Microbial Resistance to Carbapenems in Effluents from Gynaecological, Paediatric and Surgical Hospital Units
Abstract
:1. Introduction
2. Results
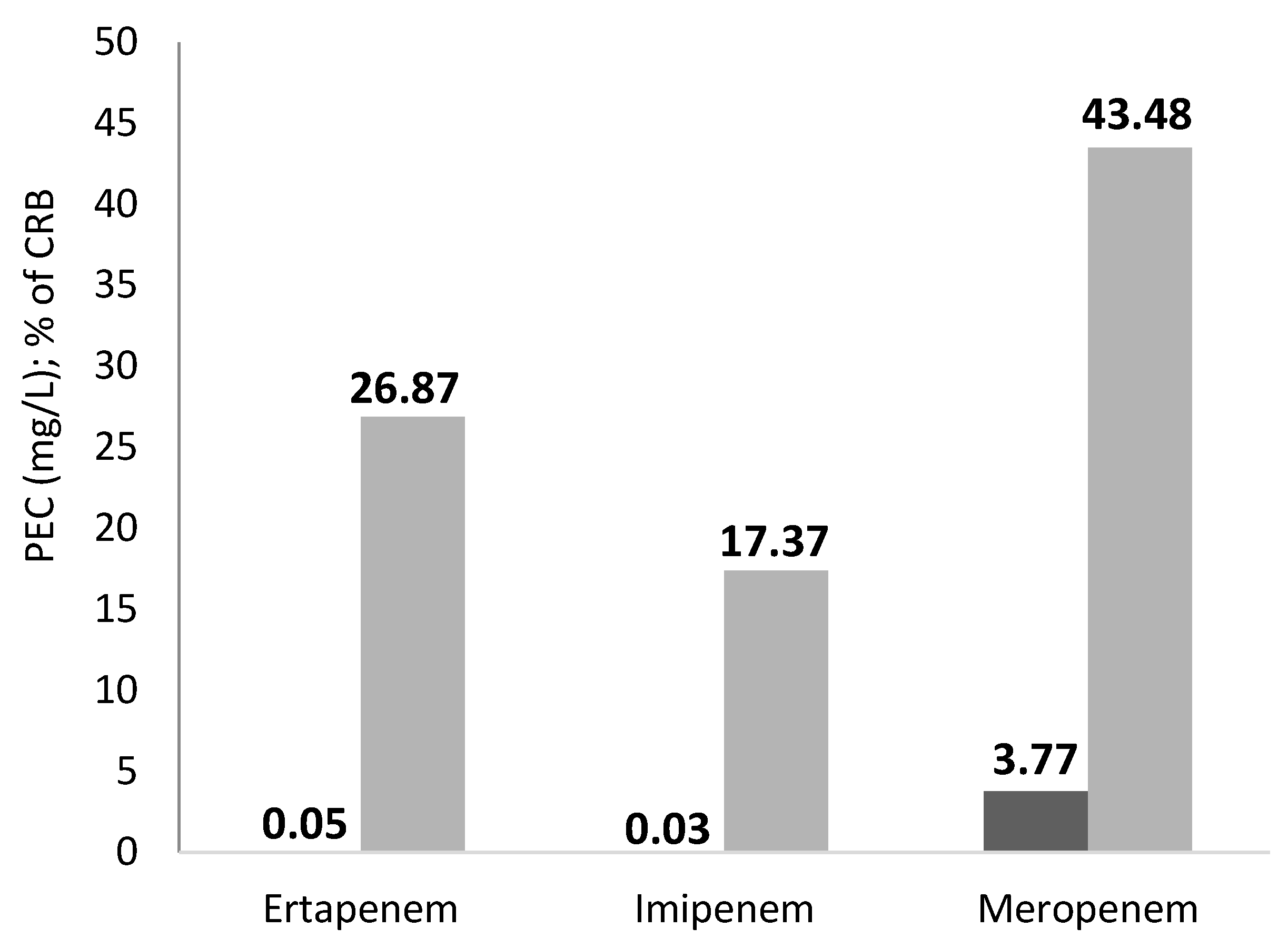
2.1. Effluent from the ArzH
- 6 species are Enterobacteriaceae and represent 40% of the bacterial strains isolated: Escherichia coli, Pantaea agglomerans, Hafnea alvii, Enterobacter aerogenes, Enterobacter cloacae, and Klebsiella pneumoniae;
- 4 are aeromonadacea. and represent 26.66% of the bacterial strains isolated: Aeromonas caviae, Aeromonas hydrophila, Aeromonas veronii bv sorbia, Aeromonas salmonicidas;
- 4 are staphylococci and represent 26.66% of the bacterial strains isolated: Staphylococcus epidermidis, Staphylococc aureus, Staphylococcus huminis, and Staphylococcus equorum.
