Thymol as an Adjuvant to Restore Antibiotic Efficacy and Reduce Antimicrobial Resistance and Virulence Gene Expression in Enterotoxigenic Escherichia coli Strains
Abstract
1. Introduction
2. Results
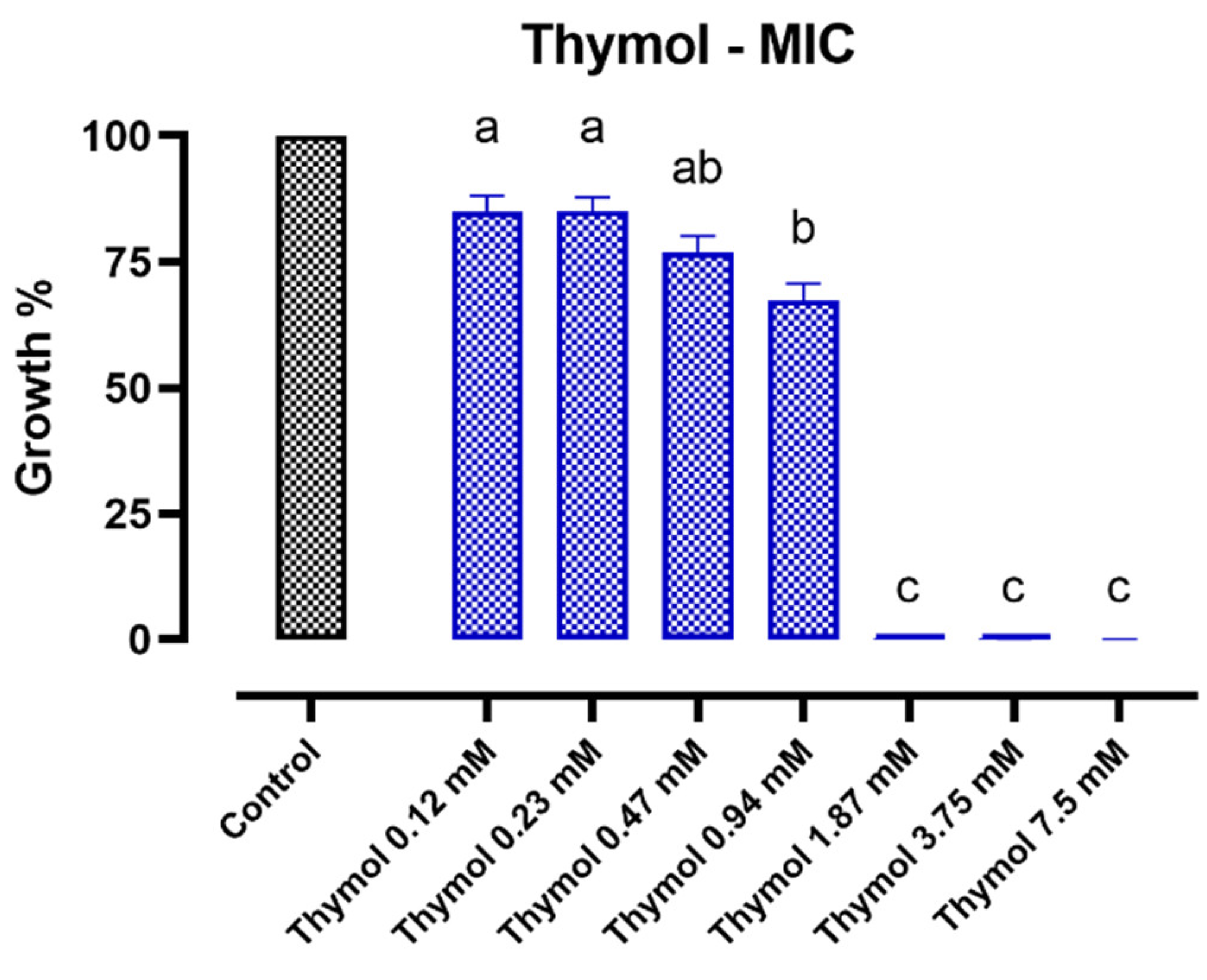
2.1. Minimal Inhibitory Concentration (MIC) Assay—Individual Compounds
2.1.1. Antibiotics
2.1.2. Thymol
2.2. Minimal Inhibitory Concentration (MIC) Assay—Combinations
2.3. Whole-Genome Sequencing (WGS) of ETEC 95 and ETEC 97
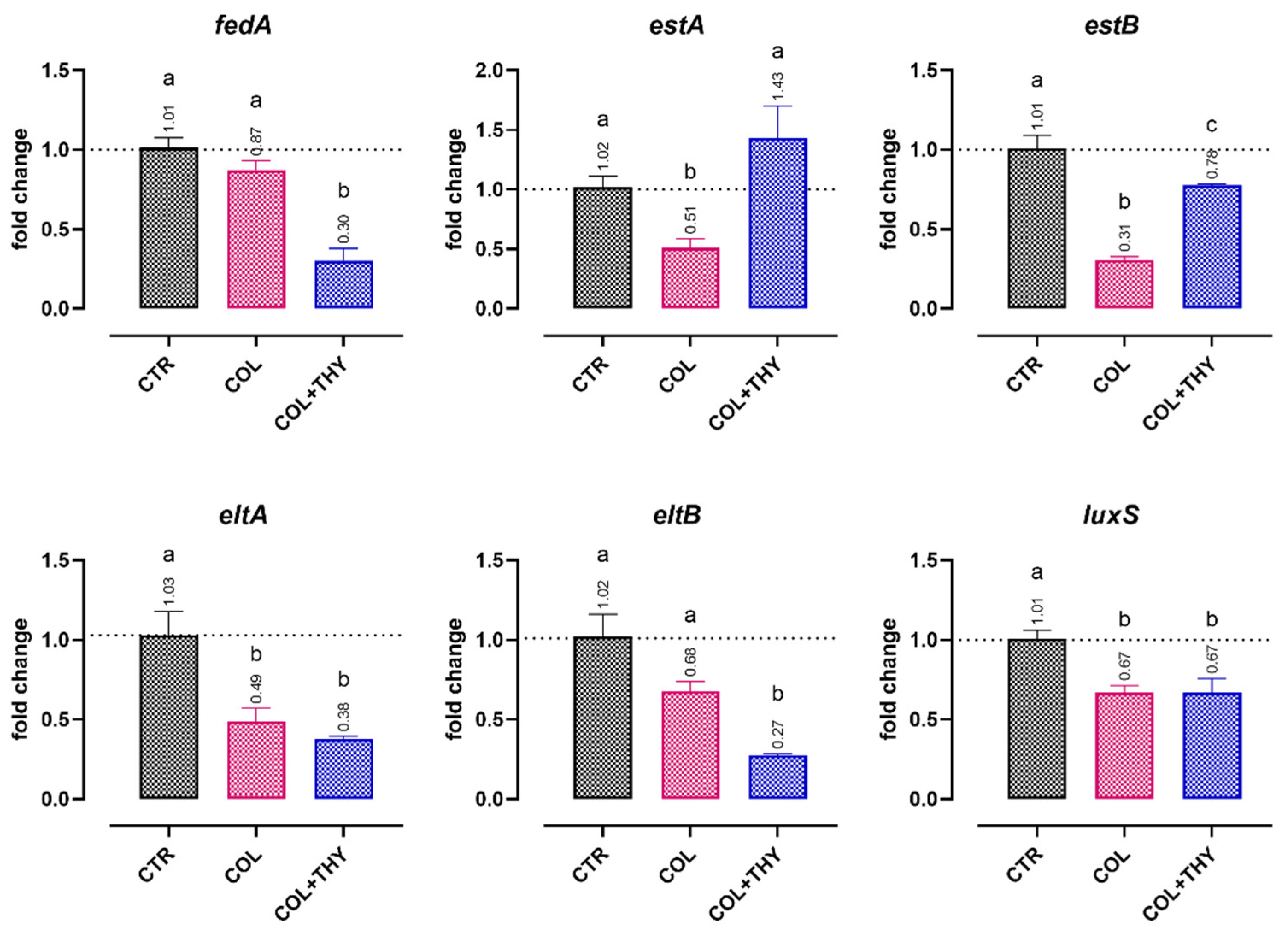
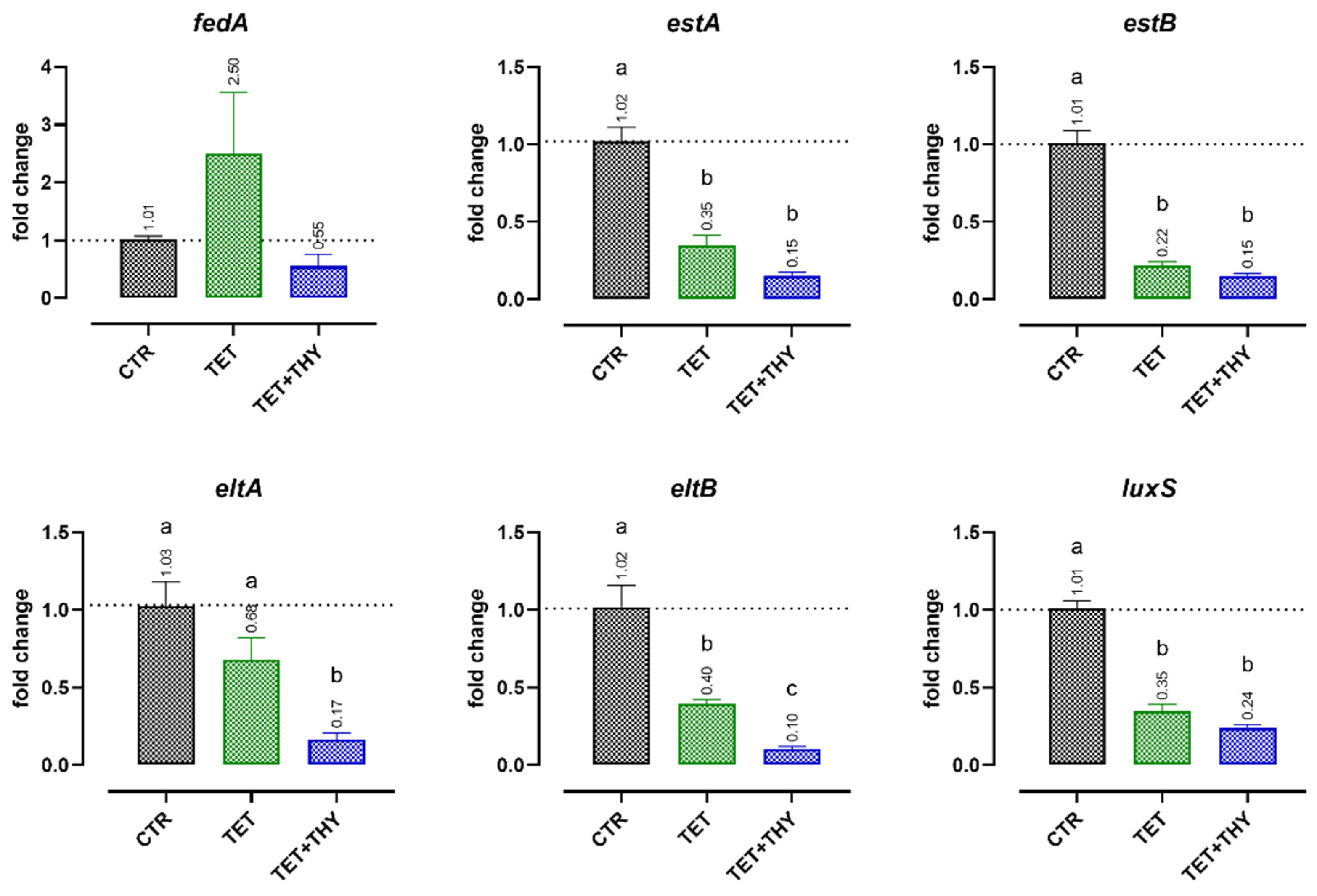
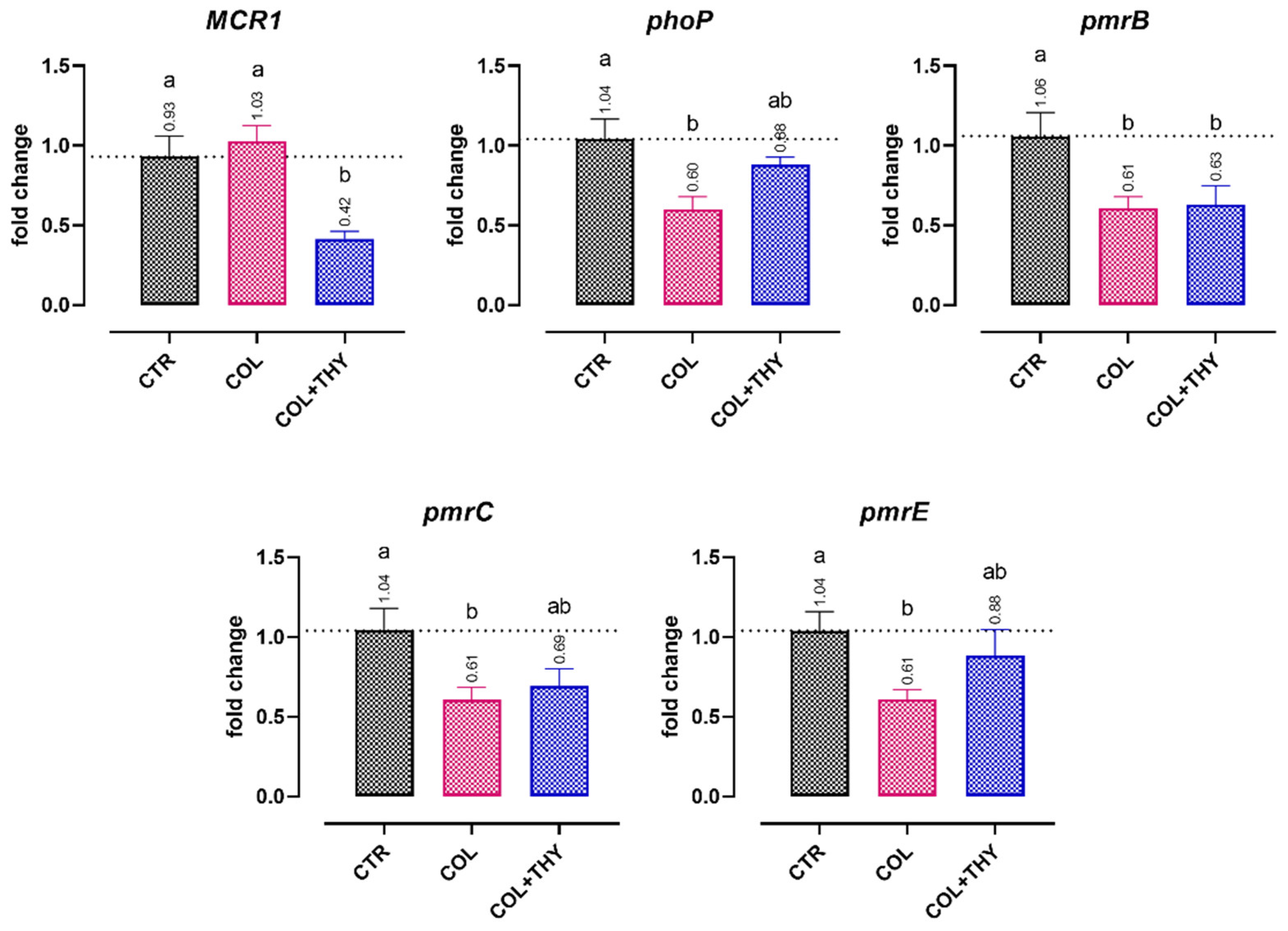
2.4. Gene Expression Analysis
2.4.1. Virulence Genes
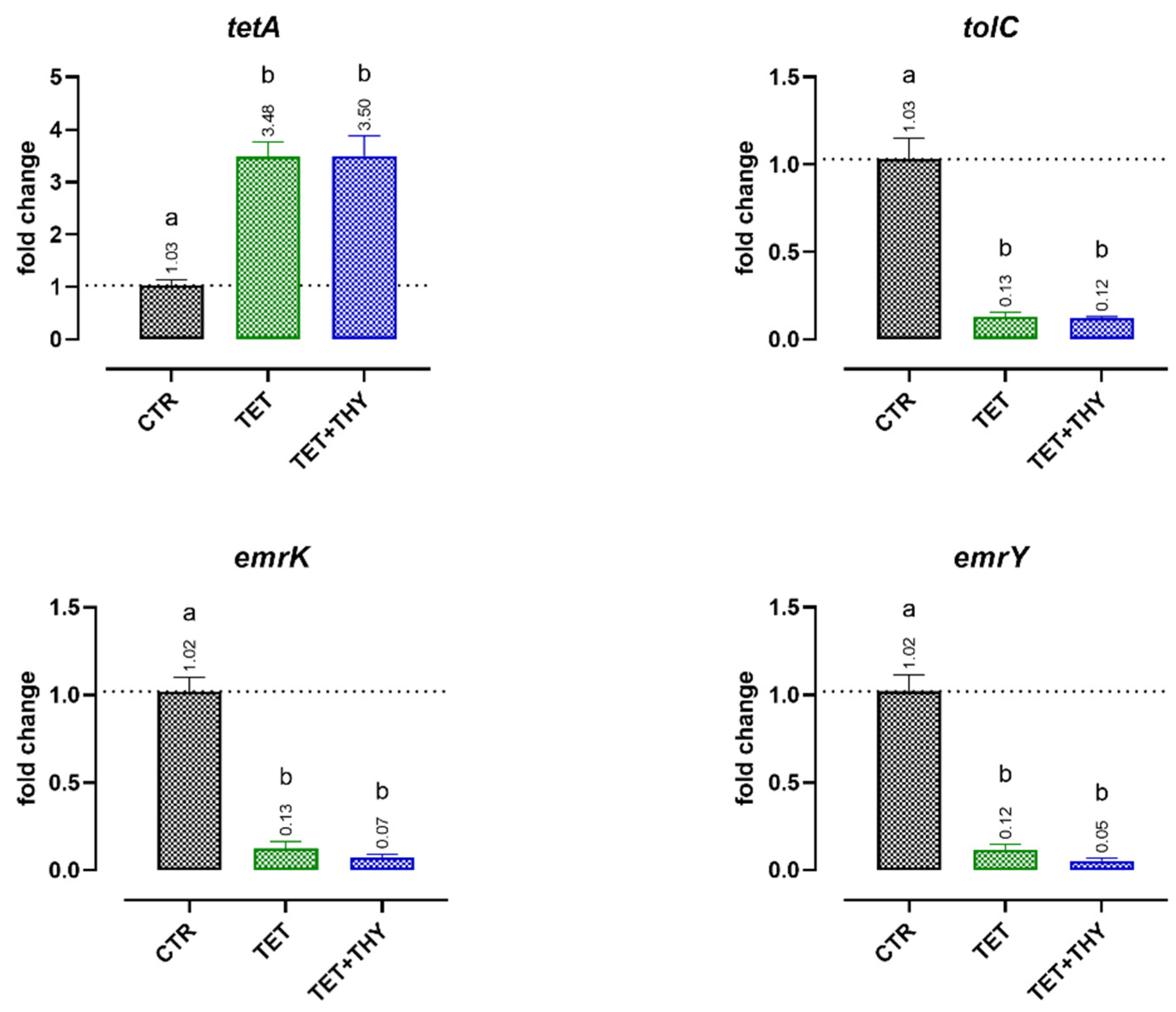
2.4.2. Antibiotic Resistance Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Chemicals and Stock Solutions
4.3. Minimal Inhibitory Concentration (MIC) Assay—Individual Compounds
4.4. Minimal Inhibitory Concentration (MIC) Assay—Combinations
4.5. Whole-Genome Sequencing (WGS) and Sequence Analysis of ETEC 95 and ETEC 97
4.6. Gene Expression Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD/FAO. Meat. In Agricultural Outlook 2021–2030; OECD-FAO Agricultural Outlook; OECD: Paris, France, 2021; pp. 163–177. ISBN 9789264436077. [Google Scholar]
