Positive Cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, T.; Anupurba, S. Risk Factors Associated with Fluoroquinolone-Resistant Enterococcal Urinary Tract Infections in a Tertiary Care University Hospital in North India. Indian J. Med. Res. 2016, 144, 604. [Google Scholar]
- Chan, Y. Low Prevalence of Vancomycin- and Bifunctional Aminoglycoside-Resistant Enterococci Isolated from Poultry Farms in Malaysia. Int. J. Food Microbiol. 2008, 122, 221–226. [Google Scholar] [CrossRef]
- Daniel, D.S.; Lee, S.M.; Gan, H.M.; Dykes, G.A.; Rahman, S. Genetic Diversity of Enterococcus Faecalis Isolated from Environmental, Animal and Clinical Sources in Malaysia. J. Infect. Public Health 2017, 10, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Wijesekara, P.; Kumbukgolla, W.; Jayaweera, J.; Rawat, D. Review on Usage of Vancomycin in Livestock and Humans: Maintaining Its Efficacy, Prevention of Resistance and Alternative Therapy. Vet. Sci. 2017, 4, 6. [Google Scholar] [CrossRef]
- Feed (Prohibited Antibiotics, Hormones and Other Chemicals) Regulations 2012. 2012. Available online: https://www.fao.org/faolex/results/details/en/c/LEX-FAOC189891/ (accessed on 11 March 2022).
- Kang, H.-K.; Park, Y. Glycopeptide Antibiotics: Structure and Mechanisms of Action. J. Bacteriol. Virol. 2015, 45, 67. [Google Scholar] [CrossRef]
- Gold, H.S. Vancomycin-Resistant Enterococci: Mechanisms and Clinical Observations. Clin. Infect. Dis. 2001, 33, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.M.; Lee, J.Y.H.; Gorrie, C.L.; Howden, B.P.; Carter, G.P. Genomic Insights Into Last-Line Antimicrobial Resistance in Multidrug-Resistant Staphylococcus and Vancomycin-Resistant Enterococcus. Front. Microbiol. 2021, 12, 637656. [Google Scholar] [CrossRef] [PubMed]
- Enterococcus Differential Agar Base (TITG Agar Base): Technical Data 2019. Available online: https://himedialabs.com/TD/M1896.pdf (accessed on 14 February 2022).
- Funke, G.; Funke-Kissling, P. Performance of the New VITEK 2 GP Card for Identification of Medically Relevant Gram-Positive Cocci in a Routine Clinical Laboratory. J. Clin. Microbiol. 2005, 43, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Package Insert VITEK® 2 AST-P592. 2018. Available online: https://www.biolab-srl.com/products/vitek-2-ast-p592/#:~:text=Vitek%202%20AST%2DP592%20 (accessed on 14 February 2022).
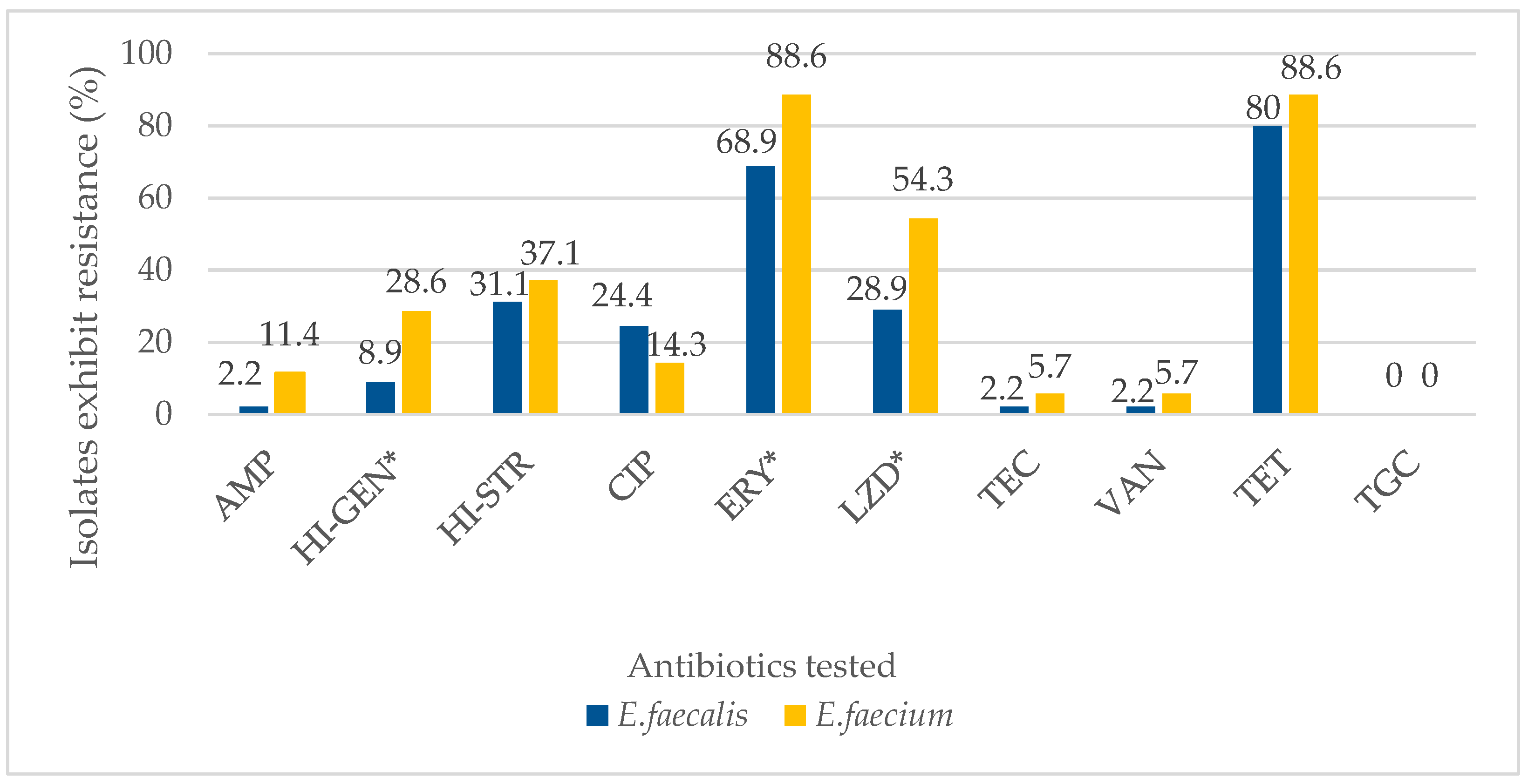

| Antibiotics Tested | Number of Isolates Tested | Antibiogram of Enterococcal Isolates | ||
|---|---|---|---|---|
| Resistant (%) | Intermediate (%) | Susceptible (%) | ||
