Transcriptomic and Metabolomic Analysis of a Fusidic Acid-Selected fusA Mutant of Staphylococcus aureus
Abstract
1. Introduction
2. Results and Discussion
2.1. FA MICs and Comparative Genomic Sequencing (CGS)
2.2. Phenotypic Characterization of SH10001st-2
2.3. Overview of the SH10001st-2 Transcriptome
2.4. Highly Upregulated and Downregulated Genes in SH10001st-2
2.5. SH10001st-2 Expression of Virulence Factors
2.6. Metabolomics of SH1000 vs SH10001st-2
3. Materials and Methods
3.1. Culture Conditions, Growth Curves and Antibiotic Susceptibility
3.2. Metabolic Activity
3.3. Hemolysis and Coagulase Test
3.4. DNA and RNA Purification and cDNA Synthesis
3.5. Comparative Genomic Sequencing
3.6. DNA Microarray and Quantitative Real-Time PCR Analyses (qRT-PCR)
3.7. Metabolite Extraction and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G.; Petit, S.; et al. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections—United States. Morb. Mortal. Wkly. Rep. 2019, 68, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Katayama, Y.; Matsuo, M.; Sasaki, T.; Morimoto, Y.; Sekiguchi, A.; Baba, T. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J. Infect. Chemother. 2014, 20, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Hajikhani, B.; Goudarzi, M.; Kakavandi, S.; Amini, S.; Zamani, S.; van Belkum, A.; Goudarzi, H.; Dadashi, M. The global prevalence of fusidic acid resistance in clinical isolates of Staphylococcus aureus: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 2021, 10, 75. [Google Scholar] [CrossRef]
- Guo, X.; Peisker, K.; Backbro, K.; Chen, Y.; Koripella, R.K.; Mandava, C.S.; Sanyal, S.; Selmer, M. Structure and function of FusB: An elongation factor G-binding fusidic acid resistance protein active in ribosomal translocation and recycling. Open Biol. 2012, 2, 120016. [Google Scholar] [CrossRef]
- O’Brien, F.G.; Price, C.; Grubb, W.B.; Gustafson, J.E. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 2002, 50, 313–321. [Google Scholar] [CrossRef]
- Chopra, I. Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J. Gen. Microbiol. 1976, 96, 229–238. [Google Scholar] [CrossRef]
- Laurberg, M.; Kristensen, O.; Martemyanov, K.; Gudkov, A.T.; Nagaev, I.; Hughes, D.; Liljas, A. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 2000, 303, 593–603. [Google Scholar] [CrossRef]
- Chen, C.M.; Huang, M.; Chen, H.F.; Ke, S.C.; Li, C.R.; Wang, J.H.; Wu, L.T. Fusidic acid resistance among clinical isolates of methicillin-resistant Staphylococcus aureus in a Taiwanese hospital. BMC Microbiol. 2011, 11, 98. [Google Scholar] [CrossRef]
- Norstrom, T.; Lannergard, J.; Hughes, D. Genetic and phenotypic identification of fusidic acid-resistant mutants with the small-colony-variant phenotype in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 4438–4446. [Google Scholar] [CrossRef]
- Huang, J.; O’Toole, P.W.; Shen, W.; Amrine-Madsen, H.; Jiang, X.; Lobo, N.; Palmer, L.M.; Voelker, L.; Fan, F.; Gwynn, M.N.; et al. Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 909–917. [Google Scholar] [CrossRef]
- EUCAST: Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 8 May 2022).