- Campbell, J.M.; Crenshaw, J.D.; Polo, J. The Biological Stress of Early Weaned Piglets. J. Anim. Sci. Biotechnol. 2013, 4, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.; Clark, J.E.; Overman, B.L.; Tozel, C.C.; Huang, J.H.; Rivier, J.E.F.; Blisklager, A.T.; Moeser, A.J. Early Weaning Stress Impairs Development of Mucosal Barrier Function in the Porcine Intestine. Am. J. Physiol. -Gastrointest. Liver Physiol. 2010, 298, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Luppi, A. Swine Enteric Colibacillosis: Diagnosis, Therapy and Antimicrobial Resistance. Porc. Health Manag. 2017, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post Weaning Diarrhea in Pigs: Risk Factors and Non-Colistin-Based Control Strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef]
- Heo, J.M.; Opapeju, F.O.; Pluske, J.R.; Kim, J.C.; Hampson, D.J.; Nyachoti, C.M. Gastrointestinal Health and Function in Weaned Pigs: A Review of Feeding Strategies to Control Post-Weaning Diarrhoea without Using in-Feed Antimicrobial Compounds. J. Anim. Physiol. Anim. Nutr. 2013, 97, 207–237. [Google Scholar] [CrossRef]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal Enterotoxigenic Escherichia Coli. EcoSal Plus 2016, 7, 1–47. [Google Scholar] [CrossRef]
- Carlson, M.S.; Hill, G.M.; Link, J.E. Early- and Traditionally Weaned Nursery Pigs Benefit from Phase-Feeding Pharmacological Concentrations of Zinc Oxide: Effect on Metallothionein and Mineral Concentrations. J. Anim. Sci. 1999, 77, 1199. [Google Scholar] [CrossRef]
