Macromolecular Structure Assembly as a Novel Antibiotic Target
Abstract
1. Introduction
2. Penicillin-Binding Protein Structures as a New Antimicrobial Target
3. DNA Polymerase Structure Formation as Another Antimicrobial Target
4. RNA Polymerase Structure Assembly as a Novel Drug Target
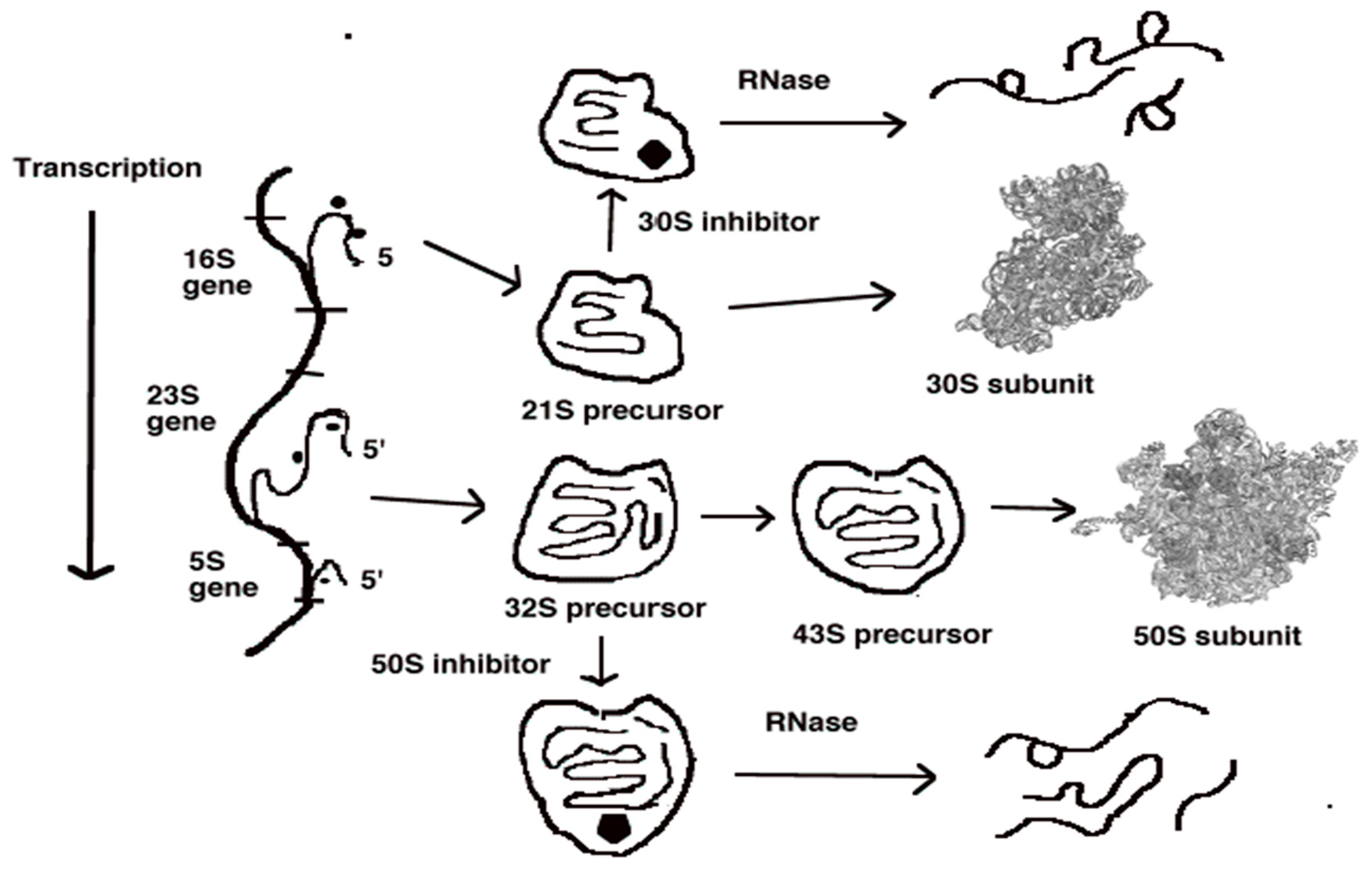
5. Ribosomal Subunit Assembly as a Fourth Antibiotic Target
6. Methods for the Identification of Antimicrobial Agents That Inhibit Macromolecular Target Formation
Funding
Conflicts of Interest
References
- Patel, A. Tackling Antimicrobial Resistance in the Shadow of COVID-19. mBio 2021, 12, e0047321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-induced horizontal transfer of antimicrobial resistance genes in bacteria: A mini-review. J. Antimicrob. Chemother. 2020, 77, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Gao, J.; Tang, W. Nosocomial infection and its molecular mechanisms of antibiotic resistance. Biosci. Trends 2016, 10, 14–21. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef]
- Monserrat-Martinez, A.; Gambin, Y.; Sierecki, E. Thinking outside the bug: Molecular targets and strategies to overcome antibiotic resistance. Int. J. Mol. Sci. 2019, 20, 1255. [Google Scholar] [CrossRef]
- Champney, W.S. Antibiotics which target bacterial ribosomal subunit biogenesis. J. Antimicrob. Chemother. 2020, 75, 787–806. [Google Scholar] [CrossRef]
- Bandyopadhyay, M.; Muthuirulan, P. Promising targets for prospective antibacterial therapy. EC Microbiol. 2018, 14, 351–360. [Google Scholar]
- Silver, L.L.; Bush, K. Beta-lactams and beta-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2017, 7, a025171. [Google Scholar]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef] [PubMed]
- Turecka, K.; Waleron, K. Inhibitors of bacterial transcription are compounds for potent antimicrobial drugs. Curr. Pharm. Biotechnol. 2013, 14, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef] [PubMed]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Terrak, M. Glycosyltransferases and Transpeptidase Proteins: Valuable Targets for New Antibacterials. Antibiotics 2016, 5, 12. [Google Scholar] [CrossRef]
- Sauvage, E.; Derouaux, A.; Fraipont, C.; Joris, M.; Herman, R.; Rocaboy, M.; Schloesser, M.; Dumas, J.; Kerff, F.; Nguyen-Distèche, M.; et al. Crystal Structure of Penicillin-Binding Protein 3 (PBP3) from Escherichia coli. PLoS ONE 2014, 9, e98042. [Google Scholar] [CrossRef]
- Zervosen, A.; Sauvage, E.; Frère, J.-M.; Charlier, P.; Luxen, A. Development of New Drugs for an Old Target—The Penicillin Binding Proteins. Molecules 2012, 17, 12478–12505, ISSN 1420-3049. [Google Scholar] [CrossRef]
- Schaeffer, P.M.; Headlam, M.J.; Dixon, N.E. Protein—Protein Interactions in the Eubacterial Replisome. IUBMB Life 2005, 57, 5–12. [Google Scholar] [CrossRef]