- Delgado, A.; Zaman, S.; Muthaiyan, A.; Nagarajan, V.; Elasri, M.O.; Wilkinson, B.J.; Gustafson, J.E. The fusidic acid stimulon of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 62, 1207–1214. [Google Scholar] [CrossRef]
- Besier, S.; Ludwig, A.; Brade, V.; Wichelhaus, T.A. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 2003, 47, 463–469. [Google Scholar] [CrossRef]
- Besier, S.; Ludwig, A.; Brade, V.; Wichelhaus, T.A. Compensatory adaptation to the loss of biological fitness associated with acquisition of fusidic acid resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 1426–1431. [Google Scholar] [CrossRef]
- Nagaev, I.; Bjorkman, J.; Andersson, D.I.; Hughes, D. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 2001, 40, 433–439. [Google Scholar] [CrossRef]
- Mikuni, O.; Ito, K.; Moffat, J.; Matsumura, K.; McCaughan, K.; Nobukuni, T.; Tate, W.; Nakamura, Y. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc. Natl. Acad. Sci. USA 1994, 91, 5798–5802. [Google Scholar] [CrossRef]
- Zaher, H.S.; Green, R. A primary role for release factor 3 in quality control during translation elongation in Escherichia coli. Cell 2011, 147, 396–408. [Google Scholar] [CrossRef]
- Peng, H.L.; Novick, R.P.; Kreiswirth, B.; Kornblum, J.; Schlievert, P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 1988, 170, 4365–4372. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Xue, T.; You, Y.; Hong, D.; Sun, H.; Sun, B. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect. Immun. 2011, 79, 2154–2167. [Google Scholar] [CrossRef]
- Dutta, A.; Batish, M.; Parashar, V. Structural basis of KdpD histidine kinase binding to the second messenger c-di-AMP. J. Biol. Chem. 2021, 296, 100771. [Google Scholar] [CrossRef]
- Zhao, L.; Xue, T.; Shang, F.; Sun, H.; Sun, B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 2010, 78, 3506–3515. [Google Scholar] [CrossRef] [PubMed]
- Crosby, H.A.; Tiwari, N.; Kwiecinski, J.M.; Xu, Z.; Dykstra, A.; Jenul, C.; Fuentes, E.J.; Horswill, A.R. The Staphylococcus aureus ArlRS two- component system regulates virulence factor expression through MgrA. Mol. Microbiol. 2020, 113, 103–122. [Google Scholar] [CrossRef]
- O’Leary, J.O.; Langevin, M.J.; Price, C.T.; Blevins, J.S.; Smeltzer, M.S.; Gustafson, J.E. Effects of sarA inactivation on the intrinsic multidrug resistance mechanism of Staphylococcus aureus. FEMS Microbiol. Lett. 2004, 237, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, J.H.; Runager, K.; Andersen, C.B.F. The human protein haptoglobin inhibits IsdH-mediated heme-sequestering by Staphylococcus aureus. J. Biol. Chem. 2020, 295, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Radka, C.D.; Batte, J.L.; Frank, M.W.; Rosch, J.W.; Rock, C.O. Oleate Hydratase (OhyA) Is a Virulence Determinant in Staphylococcus aureus. Microbiol. Spectr. 2021, 9, e0154621. [Google Scholar] [CrossRef]
- Sadykov, M.R.; Mattes, T.A.; Luong, T.T.; Zhu, Y.; Day, S.R.; Sifri, C.D.; Lee, C.Y.; Somerville, G.A. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J. Bacteriol. 2010, 192, 1459–1462. [Google Scholar] [CrossRef]
- Kredich, N.M. Biosynthesis of Cysteine. EcoSal Plus 2008, 3. [Google Scholar] [CrossRef]