- Grilli, E.; Tugnoli, B.; Vitari, F.; Domeneghini, C.; Morlacchini, M.; Piva, A.; Prandini, A. Low Doses of Microencapsulated Zinc Oxide Improve Performance and Modulate the Ileum Architecture, Inflammatory Cytokines and Tight Junctions Expression of Weaned Pigs. Animal 2015, 9, 1760–1768. [Google Scholar] [CrossRef]
- Bonetti, A.; Tugnoli, B.; Piva, A.; Grilli, E. Towards Zero Zinc Oxide: Feeding Strategies to Manage Post-Weaning Diarrhea in Piglets. Animals 2021, 11, 642. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Decision of 26.6.2017 Concerning, in the Framework of Article 35 of Directive 2001/82/EC of the European Parliament and of the Council, the Marketing Authorisations for Veterinary Medicinal Products Containing “Zinc Oxide” to Be Administered Orally to Food Producing Species; European Commission: Brussels, Belgium, 2017. [Google Scholar]
- Eriksen, E.Ø.; Kudirkiene, E.; Christensen, A.E.; Agerlin, M.V.; Weber, N.R.; Nødtvedt, A.; Nielsen, J.P.; Hartmann, K.T.; Skade, L.; Larsen, L.E.; et al. Post-Weaning Diarrhea in Pigs Weaned without Medicinal Zinc: Risk Factors, Pathogen Dynamics, and Association to Growth Rate. Porc. Health Manag. 2021, 7, 54. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials (CIA) for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2018; ISBN 978-92-4-151552-8. [Google Scholar]
- Palma, E.; Tilocca, B.; Roncada, P. Antimicrobial Resistance in Veterinary Medicine: An Overview. Int. J. Mol. Sci. 2020, 21, 1914. [Google Scholar] [CrossRef] [PubMed]