- Erzberger, J.P.; Pirruccello-Straub, M.M.; Berger, J.M. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002, 21, 4763–4773. [Google Scholar] [CrossRef]
- Grimwade, J.E.; Leonard, A.C. Targeting the Bacterial Orisome in the Search for New Antibiotics. Front. Microbiol. 2017, 8, 2352. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S. Structural Biology of Bacterial RNA Polymerase. Biomolecules 2015, 5, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, C.; Zhuang, N.; Wang, T.; Zhang, Y. Structural basis for transcription initiation by bacterial ECF σ factors. Nat. Commun. 2019, 10, 1153. [Google Scholar] [CrossRef]
- Srivastava, A.; Talaue, M.; Liu, S.; Degen, D.; Ebright, R.Y.; Sineva, E.; Chakraborty, A.; Druzhinin, S.Y.; Chatterjee, S.; Mukhopadhya, J.; et al. New target for inhibition of bacterial RNA polymerase: The switch region. Curr. Opin. Microbiol. 2011, 14, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Watson, Z.; Ward, F.R.; Méheust, R.; Ad, O.; Schepartz, A.; Banfield, J.F.; Cate, J.H.D. Structure of the bacterial ribosome at 2 Å resolution. eLife 2020, 9, e60482D. [Google Scholar] [CrossRef]
- Noeske, J.; Wasserman, M.R.; Terry, D.S.; Altman, R.; Blanchard, S.C.; Cate, J.H.D. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015, 22, 336–341. [Google Scholar] [CrossRef]
- Kaczanowska, M.; Ryden-Aulin, M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef]
- Davis, J.H.; Williamson, J.R. Structure and dynamics of bacterial ribosome biogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160181. [Google Scholar] [CrossRef]
- Semrad, K. Proteins with RNA Chaperone Activity: A World of Diverse Proteins with a Common Task—Impediment of RNA Misfolding. Biochem. Res. Int. 2010, 2011, 532908. [Google Scholar] [CrossRef]
- Deutscher, M.P. How bacterial cells keep ribonucleases under control. FEMS Microbiol. Rev. 2015, 39, 350–361. [Google Scholar] [CrossRef]
- Auerbach-Nevo, T.; Baram, D.; Bashan, A.; Belousoff, M.; Breiner, E.; Davidovich, C.; Cimicata, G.; Eyal, Z.; Halfon, Y.; Krupkin, M.; et al. Ribosomal Antibiotics: Contemporary Challenges. Antibiotics 2016, 5, 24. [Google Scholar] [CrossRef]
- Nikolay, R.; Schmidt, S.; Schlömer, R.; Deuerling, E.; Nierhaus, K.H. Ribosome Assembly as an Antimicrobial Target. Antibiotics 2016, 5, 18. [Google Scholar] [CrossRef]
- Mehta, R.; Champney, W.S. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 2003, 47, 237–243. [Google Scholar] [CrossRef]
- Foster, C.; Champney, W.S. Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch. Microbiol. 2008, 189, 441–449. [Google Scholar] [CrossRef]
- Chittum, H.S.; Champney, W.S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr. Microbiol. 1995, 30, 273–279. [Google Scholar] [CrossRef]