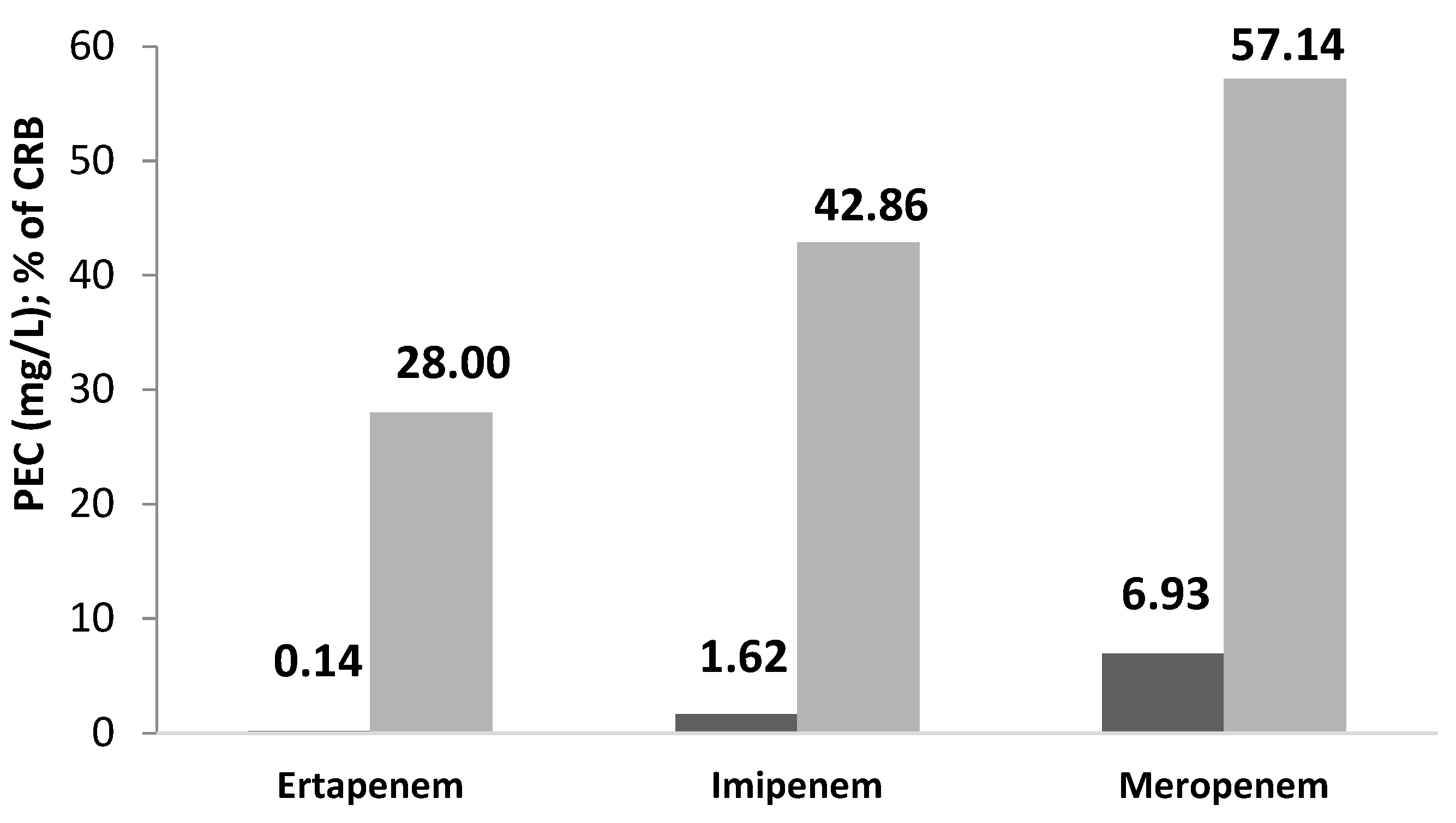
2.2. Effluent from the MCH
- Klebsiella pneumoniae: resistant to all carbapenems with an average percentage of resistance of 55.56% and resistance to other ABs tested of 40.03%.
- Klebsiella oxytoca: resistant to meropenem and imipenem-sensitive ertapenem, and resistant to other ABs: 47.61%.
- Escherichia coli: resistant to all 3 carbapenems (44.44%), with high resistance to other ABs: tested: 90.47%.
- Serratia odorifera: resistant to ertapenem (minimum inhibitory concentration: MIC > 0.13), sensitive to the other two carbapenems: meropenem and imipenem; and resistance to other ABs tested: 61.90%.