- Rossi, B.; Toschi, A.; Piva, A.; Grilli, E. Single Components of Botanicals and Nature-Identical Compounds as a Non-Antibiotic Strategy to Ameliorate Health Status and Improve Performance in Poultry and Pigs. Nutr. Res. Rev. 2020, 33, 218–234. [Google Scholar] [CrossRef] [PubMed]
- Nagoor Meeran, M.F.; Javed, H.; Al Taee, H.; Azimullah, S.; Ojha, S.K. Pharmacological Properties and Molecular Mechanisms of Thymol: Prospects for Its Therapeutic Potential and Pharmaceutical Development. Front. Pharmacol. 2017, 8, 380. [Google Scholar] [CrossRef] [PubMed]
- Kachur, K.; Suntres, Z. The Antibacterial Properties of Phenolic Isomers, Carvacrol and Thymol. Crit. Rev. Food Sci. Nutr. 2019, 16, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Ezzat Abd El-Hack, M.; Alagawany, M.; Ragab Farag, M.; Tiwari, R.; Karthik, K.; Dhama, K.; Zorriehzahra, J.; Adel, M. Beneficial Impacts of Thymol Essential Oil on Health and Production of Animals, Fish and Poultry: A Review. J. Essent. Oil Res. 2016, 28, 365–382. [Google Scholar] [CrossRef]
- Marchese, A.; Orhan, I.E.; Daglia, M.; Barbieri, R.; Di Lorenzo, A.; Nabavi, S.F.; Gortzi, O.; Izadi, M.; Nabavi, S.M. Antibacterial and Antifungal Activities of Thymol: A Brief Review of the Literature. Food Chem. 2016, 210, 402–414. [Google Scholar] [CrossRef]
- Bonetti, A.; Tugnoli, B.; Rossi, B.; Giovagnoni, G.; Piva, A.; Grilli, E. Nature-Identical Compounds and Organic Acids Reduce E. Coli K88 Growth and Virulence Gene Expression In Vitro. Toxins 2020, 12, 468. [Google Scholar] [CrossRef]