- Farrow, J.M., 3rd; Hudson, L.L.; Wells, G.; Coleman, J.P.; Pesci, E.C. CysB Negatively Affects the Transcription of pqsR and Pseudomonas Quinolone Signal Production in Pseudomonas aeruginosa. J. Bacteriol. 2015, 197, 1988–2002. [Google Scholar] [CrossRef][Green Version]
- Xu, T.; Wang, X.Y.; Cui, P.; Zhang, Y.M.; Zhang, W.H.; Zhang, Y. The Agr Quorum Sensing System Represses Persister Formation through Regulation of Phenol Soluble Modulins in Staphylococcus aureus. Front. Microbiol. 2017, 8, 2189. [Google Scholar] [CrossRef]
- Herbert, S.; Ziebandt, A.K.; Ohlsen, K.; Schafer, T.; Hecker, M.; Albrecht, D.; Novick, R.; Gotz, F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010, 78, 2877–2889. [Google Scholar] [CrossRef]
- Riordan, J.T.; Muthaiyan, A.; Van Voorhies, W.; Price, C.T.; Graham, J.E.; Wilkinson, B.J.; Gustafson, J.E. Response of Staphylococcus aureus to salicylate challenge. J. Bacteriol. 2007, 189, 220–227. [Google Scholar] [CrossRef]
- Riordan, J.T.; O’Leary, J.O.; Gustafson, J.E. Contributions of sigB and sarA to distinct multiple antimicrobial resistance mechanisms of Staphylococcus aureus. Int. J. Antimicrob. Agents 2006, 28, 54–61. [Google Scholar] [CrossRef]
- Riordan, J.T.; Dupre, J.M.; Cantore-Matyi, S.A.; Kumar-Singh, A.; Song, Y.; Zaman, S.; Horan, S.; Helal, N.S.; Nagarajan, V.; Elasri, M.O.; et al. Alterations in the transcriptome and antibiotic susceptibility of Staphylococcus aureus grown in the presence of diclofenac. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 30. [Google Scholar] [CrossRef]
- Albert, T.J.; Dailidiene, D.; Dailide, G.; Norton, J.E.; Kalia, A.; Richmond, T.A.; Molla, M.; Singh, J.; Green, R.D.; Berg, D.E. Mutation discovery in bacterial genomes: Metronidazole resistance in Helicobacter pylori. Nat. Methods 2005, 2, 951–953. [Google Scholar] [CrossRef]
- Hattangady, D.S.; Singh, A.K.; Muthaiyan, A.; Jayaswal, R.K.; Gustafson, J.E.; Ulanov, A.V.; Li, Z.; Wilkinson, B.J.; Pfeltz, R.F. Genomic, Transcriptomic and Metabolomic Studies of Two Well-Characterized, Laboratory-Derived Vancomycin-Intermediate Staphylococcus aureus Strains Derived from the Same Parent Strain. Antibiotics 2015, 4, 76–112. [Google Scholar] [CrossRef]
- Macvanin, M.; Johanson, U.; Ehrenberg, M.; Hughes, D. Fusidic acid- resistant EF-G perturbs the accumulation of ppGpp. Mol. Microbiol. 2000, 37, 98–107. [Google Scholar] [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Delekta, P.C.; Shook, J.C.; Lydic, T.A.; Mulks, M.H.; Hammer, N.D. Staphylococcus aureus Utilizes Host-Derived Lipoprotein Particles as Sources of Fatty Acids. J. Bacteriol. 2018, 200, e00728-17. [Google Scholar] [CrossRef]
- Anderson, K.L.; Roberts, C.; Disz, T.; Vonstein, V.; Hwang, K.; Overbeek, R.; Olson, P.D.; Projan, S.J.; Dunman, P.M. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006, 188, 6739–6756. [Google Scholar] [CrossRef]
 ,
,  ) 37 °C (
) 37 °C ( ,
,  ), or 42 °C (
), or 42 °C ( ,
,  ). All data shown are the mean of triplicate experiments and error bars represent the standard deviation.
). All data shown are the mean of triplicate experiments and error bars represent the standard deviation.
 ,
,  ) 37 °C (
) 37 °C ( ,
,  ), or 42 °C (
), or 42 °C ( ,
,  ). All data shown are the mean of triplicate experiments and error bars represent the standard deviation.