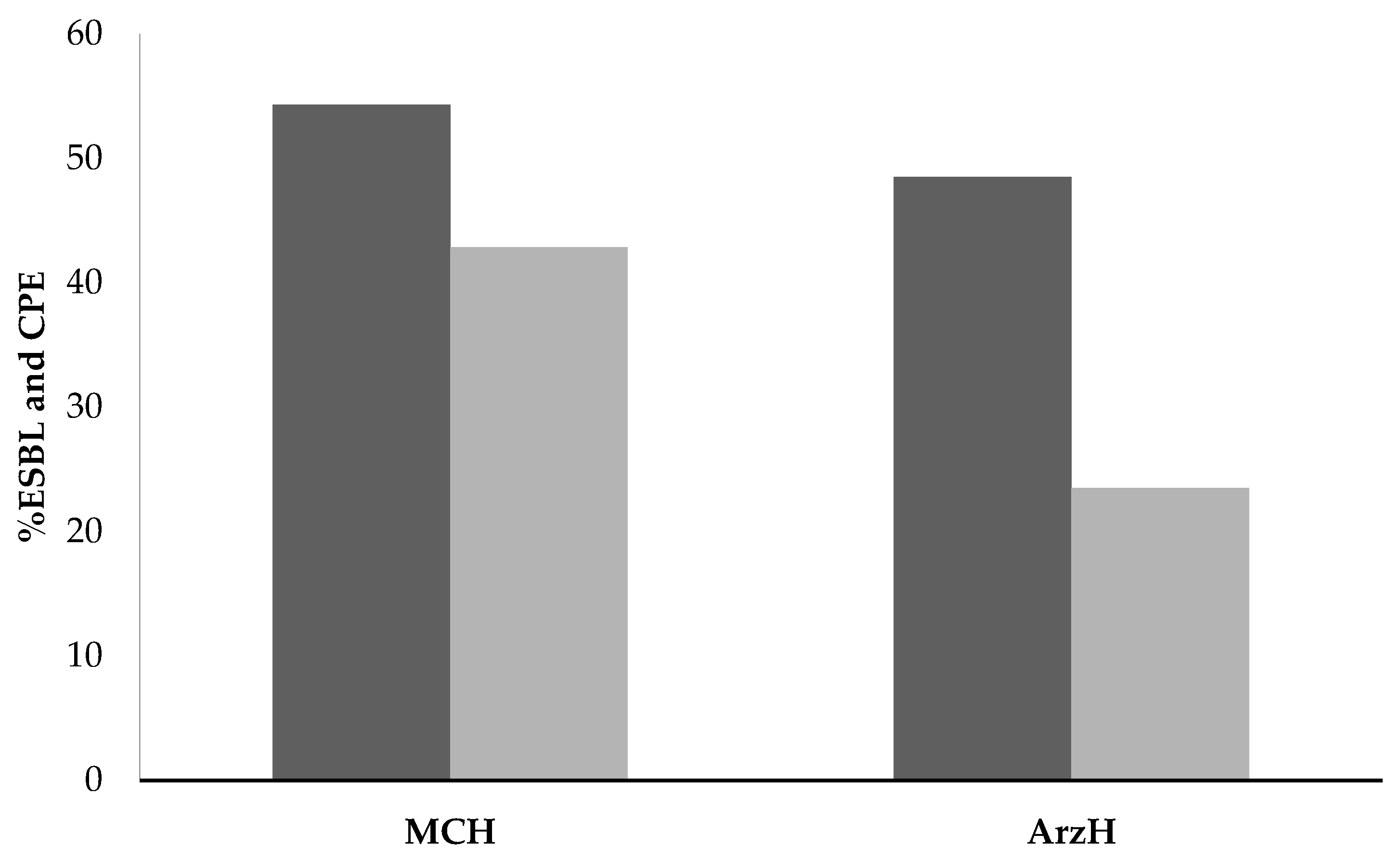
2.3. Effluents from Both Sites
3. Discussion
3.1. Ar-Razi Surgical Hospital
3.2. Mother and Child Hospital
3.3. The 2 Sites Effluents
- −
- strengthening surveillance, antibiotic consumption and monitoring of antibiotics that generate bacterial resistance (carbapenems, cephalosporins, fluoroquinolones, amoxicillin-clavulanic acid combination);
- −
- control and limitation of the prescription of antibacterial agents which allow savings to be made on last resort antibiotics such as carbapenems, tigecycline, colistin, fosfomycin, daptomycin, linezolid and phenicols) [38];
- −
- validation of ABs that can be used in hospital;
- −
- the establishment of the list of ABs for controlled distribution and the modalities of this distribution;
- −
- setting up and evaluating antibiotic therapy protocols in clinical departments by defining the priority departments in which these protocols must be set up (e.g., emergency department, operating theatre);
- −
- conducting prescription audits;
- −
- providing information on consumption, costs and new approved ABs.