- Palaniappan, K.; Holley, R.A. Use of Natural Antimicrobials to Increase Antibiotic Susceptibility of Drug Resistant Bacteria. Int. J. Food Microbiol. 2010, 140, 164–168. [Google Scholar] [CrossRef]
- Hamoud, R.; Zimmermann, S.; Reichling, J.; Wink, M. Synergistic Interactions in Two-Drug and Three-Drug Combinations (Thymol, EDTA and Vancomycin) against Multi Drug Resistant Bacteria Including E. coli. Phytomedicine 2014, 21, 443–447. [Google Scholar] [CrossRef]
- Liu, Q.; Niu, H.; Zhang, W.; Mu, H.; Sun, C.; Duan, J. Synergy among Thymol, Eugenol, Berberine, Cinnamaldehyde and Streptomycin against Planktonic and Biofilm-Associated Food-Borne Pathogens. Lett. Appl. Microbiol. 2015, 60, 421–430. [Google Scholar] [CrossRef]
- Giovagnoni, G.; Tugnoli, B.; Piva, A.; Grilli, E. Organic Acids and Nature Identical Compounds Can Increase the Activity of Conventional Antibiotics Against Clostridium Perfringens and Enterococcus Cecorum In Vitro. J. Appl. Poult. Res. 2019, 28, 1398–1407. [Google Scholar] [CrossRef]
- CLSI VET01S ED5:2020; Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 5th ed. Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2020; ISBN 978-1-68440-093-5.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, 12th ed.; EUCAST: Växjö, Sweden, 2022. [Google Scholar]
- CLSI M100; Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022; ISBN 978-1-68440-134-5.