). All data shown are the mean of triplicate experiments and error bars represent the standard deviation.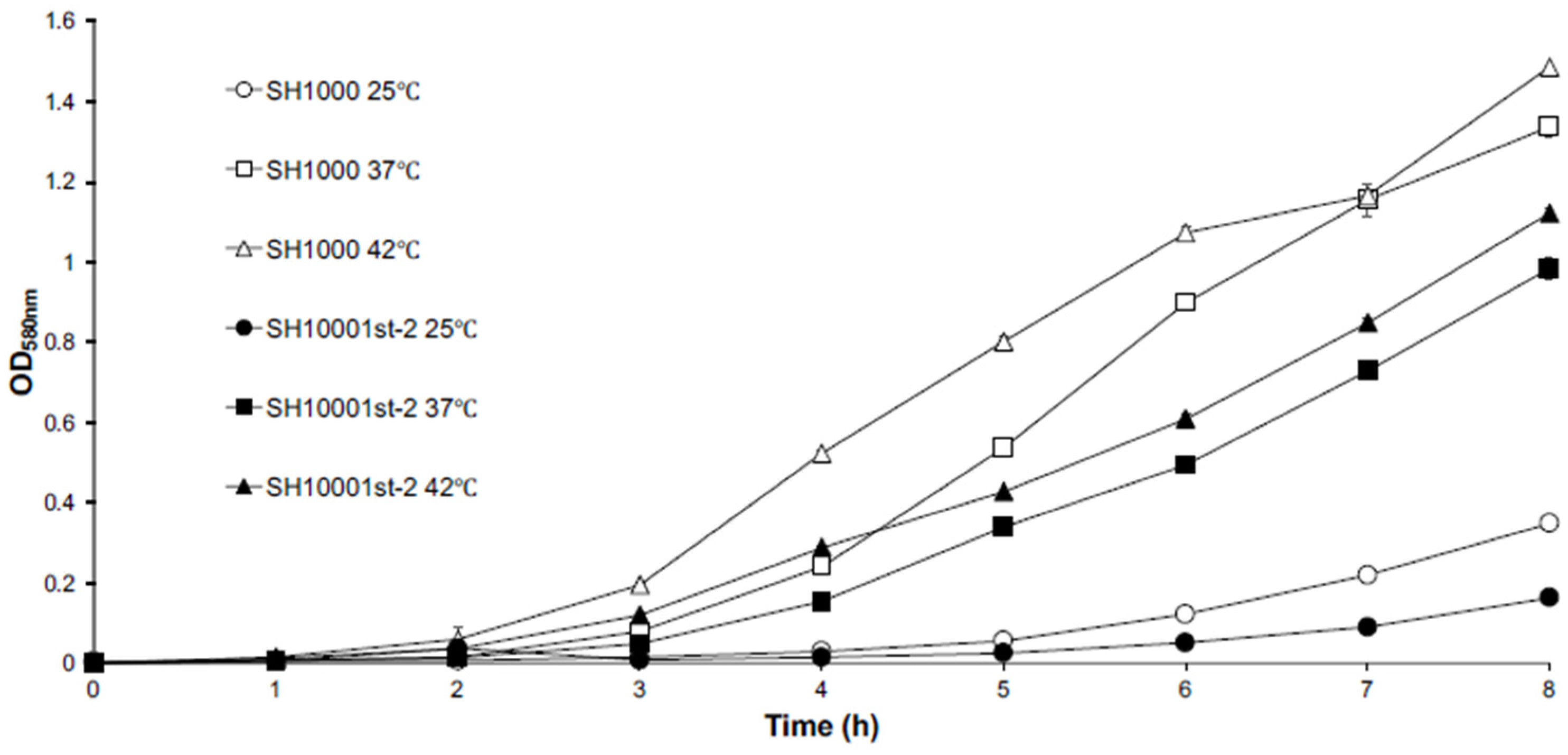
| Gene | Protein Encoded | Locus ID | SNP * | Amino Acid Change |
|---|---|---|---|---|
| fusA | Elongation factor G | SACOL0593 | C617,228 → T617,228 | H457 → Y457 |
| DUF1381 superfamily | SACOL0358 | A371,671 → T371,671 | N36 → I36 | |
| T317,672 → A371,672 | N36 → I36 | |||
| G371,676 → A371,676 | E37 → K37 | |||
| A371,685 → C371,685 | K40 → Q40 |
| Fold-Change in Gene Expression | |||
|---|---|---|---|
| Gene | Locus | Microarray | RT-PCR |
| cap5E | SACOL0140 | 10.4 | 2.1 |
| adh1 | SACOL0660 | −7.0 | −4.3 |
| crtN | SACOL2576 | −2.4 | −2.4 |
| ddh | SACOL2535 | −12.2 | −23.6 |
| spa | SACOL0095 | −8.9 | −1448.0 |
| Functional Category | Upregulated Genes | Downregulated Genes | ||||
|---|---|---|---|---|---|---|
| Number of Genes | % of Genes | >5% of Total | Number of Genes | % of Genes | >5% of Total | |
| Amino acid biosynthesis | 2 | 0.6% | 8 | 2.8% | ||
| Biosynthesis of cofactors, prosthetic groups, and carriers | 5 | 1.5% | 17 | 5.9% | 5.9% | |
| Cell envelope | 66 | 20.2% | 20.2% | 46 | 16.0% | 16.0% |
| Cellular processes | 3 | 0.9% | 9 | 3.1% | ||
| Central intermediary metabolism | 3 | 0.9% | 13 | 4.5% | ||
| DNA metabolism | 9 | 2.8% | 17 | 5.9% | 5.9% | |
| Fatty acid and phospholipid metabolism | 8 | 2.5% | 2 | 0.7% | ||
| Mobile and extrachromosomal element functions | 2 | 0.6% | 1 | 0.3% | ||
| Protein fate | 7 | 2.1% | 7 | 2.4% | ||
| Protein synthesis | 10 | 3.1% | 9 | 3.1% | ||
| Purines, pyrimidines, nucleosides, and nucleotides | 7 | 2.1% | 8 | 2.8% | ||
| Regulatory functions | 29 | 8.9% | 8.9% | 9 | 3.1% | |
| Signal transduction | 0 | 0.0% | 0 | 0.0% | ||
| Transcription | 0 | 0.0% | 2 | 0.7% | ||
| Transport and binding proteins | 52 | 16.0% | 16.0% | 33 | 11.5% | 11.5% |
| Hypothetical proteins/Unknown function/Unclassified | 123 | 37.7% | 37.7% | 106 | 36.9% | 36.9% |
| Totals | 326 | 100.0% | 82.8% | 287 | 100.0% | 76.2% |
| Locus ID | Gene | Protein Encoded | Fold Increase |
|---|---|---|---|
| Cell envelope | |||
| SACOL0136 | cap5A | capsular polysaccharide synthesis Cap5A | 18.31 |
| SACOL0137 | cap5B | capsular polysaccharide synthesis Cap5B | 13.74 |
| SACOL0138 | cap5C | capsular polysaccharide synthesis Cap5C | 21.30 |