4. Materials and Methods
4.1. Sites of Study
4.2. Sampling
4.3. Strain Identification and ARM Determination
4.4. Predicted Environmental Values (PEC)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nasri, E.; Subirats, J.; Sànchez-Melsió, A.; Ben Mansour, H.C.; Borrego, M.; Balcázar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Huijbers, P.M.C.; Flach, C.F.; Larsson, D.G.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef] [PubMed]
- Mills, A.; Bingham, M.; O’Rourke, C.; Bowker, M. Modelled kinetics of the rate of hydrogen evolution as a function of metal catalyst loading in the photocatalysed reforming of methanol by Pt (or Pd)/TiO2. J. Photochem. Photobiol. A Chem. 2019, 373, 122–130. [Google Scholar] [CrossRef]
- Kelvin, A.A.; Halperin, S. COVID-19 in children: The link in the transmission chain. Lancet Infect. Dis. 2020, 20, 633–634. [Google Scholar] [CrossRef]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Iovleva, A.; Doi, Y. Carbapenem-resistant enterobacteriaceae. Clin. Lab. Med. 2017, 37, 303–315. [Google Scholar] [CrossRef]
- US Department of Health and Human Services. Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Ory, J.; Bricheux, G.; Robin, F.; Togola, A.; Forestier, C.; Traore, O. Biofilms in hospital effluents as a potential crossroads for carbapenemase-encoding strains. Sci. Total Environ. 2019, 657, 7–15. [Google Scholar] [CrossRef]
- Savard, P.; Perl, T.M. Combating the spread of carbapenemases in E nterobacteriaceae: A battle that infection prevention should not lose. Clin. Microbiol. Infect. 2014, 20, 854–861. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, Á.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef]
- Queenan, A.M.; Karen, B. Carbapenemases: The versatile β-lactamases. Clinical. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Livorsi, D.J.; Chorazy, M.L.; Schweizer, M.L.; Balkenende, E.C.; Blevins, A.E.; Nair, R.; Samore, M.H.; Nelson, R.E.; Khader, K.; Perencevich, E.N. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control 2018, 7, 55. [Google Scholar] [CrossRef]
- El-Ogri, F.; Ouazzani, N.; Boraâm, F.; Mandi, L. A survey of wastewaters generated by a hospital in Marrakech city and their characterization. Desalin. Water Treat. 2016, 57, 17061–17074. [Google Scholar] [CrossRef]
- Saliba, E.D.O.S.; Faria, E.P.; Rodriguez, N.M.; Moreira, G.R.; Sampaio, I.B.M.; Saliba, J.S.; Gonçalves, L.C.; Borges, I.; Borges, A.L.C.C. Use of infrared spectroscopy to estimate fecal output with marker Lipe®. Int. J. Food Sci. Nutr. Diet. 2015, 4, 1–10. [Google Scholar] [CrossRef]
- Picão, R.C.; Cardoso, J.P.; Campana, E.H.; Nicoletti, A.G.; Petrolini, F.V.; Assis, D.M.; Juliano, L.; Gales, A.C. The route of antimicrobial resistance from the hospital effluent to the environment: Focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 2013, 76, 80–85. [Google Scholar] [CrossRef]
- Pärnänen, K.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, 9124. [Google Scholar] [CrossRef]