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2019–2020. EFSA J. 2022, 20, 7209. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Keiffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 14. [Google Scholar] [CrossRef]
- Cassir, N.; Rolain, J.M.; Brouqui, P. A New Strategy to Fight Antimicrobial Resistance: The Revival of Old Antibiotics. Front. Microbiol. 2014, 5, 551. [Google Scholar] [CrossRef] [PubMed]
- Scarpignato, C.; Rampal, P. Prevention and Treatment of Traveler’s Diarrhea: A Clinical Pharmacological Approach (Part 1 of 2). Chemotherapy 1995, 41, 48–62. [Google Scholar] [CrossRef]
- Burch, D.G.S.; Sperling, D. Amoxicillin—Current Use in Swine Medicine. J. Vet. Pharmacol. Ther. 2018, 41, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Song, H.J.; Kim, S.J.; Moon, D.C.; Mechesso, A.F.; Choi, J.H.; Kang, H.Y.; Boby, N.; Yoon, S.S.; Lim, S.K. Antimicrobial Resistance in Escherichia Coli Isolates from Healthy Food Animals in South Korea, 2010–2020. Microorganisms 2022, 10, 524. [Google Scholar] [CrossRef]
- Stannarius, C.; Bürgi, E.; Regula, G.; Zychowska, M.A.; Zweifel, C.; Stephan, R. Antimicrobial Resistance in Escherichia Coli Strains Isolated from Swiss Weaned Pigs and Sows. Schweiz. Arch. Tierheilkd. 2009, 151, 119–125. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Wei, S.H.; Hobman, J.L.; Dodd, C.E.R. Antibiotic and Metal Resistance in Escherichia Coli Isolated from Pig Slaughterhouses in the United Kingdom. Antibiotics 2020, 9, 746. [Google Scholar] [CrossRef]
- Belmar-Liberato, R.; Gonzalez-Canga, A.; Tamame-Martin, P.; Escribano-Salazar, M. Amoxicillin and Amoxicillin-Clavulanic Acid Resistance in Veterinary Medicine—The Situation in Europe: A Review. Vet. Med. 2011, 56, 473–485. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin Update on Its Mechanism of Action and Resistance, Present and Future Challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef] [PubMed]
- Tiku, A.R. Antimicrobial Compounds and Their Role in Plant Defense. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Berlin/Heidelberg, Germany, 2018; pp. 283–307. [Google Scholar] [CrossRef]
- Savoia, D. Plant-Derived Antimicrobial Compounds: Alternatives to Antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Mickymaray, S. Efficacy and Mechanism of Traditional Medicinal Plants and Bioactive Compounds against Clinically Important Pathogens. Antibiotics 2019, 8, 257. [Google Scholar] [CrossRef]
- Salehi, B.; Mishra, A.P.; Shukla, I.; Sharifi-Rad, M.; del Mar Contreras, M.; Segura-Carretero, A.; Fathi, H.; Nasrabadi, N.N.; Kobarfard, F.; Sharifi-Rad, J. Thymol, Thyme, and Other Plant Sources: Health and Potential Uses. Phyther. Res. 2018, 32, 1688–1706. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, F.; Ji, B.P.; Pei, R.S.; Xu, N. The Antibacterial Mechanism of Carvacrol and Thymol against Escherichia coli. Lett. Appl. Microbiol. 2008, 47, 174–179. [Google Scholar] [CrossRef]
- Al-Kandari, F.; Al-Temaimi, R.; Van Vliet, A.H.M.; Woodward, M.J. Thymol Tolerance in Escherichia Coli Induces Morphological, Metabolic and Genetic Changes. BMC Microbiol. 2019, 19, 294. [Google Scholar] [CrossRef]
- Yao, Z.; Feng, L.; Zhao, Y.; Zhang, X.; Chen, L.; Wang, L.; Zhang, Y.; Sun, Y.; Zhou, T.; Cao, J. Thymol Increases Sensitivity of Clinical Col-R Gram-Negative Bacteria to Colistin. Microbiol. Spectr. 2022, 12, e00184-22. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and Its Role in the Era of Antibiotic Resistance: An Extended Review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, L.; Liu, J.; Ma, H. Synergistic Effect of Eugenol with Colistin against Clinical Isolated Colistin-Resistant Escherichia Coli Strains. Antimicrob. Resist. Infect. Control 2018, 7, 17. [Google Scholar] [CrossRef]
- Chen, H.D.; Groisman, E.A. The Biology of the PmrA/PmrB Two-Component System: The Major Regulator of Lipopolysaccharide Modifications. Annu. Rev. Microbiol. 2013, 67, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular Mechanisms Related to Colistin Resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.L.; Worthington, R.J.; Hittle, L.E.; Zurawski, D.V.; Ernst, R.K.; Melander, C. Small Molecule Downregulation of PmrAB Reverses Lipid A Modification and Breaks Colistin Resistance. ACS Chem. Biol. 2014, 9, 122–127. [Google Scholar] [CrossRef]
- Duan, Q.; Yao, F.; Zhu, G. Major Virulence Factors of Enterotoxigenic Escherichia Coli in Pigs. Ann. Microbiol. 2012, 62, 7–14. [Google Scholar] [CrossRef]
- Fleckenstein, J.M.; Hardwidge, P.R.; Munson, G.P.; Rasko, D.A.; Sommerfelt, H.; Steinsland, H. Molecular Mechanisms of Enterotoxigenic Escherichia Coli Infection. Microbes Infect. 2010, 12, 89–98. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline Antibiotics and Resistance Mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Miladi, H.; Zmantar, T.; Kouidhi, B.; Al Qurashi, Y.M.A.; Bakhrouf, A.; Chaabouni, Y.; Mahdouani, K.; Chaieb, K. Synergistic Effect of Eugenol, Carvacrol, Thymol, p-Cymene and γ-Terpinene on Inhibition of Drug Resistance and Biofilm Formation of Oral Bacteria. Microb. Pathog. 2017, 112, 156–163. [Google Scholar] [CrossRef]
- Miladi, H.; Zmantar, T.; Chaabouni, Y.; Fedhila, K.; Bakhrouf, A.; Mahdouani, K.; Chaieb, K. Antibacterial and Efflux Pump Inhibitors of Thymol and Carvacrol against Food-Borne Pathogens. Microb. Pathog. 2016, 99, 95–100. [Google Scholar] [CrossRef]
- Cirino, I.C.S.; Menezes-Silva, S.M.P.; Silva, H.T.D.; de Souza, E.L.; Siqueira-Júnior, J.P. The Essential Oil from Origanum vulgare L. and Its Individual Constituents Carvacrol and Thymol Enhance the Effect of Tetracycline against Staphylococcus aureus. Chemotherapy 2014, 60, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Pasqua, M.; Bonaccorsi di Patti, M.C.; Fanelli, G.; Utsumi, R.; Eguchi, Y.; Trirocco, R.; Prosseda, G.; Grossi, M.; Colonna, B. Host-Bacterial Pathogen Communication: The Wily Role of the Multidrug Efflux Pumps of the MFS Family. Front. Mol. Biosci. 2021, 8, 723274. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Deng, Z.; Yan, A. Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Agreles, M.A.A.; Cavalcanti, I.D.L.; Cavalcanti, I.M.F. The Role of Essential Oils in the Inhibition of Efflux Pumps and Reversion of Bacterial Resistance to Antimicrobials. Curr. Microbiol. 2021, 78, 3609–3619. [Google Scholar] [CrossRef] [PubMed]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Karami, N.; Nowrouzian, F.; Adlerberth, I.; Wold, A.E. Tetracycline Resistance in Escherichia Coli and Persistence in the Infantile Colonic Microbiota. Antimicrob. Agents Chemother. 2006, 50, 156–161. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-Virulence Strategies to Combat Bacteria-Mediated Disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Center for Genomic Epidemiology. Available online: http://www.genomicepidemiology.org/services/ (accessed on 7 June 2022).