| SACOL0140 | cap5E | capsular polysaccharide synthesis Cap5E | 10.40 |
| SACOL0146 | cap5K | capsular polysaccharide synthesis Cap5K | 14.59 |
| SACOL0147 | cap5L | capsular polysaccharide synthesis Cap5L | 10.73 |
| SACOL2022 | hld | delta-hemolysin precursor | 12.60 |
| Fatty acid and phospholipid metabolism | |||
| SACOL0390 | NA | lipase-2 precursor, interruption-C | 11.32 |
| SACOL2694 | gehA | triacylglycerol extracellular lipase-1 precursor | 19.27 |
| SACOL0212 | NA | putative 3-hydroxyacyl-CoA dehydrogenase | 11.99 |
| SACOL0214 | NA | putative long-chain-fatty-acid-acetyl-CoA ligase | 10.51 |
| Regulatory functions | |||
| SACOL2026 | agrA | accessory gene regulator A | 20.27 |
| SACOL2023 | agrB | accessory gene regulator B | 17.15 |
| SACOL2024 | agrD | accessory gene regulator D | 14.35 |
| SACOL2025 | argC2 | accessory gene regulator C | 22.63 |
| SACOL1032 | NA | competence transcription factor ComK | 13.76 |
| SACOL2070 | kdpD | two-component system sensor histidine kinase KdpD | 10.50 |
| SACOL2071 | kdpE | two-component system response regulator KdpE | 11.24 |
| Transport and binding proteins | |||
| SACOL2068 | kdpA | potassium-transporting ATPase subunit A | 26.07 |
| SACOL2066 | kdpC | potassium-transporting ATPase, C subunit | 16.28 |
| SACOL2069 | kdpF | potassium-transporting ATPase, F subunit | 29.64 |
| SACOL1993 | NA | putative ABC-2 type transport system permease | 12.24 |
| Hypothetical proteins/unknown function/unclassified | |||
| SACOL1187 | NA | phenol-soluble modulin beta antibacterial protein | 10.36 |
| SACOL0492 | NA | hypothetical protein | 18.67 |
| SACOL0493 | NA | hypothetical protein | 12.20 |
| SACOL2065 | NA | hypothetical protein | 13.81 |
| Locus ID | Gene | Protein Encoded | Fold Decrease |
|---|---|---|---|
| Cell envelope | |||
| SACOL2660 | isaB | immunodominant surface antigen B | −12.24 |
| SACOL0089 | NA | oleate hydratase (putative myosin-cross-reactive antigen) | −12.27 |
| SACOL1056 | sspB | cysteine protease precursor staphopain B | −10.80 |
| Central intermediary metabolism | |||
| SACOL2535 | ddh | D-lactate dehydrogenase | −12.16 |
| SACOL2395 | narG | respiratory nitrate reductase, alpha subunit | −13.51 |
| Regulatory functions | |||
| SACOL2399 | nirR | nitrite reductase transcriptional regulator NirR | −10.86 |