- Proia, L.; Anzil, A.; Borrego, C.; Farrè, M.; Llorca, M.; Sanchis, J.; Bogaerts, P.; Balcázar, J.L.; Servais, P. Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J. Hazard. Mater. 2018, 358, 33–43. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming–Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499. [Google Scholar] [CrossRef]
- Caucci, S.; Karkman, A.; Cacace, D.; Rybicki, M.; Timpel, P.; Voolaid, V.; Gurke, R.; Virta, M.; Berendonk, T.U. Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef]
- Marx, A.; Chan, J.K.; Coindre, J.M.; Detterbeck, F.; Girard, N.; Harris, N.L.; Jaffe, E.S.; Kurrer, M.O.; Marom, E.M.; Moreira, A.L.; et al. The 2015 World Health Organization classification of tumors of the thymus: Continuity and changes. J. Thorac. Oncol. 2015, 10, 1383–1395. [Google Scholar] [CrossRef]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Alsharapy, S.A.; Gharout-Sait, A.; Muggeo, A.; Guillard, T.; Cholley, P.; Brasme, L.; Bertrand, X.; Moghram, G.S.; Touati, A.; De Champs, C. Characterization of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates in Al Thawra University Hospital, Sana’a, Yemen. Microb. Drug Resist. 2020, 26, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Loncaric, I.; Heigl, H.; Licek, E.; Moosbeckhofer, R.; Busse, H.J.; Rosengarten, R. Typing of Pantoea agglomerans isolated from colonies of honey bees (Apis mellifera) and culturability of selected strains from honey. Apidologie 2009, 40, 40–54. [Google Scholar] [CrossRef]
- Rosso, F.; Cedano, J.A.; Parra-Lara, L.G.; Sanz, A.M.; Toala, A.; Velez, J.F.; Hormaza, M.P.; Moncada, P.A.; Correa, A. Emerging carbapenem-resistant Aeromonas spp. infections in Cali, Colombia. Braz. J. Infect. Dis. 2019, 23, 336–342. [Google Scholar] [CrossRef]
- Ghatak, S.; Blom, J.; Das, S.; Sanjukta, R.; Puro, K.; Mawlong, M.; Shakuntala, I.; Sen, A.; Goesmann, A.; Kumar, A.; et al. Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila. Antonie Van Leeuwenhoek 2016, 109, 945–956. [Google Scholar] [CrossRef]
- Anandan, S.; Gopi, R.; Ragupathi, N.K.D.; Sethuvel, D.P.M.; Gunasekaran, P.; Walia, K.; Veeraraghavan, B. First report of blaOXA-181-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: Threat for rapid dissemination. J. Glob. Antimicrob. Resist. 2017, 10, 310–314. [Google Scholar] [CrossRef]
- Hafiane, F.Z.; Tahri, L.; Ameur, N.; Rochdi, R.; Arifi, K.; Fekhaui, M. Antibiotic Resistance of Pseudomonas aeruginosa in Well Waters in Irrigated Zone (Middle Atlas-Morocco). Nat. Environ. Pollut. Technol. 2019, 18, 1193–1200. [Google Scholar]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, 3463–3470. [Google Scholar] [CrossRef]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y.H. Occurrence and characteristics of extended-spectrum β-lactamase-and carbapenemase-producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef]