- Giovagnoni, G.; Tugnoli, B.; Piva, A.; Grilli, E. Dual Antimicrobial Effect of Medium-Chain Fatty Acids against an Italian Multidrug Resistant Brachyspira Hyodysenteriae Strain. Microorganisms 2022, 10, 301. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Enterotoxigenic Escherichia coli (ETEC) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibiotics | MIC (mg/L) | ||||||||
| >64 | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | |
| Amoxicillin | 5 | 1 | |||||||
| Ceftiofur | 1 | 4 | 1 | ||||||
| Colistin | 2 | 2 | 2 | ||||||
| Nalidixic acid | 3 | 3 | |||||||
| Enrofloxacin | 3 | 3 | |||||||
| Florfenicol | 1 | 3 | 1 | 1 | |||||
| Neomycin | 3 | 1 | 2 | ||||||
| Trimethoprim | 2 | 2 | 2 | ||||||
| Tetracycline | 4 | 2 | |||||||
| >608 | 608 | 304 | 152 | 76 | 38 | 19 | 9.5 | 4.75 | |
| Sulfamethoxazole | 4 | 1 | 1 | ||||||
| >64/32 | 64/32 | 32/16 | 16/8 | 8/4 | 4/2 | 2/1 | 1/0.5 | 0.5/0.25 | |
| Amox./Clav. (2:1) 1 | 1 | 2 | 2 | 1 | |||||
| >32/608 | 32/608 | 16/304 | 8/152 | 4/76 | 2/38 | 1/19 | 0.5/9.5 | 0.25/4.75 | |
| Trim./Sulf. (1:19) 2 | 4 | 2 | |||||||
| Inhibition | COL 16 | COL 8 | COL 4 | |
|---|---|---|---|---|
| % | 35 | 10 | 7 | |
| THY 0.94 | 15 | 100 *§ | 54 *§ | 35 |
| THY 0.47 | 8 | 100 *§ | 47 *§ | 38 |
| Inhibition | TET 64 | TET 32 | TET 16 | |
|---|---|---|---|---|
| % | 53 | 35 | 28 | |
| THY 0.94 | 47 | 95 *§ | 77 *§ | 61 * |
| THY 0.47 | 32 | 61 | 47 | 39 |
| Inhibition | T + S 32/608 | T + S 16/304 | T + S 8/152 | |
|---|---|---|---|---|
| % | 39 | 25 | 19 | |
| THY 0.94 | 25 | 45 § | 34 | 30 |
| THY 0.47 | 22 | 41 § | 30 | 25 |
| Inhibition | ENR 1 | ENR 0.5 | ENR 0.25 | |
|---|---|---|---|---|
| % | 53 | 24 | 17 | |
| THY 0.94 | 38 | 79 § | 46 | 38 |
| THY 0.47 | 25 | 48 § | 30 | 25 |
| Inhibition | AMO 64 | AMO 32 | AMO 16 | |
|---|---|---|---|---|
| % | 9 | 14 | 16 | |
| THY 0.94 | 25 | 15 | 26 | 27 |
| THY 0.47 | 21 | 15 | 24 | 25 |
| Inhibition | NEO 64 | NEO 32 | NEO 16 | |
|---|---|---|---|---|
| % | 6 | 8 | 8 | |
| THY 0.94 | 16 | 11 | 10 | 10 |
| THY 0.47 | 10 | 9 | 13 | 12 |
| Strain | ETEC 95 | ETEC 97 | |
|---|---|---|---|
| Serotype | O131:H4 | O138:H14 | |
| Adhesin | F18 | F18 | |
| Toxins | STa, STb, LT | STa, STb, LT | |
| Other virulence genes | fedA, fedF, gad, hra, iha, iss, neuC, ompT, terC, traT | air, astA, cba, chuA, cma, fedA, fedF, iss, lpfA, ltcA, ompT, terC, traT | |
| Plasmids | IncFII(29), IncHI2, IncHI2A, IncQ1, IncX1 | IncFIB, IncFII, IncI1-I, IncX1, IncX4 | |
| Resistance genes | Aminoglycosides | aadA1, aph(3′)-Ia, aph(3′’)-Ib, aac(3)-IIa, aph(6)-Id | aac(3)-IV, aph(3′)-Ia, aph(4)-Ia, aadA2, aadA1 |
| Polymyxins | MCR-1, pmrC, pmrE, pmrF | MCR-1, pmrC, pmrE, pmrF | |
| Folate pathway | sul1, sul2, sul3, drfA1 | sul3, drfA12 | |
| Tetracyclines | tet(A), emrK, emrY, tolC | tet(A), emrK, emrY, tolC | |
| Quinolones | gyrA, emrK, emrY, tolC | gyrA, emrK, emrY, tolC | |
| Amphenicols | marA, marB, marR | cmlA1, floR | |
| Beta-lactams | blaTEM-1A | blaTEM-1A | |
| Observed antimicrobial resistance (MIC in mg/L) 1 | AMO (>64), AMO + CLA (16/8), COL (32), TRI (>64), SUL (>608), TRI + SUL (>32/608), NAL (>64), ENR (2), FLO (16), NEO (>64), TET (>64) | AMO (>64), AMO + CLA (32/16), COL (32), TRI (>64), SUL (>608), TRI + SUL (>32/608), NAL (>64), ENR (2), FLO (>64), NEO (>64), TET (>64) | |