| Transport and binding proteins | |||
| SACOL1144 | smpB | probable transmembrane protein SmpB iron compound ABC transporter | −23.21 |
| SACOL0310 | NA | nucleoside permease NupC, putative | −11.35 |
| SACOL1476 | NA | basic amino acid/polyamine antiporter, APA family | −10.63 |
| SACOL2525 | NA | lantibiotic ABC transporter ATP-binding protein | −12.61 |
| SACOL2386 | narK | nitrite extrusion protein | −14.55 |
| Metabolite Class | Metabolite | Metabolite Relative Concentration/Gram Dry Weight (Mean ± SE) | Fold Increase SH10001st-2/SH1000 | |
|---|---|---|---|---|
| SH1000 | SH10001st-2 | |||
| Amines & polyamines | glucosamine | 4.5 ± 0.3 | 49.1 ± 2.4 | 10.91 |
| N-acetylglucosamine | 12.3 ± 0.9 | 39.4 ± 2.6 | 3.20 | |
| tyramine | 4.6 ± 0.2 | 8.9 ± 0.2 | 1.93 | |
| 5-methylthioadenosine | 11.4 ± 1.9 | 20.0 ± 2.3 | 1.75 | |
| Amino acids | asparagine | ND * | 15.7 ± 1.7 | |
| aspartic acid | 3488.5 ± 215.9 | 9848.3 ± 298.4 | 2.82 | |
| cysteine | 2.2 ± 0.2 | 6.8 ± 0.8 | 3.09 | |
| glutamine | 31.5 ± 2.3 | 408.1 ± 19.2 | 12.95 | |
| glycine | 116.4 ± 10.8 | 1252.9 ± 244.9 | 10.76 | |
| homocysteine | ND | 1.9 ± 0.1 | ||
| homoserine | 6.3 ± 0.3 | 22.3 ± 1.4 | 3.53 | |
| isoleucine | 374.8 ± 38.4 | 617.3 ± 69.0 | 1.64 | |
| leucine | 475.5 ± 28.9 | 800.6 ± 61.6 | 1.68 | |
| N-acetyl-serine | 12.2 ± 1.5 | 858.3 ± 73.8 | 70.35 | |
| phenylalanine | 182.7 ± 12.8 | 301.0 ± 21.9 | 1.64 | |
| proline | 8544.3 ± 579.9 | 19,899.2 ± 1871.3 | 2.32 | |
| proline-like | 139.9 ± 19.4 | 262.3 ± 14.6 | 1.87 | |
| serine | 11.5 ± 0.7 | 2,551.2 ± 196.7 | 221.84 | |
| threonine | ND | 101.4 ± 4.2 | ||
| tryptophan | 1.0 ± 0.2 | 5.7 ± 0.3 | 5.7 | |
| valine | 560.7 ± 34.5 | 803.5 ± 72.1 | 1.43 | |
| Polar organic acids | aminomalonic acid | ND | 8.4 ± 0.4 | |
| citric acid | 7.9 ± 1.2 | 39.9 ± 3.1 | 5.05 | |
| fumaric acid | 7.2 ± 0.7 | 42.6 ± 8.4 | 5.91 | |
| malic acid | 19.6 ± 0.7 | 32.6 ± 4.1 | 1.66 | |
| phosphonic acid | 18.9 ± 1.5 | 58.9 ± 1.2 | 3.11 | |
| phosphoric acid | 18,144.9 ± 570.4 | 23,007.2 ± 1620.9 | 1.26 | |
| pyruvic acid | 2.9 ± 0.3 | 7.0 ± 0.4 | 2.41 | |
| Sugars | 1-methyl-beta-D-galactopyranoside | 3.4 ± 0.4 | 16.6 ± 1.4 | 4.88 |
| 2(1H)-Pyrimidinone, 1-ribofuranosyl-5-P | ND | 15.7 ± 1.5 | ||
| fructose | ND | 3.7 ± 0.6 | ||
| glucose | 1.4 | 3.4 ± 0.5 | 2.42 | |