| Ampicillin | 80 | 5 (6.3) | 0 | 75 (93.8) |
| High-level gentamicin | 80 | 14 (17.5) | 0 | 66 (82.5) |
| High-level Streptomycin | 80 | 27 (33.8) | 1 (1.3) | 52 (65.0) |
| Ciprofloxacin | 80 | 16 (20.0) | 22 (27.5) | 42 (52.5) |
| Erythromycin | 80 | 62 (77.5) | 14 (17.5) | 4 (5.0) |
| Linezolid | 80 | 32 (40.0) | 12 (15.0) | 36 (45.0) |
| Teicoplanin | 80 | 3 (3.8) | 0 | 77 (96.3) |
| Vancomycin | 80 | 3 (3.8) | 1 (1.3) | 76 (95.0) |
| Tetracycline | 80 | 67 (83.8) | 0 | 13 (16.3) |
| Tigecycline | 80 | 0 | 0 | 80 (100) |
| Number of Antibiotics | Phenotypes | Antibiotic Resistance Profile | E. faecalis (n = 45) | E. faecium (n = 35) |
|---|---|---|---|---|
| Resistance to 3 or more antibiotics | ||||
| 7 | P1 | TET/ERY/LZD/CIP/AMP/VAN/TEC | NA | 1 |
| 6 | P2 | TET/ERY/LZD/AMP/VAN/TEC | 1 | NA |
| P3 | TET/ERY/LZD/HI-STR/CIP/HI-GEN | NA | 2 | |
| 5 | P4 | TET/ERY/LZD/HI-STR/CIP | 1 | 1 |
| P5 | TET/ERY/LZD/HI-STR/HI-GEN | NA | 4 | |
| P6 | TET/ERY/LZD/CIP/HI-GEN | 1 | NA | |
| P7 | TET/ERY/HI-STR/CIP/HI-GEN | 2 | NA | |
| P8 | TET/ERY/HI-STR/HI-GEN/AMP | NA | 1 | |
| 4 | P9 | TET/ERY/LZD/HI-STR | 4 | 3 |
| P10 | TET/ERY/LZD/HI-GEN | NA | 2 | |
| P11 | TET/ERY/LZD/AMP | NA | 2 | |
| P12 | TET/ERY/HI-STR/CIP | 2 | NA | |
| P13 | TET/ERY/CIP/HI-GEN | 1 | NA | |
| 3 | P14 | TET/ERY/LZD | 6 | 4 |
| P15 | TET/ERY/HI-STR | 2 | 2 | |
| P16 | TET/ERY/CIP | 2 | 1 | |
| P17 | ERY/VAN/TEC | NA | 1 | |
| Resistance to one or two antibiotics | ||||
| 2 | P18 | TET/ERY | 7 | 6 |
| P19 | TET/HI-STR | 2 | NA | |
| P20 | ERY/HI-GEN | NA | 1 | |
| P21 | ERY/CIP | 2 | NA | |
| P19 | TET/HI-STR | 2 | NA | |
| 1 | P22 | TET | 5 | 2 |
| P23 | HI-STR | 1 | NA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajendiran, S.; Veloo, Y.; Thahir, S.S.A.; Shaharudin, R. Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia. Antibiotics 2022, 11, 1118. https://doi.org/10.3390/antibiotics11081118
Rajendiran S, Veloo Y, Thahir SSA, Shaharudin R. Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia. Antibiotics. 2022; 11(8):1118. https://doi.org/10.3390/antibiotics11081118
Chicago/Turabian StyleRajendiran, Sakshaleni, Yuvaneswary Veloo, Syahidiah Syed Abu Thahir, and Rafiza Shaharudin. 2022. "Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia" Antibiotics 11, no. 8: 1118. https://doi.org/10.3390/antibiotics11081118
APA StyleRajendiran, S., Veloo, Y., Thahir, S. S. A., & Shaharudin, R. (2022). Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia. Antibiotics, 11(8), 1118. https://doi.org/10.3390/antibiotics11081118