- Huang, H.; Zeng, S.; Dong, X.; Li, D.; Zhang, Y.; He, M.; Du, P. Diverse and abundant antibiotics and antibiotic resistance genes in an urban water system. J. Environ. Manag. 2019, 231, 494–503. [Google Scholar] [CrossRef]
- Ory, J. Effluents Hospitaliers: Sources de Pollution en Antibiotiques et de Résistances Bacériennes Potentiellement TRANSMISSIBLES via un Biofilm? Ph.D. Thesis, Speciality Microbiology, Clermont Auvergne University, Clermont-Ferrand, France, 2017. [Google Scholar]
- Hormatallah, F. Evaluation de la Consommation des Antibiotiques au CHU Mohammed V. Ph.D. Thesis, Medicine Cadi Ayyad University, Marrakesh, Morocco, 2019. [Google Scholar]
- Thai, P.K.; Binh, V.N.; Nhung, P.H.; Nhan, P.T.; Hieu, N.Q.; Dang, N.T.; Tam, N.K.B.; Anh, N.T.K. Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci. Total Environ. 2018, 645, 393–400. [Google Scholar] [CrossRef]
- Tran, N.H.; Hoang, L.; Nghiem, L.D.; Nguyen, N.M.H.; Ngo, H.H.; Guo, W.; Trinh, Q.T.; Mai, N.H.; Chen, H.; Nguyen, D.D.; et al. Occurrence and risk assessment of multiple classes of antibiotics in urban canals and lakes in Hanoi, Vietnam. Sci. Total Environ. 2019, 692, 157–174. [Google Scholar] [CrossRef]
- Le, T.H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef]
- Zahlane, K.; Tadlaoui Ouafi, A.; Barakate, M. The clinical and epidemiological risk factors of infections due to multi-drug resistant bacteria in an adult intensive care unit of University Hospital Center in Marrakesh-Morocco. J. Infect. Public Health 2020, 13, 637–643. [Google Scholar] [CrossRef]
- Gandolière, A. Évaluation de la Politique de bon Usage des Antibiotiques du CHR Metz-Thionville de 2007 à 2014: Confrontation au Suivi des Consommations D’antibiotiques et des Résistances Bactériennes. Ph.D. Thesis, Lorraine University, Metz, France, 2015. [Google Scholar]
- Ameziane, N.E.; Benaabidate, L. Caractérisation microbiologique des effluents de l’hôpital Mohamed V de Meknès et étude de leur impact sur l’environnement. Nat. Technol. 2014, 10, 31. [Google Scholar]
- Bonnet, R.; Caron, F.; Cavallo, J.D.; Chardon, H.; Chidiac, C.; Courvalin, P.; Drugeon, H.; Dubreuil, L.; Jarlier, V.; Jehl, F.; et al. Comité de l’Antibiogramme de la Société Française de Microbiologie. Recommandations 2013, 19, 133–142. [Google Scholar]
- Ellner, P.D.; Stoessel, C.J.; Drakeford, E.; Vasi, F. A new culture medium for medical bacteriology. Am. J. Clin. Pathol. 1966, 45, 502–504. [Google Scholar] [CrossRef]
- Perilli, M.; Bottoni, C.; Pontieri, E.; Segatore, B.; Celenza, G.; Setacci, D.; Bellio, P.; Strom, R.; Amicosante, G. Emergence of blaKPC-3–Tn4401a in Klebsiella pneumoniae ST512 in the municipal wastewater treatment plant and in the university hospital of a town in central Italy. J. Glob. Antimicrob. Resist. 2013, 1, 217–220. [Google Scholar] [CrossRef]
- Haffler, Z.J.; Kulengowski, B.; Ribes, J.A.; Burgess, D.S. Evaluation of the BD Phoenix automated system for determining antimicrobial susceptibility against carbapenem-resistant Enterobacteriaceae compared with broth microdilution. Int. J. Antimicrob. Agents 2019, 54, 249–254. [Google Scholar] [CrossRef]