| glucose-6-P | ND | 3.4 ± 0.4 | ||
| glycerol | 474.9 ± 46.9 | 2223.0 ± 108.1 | 4.68 | |
| glycerol-2-P | 4.1 ± 1.0 | 7.3 ± 0.4 | 1.78 | |
| glycerol-3-P | 185.7 ± 6.1 | 248.6 ± 10.3 | 1.33 | |
| ribitol | ND | 2.9 ± 0.1 | ||
| ribose | ND | 5.0 ± 0.2 | ||
| ribose-5-P | 2.1 ± 0.1 | 5.3 ± 0.3 | 2.52 | |
| sorbitol | 8.1 ± 0.4 | 51.5 ± 5.8 | 6.35 | |
| sorbitol-6-P | 12.6 ± 2.1 | 23.6 ± 2.3 | 1.87 | |
| Metabolite Class | Metabolite | Metabolite Relative Concentration/Gram Dry Weight (Mean ± SE) | Fold Decrease SH1000/ SH10001st-2 | |
|---|---|---|---|---|
| SH1000 | SH10001st-2 | |||
| Amines & Polyamines | adenosine | 50.8 ± 2.0 | 15.3 ± 2.1 | −3.32 |
| galactosamine | 25.6 ± 4.2 | 14.9 ± 3.6 | −1.71 | |
| hydroxycarbamic acid | 9.6 ± 0.4 | 6.8 ± 1.0 | −1.41 | |
| Amino acids | pyroglutamic acid | 2291.2 ± 68.0 | 1631.5 ± 103.1 | −1.40 |
| arginine | 3.7 ± 0.4 | ND * | ||
| Polar organic acids | 2-hydroxyglutaric acid | 67.5 ± 11.6 | 13.0 ± 0.5 | −5.19 |
| 2-hydroxyphosphinyl | 2.1 ± 0.1 | 1.1 ± 0.1 | −1.91 | |
| gluconic acid | 24.0 ± 1.6 | 9.0 ± 0.8 | −2.66 | |
| glyceric acid | 4.6 ± 0.4 | 2.8 ± 0.2 | −1.64 | |
| linoleic acid | 2.1 ± 0.1 | ND | ||
| malonic acid | 5.3 ± 0.4 | 1.8 ± 0.2 | −2.94 | |
| succinic acid | 166.0 ± 13.6 | 23.2 ± 2.8 | −7.15 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, S.K.; Pfeltz, R.F.; Wilkinson, B.J.; Gustafson, J.E. Transcriptomic and Metabolomic Analysis of a Fusidic Acid-Selected fusA Mutant of Staphylococcus aureus. Antibiotics 2022, 11, 1051. https://doi.org/10.3390/antibiotics11081051
Gupta SK, Pfeltz RF, Wilkinson BJ, Gustafson JE. Transcriptomic and Metabolomic Analysis of a Fusidic Acid-Selected fusA Mutant of Staphylococcus aureus. Antibiotics. 2022; 11(8):1051. https://doi.org/10.3390/antibiotics11081051
Chicago/Turabian StyleGupta, Sushim K., Richard F. Pfeltz, Brian J. Wilkinson, and John E. Gustafson. 2022. "Transcriptomic and Metabolomic Analysis of a Fusidic Acid-Selected fusA Mutant of Staphylococcus aureus" Antibiotics 11, no. 8: 1051. https://doi.org/10.3390/antibiotics11081051
APA StyleGupta, S. K., Pfeltz, R. F., Wilkinson, B. J., & Gustafson, J. E. (2022). Transcriptomic and Metabolomic Analysis of a Fusidic Acid-Selected fusA Mutant of Staphylococcus aureus. Antibiotics, 11(8), 1051. https://doi.org/10.3390/antibiotics11081051