| ETEC Strain | Adhesin | Toxins |
|---|---|---|
| ETEC 95 | F18+ | STa+; STb+; LT+ |
| ETEC 97 | F18+ | STa+; STb+; LT+ |
| ETEC 99 | F4+ | STa+; STb+; LT+ |
| ETEC 104 | F18+ | STa+; STb+ |
| ETEC 105 | F4+ | STa+; STb+ |
| ETEC 106 | F18+ | STa+; STb+ |
| Molecule | Range of Tested Concentrations |
|---|---|
| Single antibiotics | 0.5–64 mg/L |
| Amoxicillin/clavulanic acid (2:1) 1 | 0.5/0.25–64/32 mg/L |
| Trimethoprim/sulfamethoxazole (1:19) 1 | 0.25/4.75–32/608 mg/L |
| Sulfamethoxazole | 608–4.75 mg/L |
| Thymol | 0.12–7.5 mM |
| Antibiotic | Sub-MIC Tested Concentrations |
|---|---|
| Amoxicillin, neomycin, tetracycline | 64–32–16 mg/L |
| Enrofloxacin | 1–0.5–0.25 mg/L |
| Trimethoprim/sulfamethoxazole (1:19) 1 | 32/608–16/304–8/152 mg/L |
| Colistin | 16–8–4 mg/L |
| Functions | Gene | Sequences (5′→3′) | Product Length (bp) | Ref 1 |
|---|---|---|---|---|
| Adhesion to cells | fedA | F: GCTAATCAAGGGGGAGTGGC R: ACAGTGCTATTCGACGCCTT | 110 | This study |
| LT toxin production | eltA | F: TTGGTGATCCGGTGGGAAAC R: AGGAGGTTTCTGCGTTAGGTG | 185 | [20] |
| eltB | F: CACGGAGCTCCCCAGACTAT R: GCCTGCCATCGATTCCGTAT | 105 | ||
| STa toxin production | estA | F: CAACTGAATCACTTGACTCTT R: TTAATAACATCCAGCACAGG | 158 | |
| STb toxin production | estB | F: TGCCTATGCATCTACACAA R: CTCCAGCAGTACCATCTC | 113 | |
| Quorum sensing | luxS | F: CAGTGCCAGTTCTTCGTTGC R: TGAACGTCTACCAGTGTGGC | 116 | |
| Resistance to colistin | MCR1 | F: GGGCCTGCGTATTTTAAGCG R: CATAGGCATTGCTGTGCGTC | 184 | This study |
| phoP | F: CTGGTATTAACCGCCCGTGA R: GCGAAATGACCTGTGAAGCC | 160 | ||
| pmrB | F: ATCTGGAACTGCTGGCGAAA R: GAAAATGACTGTCCGGCACG | 121 | ||
| pmrC | F: GTACCGTGCATGTTCTCGGA R: CGCCATCGTTGTCATTCCAC | 127 | ||
| pmrE | F: TCATCATCGCCACTCCAACC R: GCGGTAAAACCAACGGGAAC | 145 | ||
| Resistance to tetracyclines | tetA | F: GCTGTTTCCTTTTGCCGGAG R: TGAAGAAGACCGCCATCAGG | 132 | This study |
| tolC | F: ACGAAGTGACCGCACGTAAT R: AGCGACAGGTTGCGTTTTTC | 169 | ||
| emrK | F: GCACAAAATGCGACAGGGAA R: GCATCTCGGCAATGTCTTCG | 172 | ||
| emrY | F: TGGGAGTCGACCTCAGAGAA R: GTCCGGCCCATATCGCATTA | 154 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonetti, A.; Tugnoli, B.; Piva, A.; Grilli, E. Thymol as an Adjuvant to Restore Antibiotic Efficacy and Reduce Antimicrobial Resistance and Virulence Gene Expression in Enterotoxigenic Escherichia coli Strains. Antibiotics 2022, 11, 1073. https://doi.org/10.3390/antibiotics11081073
Bonetti A, Tugnoli B, Piva A, Grilli E. Thymol as an Adjuvant to Restore Antibiotic Efficacy and Reduce Antimicrobial Resistance and Virulence Gene Expression in Enterotoxigenic Escherichia coli Strains. Antibiotics. 2022; 11(8):1073. https://doi.org/10.3390/antibiotics11081073
Chicago/Turabian StyleBonetti, Andrea, Benedetta Tugnoli, Andrea Piva, and Ester Grilli. 2022. "Thymol as an Adjuvant to Restore Antibiotic Efficacy and Reduce Antimicrobial Resistance and Virulence Gene Expression in Enterotoxigenic Escherichia coli Strains" Antibiotics 11, no. 8: 1073. https://doi.org/10.3390/antibiotics11081073
APA StyleBonetti, A., Tugnoli, B., Piva, A., & Grilli, E. (2022). Thymol as an Adjuvant to Restore Antibiotic Efficacy and Reduce Antimicrobial Resistance and Virulence Gene Expression in Enterotoxigenic Escherichia coli Strains. Antibiotics, 11(8), 1073. https://doi.org/10.3390/antibiotics11081073