- Carroll, K.C.; Glanz, B.D.; Borek, A.P.; Burger, C.; Bhally, H.S.; Henciak, S.; Flayhart, D. Evaluation of the BD Phoenix automated microbiology system for identification and antimicrobial susceptibility testing of Enterobacteriaceae. J. Clin. Microbiol. 2006, 44, 3506–3509. [Google Scholar] [CrossRef]
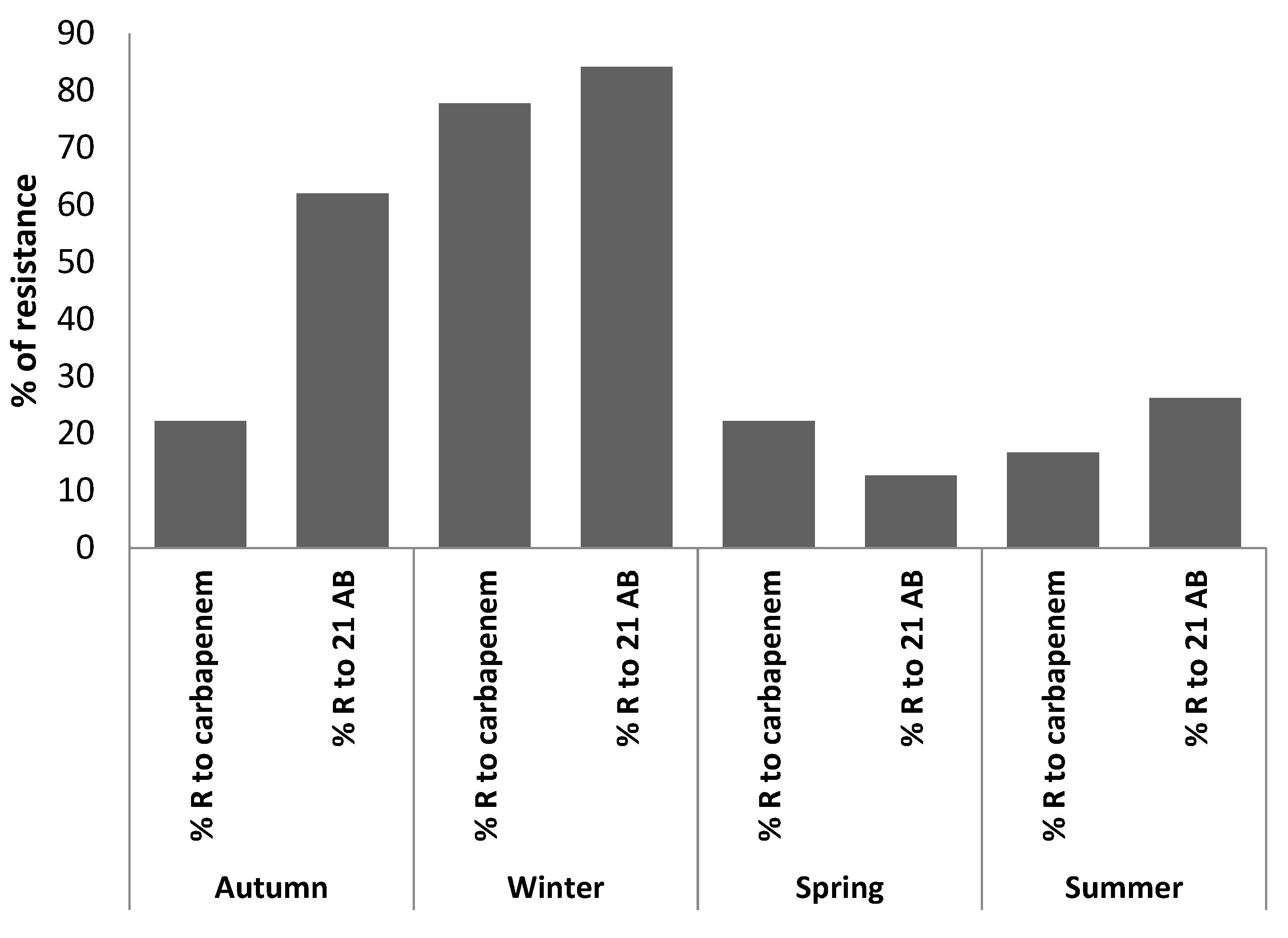
| Exam No. | Collection Season | Identified Germs | Ertapenem | Imipenem | Meropenem | % R to Carbp | Frequency of Bacterial Resistance among 21 AB | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R/S | MIC | R/S | MIC | R/S | MIC | R/N.AB | % | ||||
| 1 | Autumn | Escherichia coli | S | S | <0.25 | R | >8 | 0 | 17/21 | 80.95 | |
| 2 | Autumn | Escherichia coli | S | S | 0.25 | R | >8 | 33.33 | 10/21 | 47.62 | |
| 3 | Autumn | Escherichia coli | S | 0.25 | S | <0.25 | R | >8 | 33.33 | 16/21 | 76.19 |
| 4 | Autumn | Pantaea agglomerans | R | >1 | S | S | =0.5 | 33.33 | 9/21 | 42.86 | |
| 5 | Autumn | Hafnea alvii | S | S | S | 0 | 9/21 | 42.86 | |||
| 6 | Autumn | Aeromonas caviae | S | <0.13 | S | <0.25 | R | >8 | 33.33 | 17/21 | 80.95 |
| 7 | Winter | Escherichia coli | R | 0.25 | S | R | >8 | 66.66 | 18/21 | 85.71 | |
| 8 | Winter | Enterobacter aerogenes | R | >1 | R | >8 | R | >8 | 100 | 20/21 | 95.24 |
| 9 | Winter | Klebsiella pneumoniae | R | >1 | I | 1 | R | >8 | 66.66 | 19/21 | 90.48 |
| 10 | Winter | Enterobacter aerogenes | R | >1 | R | >2 | R | >8 | 100 | 19/21 | 90.48 |
| 11 | Winter | Aeromonas hydrophila | S | S | R | >8 | 33.33 | 13/21 | 61.91 | ||
| 12 | Winter | Aeromonas veronii bv sorbia | R | >1 | R | >2 | R | >8 | 100 | 17/21 | 80.95 |
| 13 | Spring | Escherichia coli | S | <0.13 | S | 0.25 | S | <0.13 | 0 | 00/21 | 00 |
| 14 | Spring | Escherichia coli | S | 0.25 | S | 0.25 | S | 0 | 01/21 | 4.76 | |
| 15 | Spring | Aeromonas caviae | S | S | <0.25 | S | 0 | 4/21 | 19.05 | ||
| 16 | Spring | Aeromonas caviae | S | S | <0.13 | S | <0.25 | 0 | 2/21 | 9.52 | |
| 17 | Spring | Staphylococcus epidermidis | S | S | S | 0 | 3/21 | 14.29 | |||
| 18 | Spring | Pseudomonas aeroginosa | S | S | S | 0 | 6/21 | 28.57 | |||
| 19 | Summer | Aeromonas salmonicidas | S | S | S | 0 | 7/21 | 33.33 | |||
| 20 | Summer | Staphylococcus aureus | S | S | S | 0 | 4/21 | 19.05 | |||
| 21 | Summer | Staphylococcus huminis | S | S | S | 0 | 10/21 | 47.62 | |||
| 22 | Summer | Staphylococcus equorum | S | S | S | 0 | 6/21 | 28.57 | |||
| 23 | Summer | Escherichia coli | S | S | S | 0 | 3/21 | 14.29 | |||
| 24 | Summer | Enterobacter cloacae | S | <0.13 | S | <0.25 | S | 0 | 3/21 | 14.29 | |
| Year | 2016 | 2017 | 2018 | 2019 | Standard Deviation |
|---|---|---|---|---|---|
| ESBL | 313.58 | 367.25 | 280.72 | 420.96 | 48.48 |
| Penicillins | 197.28 | 251.61 | 120.87 | 221.90 | 38.84 |
| Cephalosporins | 104.69 | 111.45 | 147.29 | 180.45 | 27.90 |
| Carbapenems | 11.61 | 4.18 | 12.55 | 18.61 | 3.84 |