- Pokkunuri, I.; Champney, W.S. Characteristics of a 50S ribosomal subunit precursor particle as a substrate for ermE methyltransferase activity and erythromycin binding in Staphylococcus aureus. RNA Biol. 2007, 4, 147–153. [Google Scholar] [CrossRef]
- Champney, W.S.; Tober, C.L. Specific inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide and streptogramin B antibiotics. Curr. Microbiol. 2000, 41, 126–135. [Google Scholar] [CrossRef]
- Silvers, J.A.; Champney, W.S. Accumulation and turnover of 23S ribosomal RNA in azithromycin-inhibited ribonuclease mutant strains of Escherichia coli. Arch. Microbiol. 2005, 184, 66–77. [Google Scholar] [CrossRef]
- Frazier, A.; Champney, W.S. Impairment of ribosomal subunit synthesis in aminoglycoside treated ribonuclease mutants of Escherichia coli. Arch. Microbiol. 2012, 194, 1033–1041. [Google Scholar] [CrossRef][Green Version]
- Carro, L. Protein–protein interactions in bacteria: A promising and challenging avenue towards the discovery of new antibiotics. Beilstein J. Org. Chem. 2018, 14, 2881–2896. [Google Scholar] [CrossRef]
- Staker, B.L.; Buchko, G.W.; Myler, P.J. Recent contributions of structure-based drug design to the development of antibacterial compounds. Curr. Opin. Microbiol. 2015, 27, 133–138. [Google Scholar] [CrossRef]
- Neville, N.; Jia, Z. Approaches to the Structure-Based Design of Antivirulence Drugs: Therapeutics for the Post-Antibiotic Era. Molecules 2019, 24, 378. [Google Scholar] [CrossRef]
- Jani, S.; Ramirez, M.S.; Tolmasky, M.E. Silencing Antibiotic Resistance with Antisense Oligonucleotides. Biomedicines 2021, 9, 416. [Google Scholar] [CrossRef]
- Fields, F.R.; Lee, S.W.; McConnell, M.J. Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 2017, 134, 74–86. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Beach, J.M.; Champney, W.S. An examination of the inhibitory effects of three antibiotics in combination on ribosome biosynthesis in Staphylococcus aureus. Arch. Microbiol. 2014, 196, 249–260. [Google Scholar] [CrossRef]
- González-Bello, C. Antibiotic adjuvants—A strategy to unlock bacterial resistance to antibiotics. Bioorg. Med. Chem. Lett. 2017, 27, 4221–4228. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Champney, S. Macromolecular Structure Assembly as a Novel Antibiotic Target. Antibiotics 2022, 11, 937. https://doi.org/10.3390/antibiotics11070937
Champney S. Macromolecular Structure Assembly as a Novel Antibiotic Target. Antibiotics. 2022; 11(7):937. https://doi.org/10.3390/antibiotics11070937
Chicago/Turabian StyleChampney, Scott. 2022. "Macromolecular Structure Assembly as a Novel Antibiotic Target" Antibiotics 11, no. 7: 937. https://doi.org/10.3390/antibiotics11070937
APA StyleChampney, S. (2022). Macromolecular Structure Assembly as a Novel Antibiotic Target. Antibiotics, 11(7), 937. https://doi.org/10.3390/antibiotics11070937