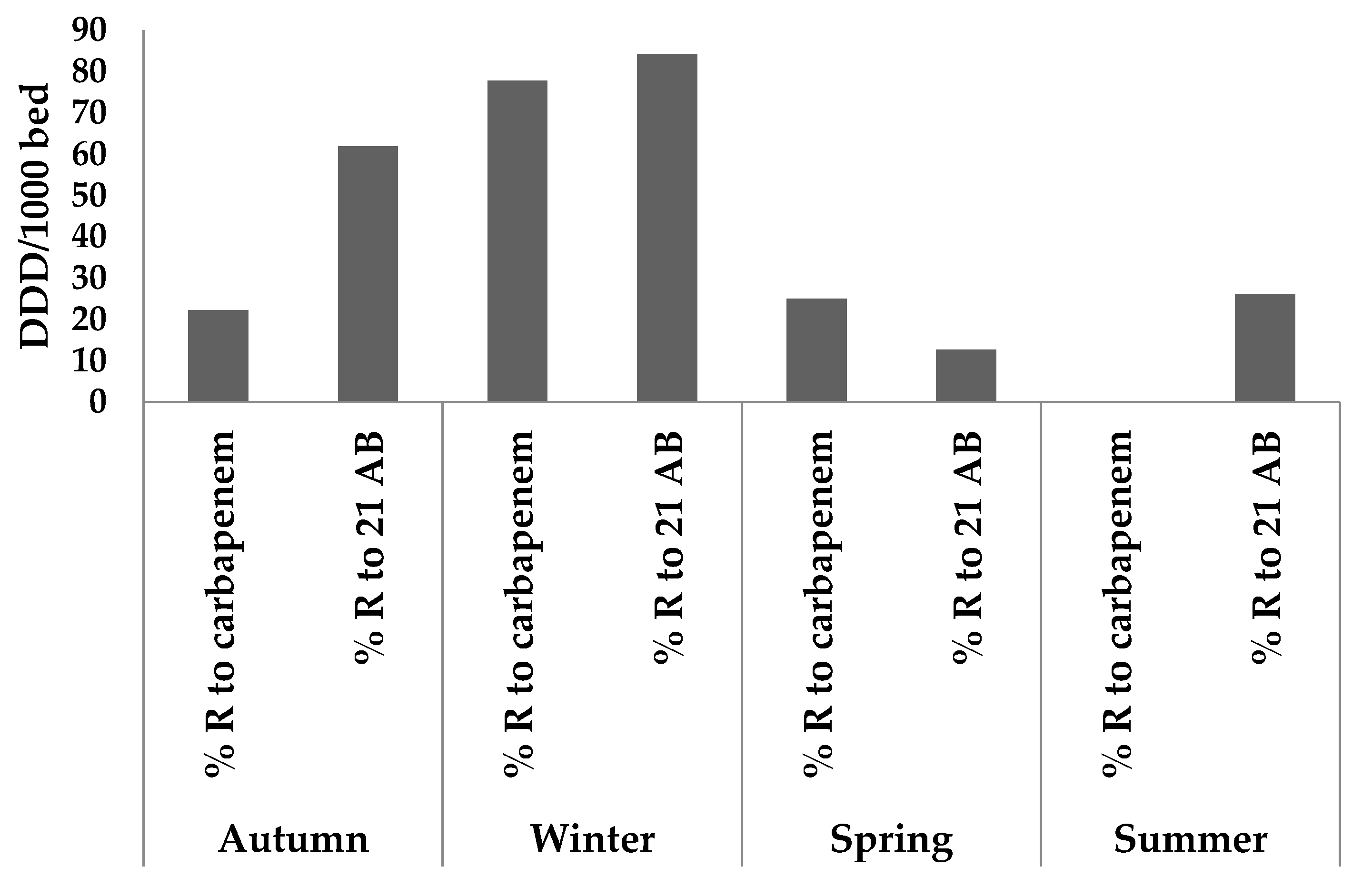
| Exam No. | Collection Season | Identified Germs | Ertapenem | Imipenem | Meropenem | % R to Carbp | Frequency of Bacterial Resistance among 21 AB | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R/S | MIC | R/S | MIC | R/S | MIC | R/N.AB | % Resistance | ||||
| 1 | Autumn | Klebsiella oxytoca | S | S | S | 0 | 10/21 | 47.61 | |||
| 2 | Autumn | Escherichia coli | S | S | S | 0 | 6/21 | 28.57 | |||
| 3 | Autumn | Klebsiella pneumoniae | S | R | >4 | R | >2 | 66.66 | 12/21 | 57.14 | |
| 4 | Winter | Klebsiella pneumoniae | R | >0.13 | R | >0.25 | R | >0.13 | 100 | 14/21 | 66.66 |
| 5 | Winter | Escherichia coli | S | R | >2 | R | >1 | 66.66 | 19/21 | 90.47 | |
| 6 | Winter | Serratia odorifera | R | >0.13 | S | S | 33.33 | 13/21 | 61.90 | ||
| 7 | Spring | Klebsiella pneumoniae | S | S | <0.25 | S | <0.13 | 0 | 3/21 | 14.28 | |
| 8 | Spring | Escherichia coli | R | S | S | 33.33 | 3/21 | 14.28 | |||
| 9 | Spring | Escherichia coli | S | S | R | 33.33 | 6/21 | 28.57 | |||
| 10 | Summer | Klebsiella oxytoca | R | S | <0.25 | R | >1 | 66.66 | 10/21 | 47.61 | |
| 11 | Summer | Aeromonas caviae | S | S | <0.13 | S | 0 | 5/21 | 23.80 | ||
| 12 | Summer | Escherichia coli | S | S | S | 0 | 4/21 | 19.00 | |||
| Year | 2016 | 2017 | 2018 | 2019 | Standard Deviation |
|---|---|---|---|---|---|
| ESBL | 539.39 | 611.71 | 651.91 | 821.02 | 82.51 |
| Penicillins | 357.99 | 467.68 | 459.50 | 577.65 | 56.97 |
| Cephalosporins | 170.32 | 138.70 | 181.50 | 219.29 | 22.94 |
| Carbapenems | 11.12 | 5.32 | 10.91 | 24.08 | 5.61 |
| Study Site | % of Ertapenem-Resistant Bacteria | % of Imipenem-Resistant Bacteria | % of Meropenem-Resistant Bacteria | % Resistance to the All Carbapenem Family |
|---|---|---|---|---|
| ArzH | 26.87 | 17.39 | 43.48 | 23.51 |
| MCH | 28.57 | 42.86 | 57.14 | 42.86 |
| Average | 18.48 | 38.42 | 33.54 | 28.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loumame, E.h.; Tounsi, A.; Amir, S.; Soraa, N.; Ouazzani, N. Microbial Resistance to Carbapenems in Effluents from Gynaecological, Paediatric and Surgical Hospital Units. Antibiotics 2022, 11, 1103. https://doi.org/10.3390/antibiotics11081103
Loumame Eh, Tounsi A, Amir S, Soraa N, Ouazzani N. Microbial Resistance to Carbapenems in Effluents from Gynaecological, Paediatric and Surgical Hospital Units. Antibiotics. 2022; 11(8):1103. https://doi.org/10.3390/antibiotics11081103
Chicago/Turabian StyleLoumame, El hassan, Abdessamad Tounsi, Soumia Amir, Nabila Soraa, and Naaila Ouazzani. 2022. "Microbial Resistance to Carbapenems in Effluents from Gynaecological, Paediatric and Surgical Hospital Units" Antibiotics 11, no. 8: 1103. https://doi.org/10.3390/antibiotics11081103
APA StyleLoumame, E. h., Tounsi, A., Amir, S., Soraa, N., & Ouazzani, N. (2022). Microbial Resistance to Carbapenems in Effluents from Gynaecological, Paediatric and Surgical Hospital Units. Antibiotics, 11(8), 1103. https://doi.org/10.3390/antibiotics11081103






