Antibiofilm Activity of Omega-3 Fatty Acids and Its Influence on the Expression of Biofilm Formation Genes on Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substances
2.2. Microorganisms and Primers
2.3. Biofilm Formation
2.4. RNA Extraction
2.5. cDNA Synthesis from Bacterial RNA
2.6. Investigation of Influence by DHA on Bacterial Biofilm Gene Expression Using Real-Time Quantitative Polymerase Chain Reaction (Real-Time qPCR)
2.7. Statistical Analysis
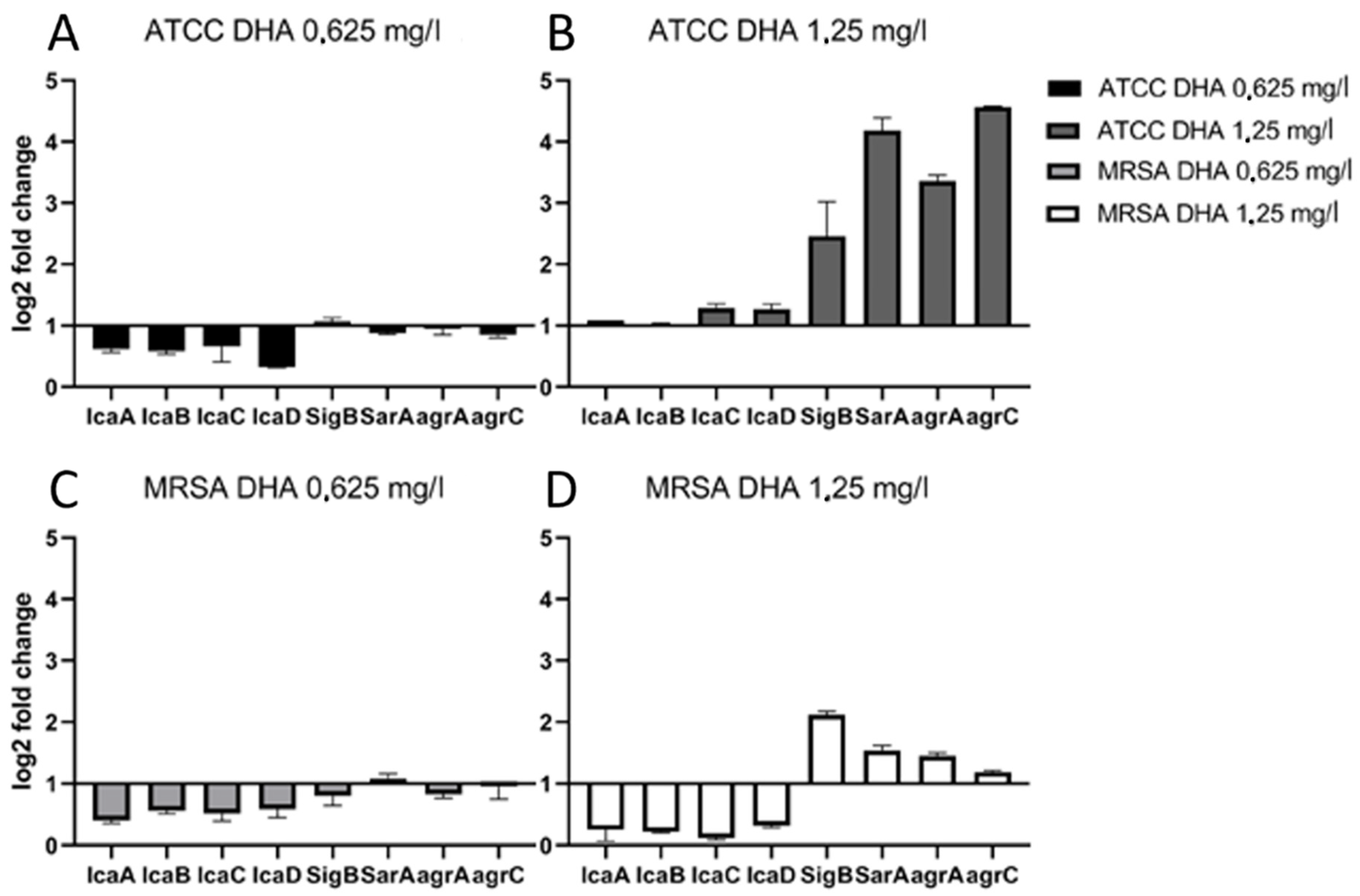
3. Results
Influence of DHA on Expression of Biofilm-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.S.; Haddad, F.S. Prosthetic joint infection. Bone Jt. Res. 2019, 8, 570–572. [Google Scholar] [CrossRef] [PubMed]
- Natsuhara, K.M.; Shelton, T.J.; Meehan, J.P.; Lum, Z.C. Mortality During Total Hip Periprosthetic Joint Infection. J. Arthroplasty 2019, 34, S337–S342. [Google Scholar] [CrossRef] [PubMed]
- Okike, K.; Bhattacharyya, T. Trends in the management of open fractures. A critical analysis. J. Bone Jt. Surg. Am. 2006, 88, 2739–2748. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Montanaro, L.; Poggi, A.; Visai, L.; Ravaioli, S.; Campoccia, D.; Speziale, P.; Arciola, C.R. Extracellular DNA in biofilms. Int. J. Artif. Organs. 2011, 34, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Rachid, S.; Cho, S.; Ohlsen, K.; Hacker, J.; Ziebuhr, W. Induction of Staphylococcus epidermidis biofilm formation by environmental factors: The possible involvement of the alternative transcription factor sigB. Adv. Exp. Med. Biol. 2000, 485, 159–166. [Google Scholar]
- Andrey, D.O.; Jousselin, A.; Villanueva, M.; Renzoni, A.; Monod, A.; Barras, C.; Kelley, W.L. Impact of the Regulators SigB, Rot, SarA and sarS on the Toxic Shock Tst Promoter and TSST-1 Expression in Staphylococcus aureus. PLoS ONE 2015, 10, e0135579. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides 2001, 22, 1603–1608. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal infections: Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Trotonda, M.P.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 2005, 187, 5790–5798. [Google Scholar] [CrossRef] [Green Version]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Campoccia, D.; Baldassarri, L.; Pirini, V.; Ravaioli, S.; Montanaro, L.; Arciola, C.R. Molecular epidemiology of Staphylococcus aureus from implant orthopaedic infections: Ribotypes, agr polymorphism, leukocidal toxins and antibiotic resistance. Biomaterials 2008, 29, 4108–4116. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhou, Z.; Dong, J.; Zhang, J.; Xia, Y.; Shu, R. Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microb. Pathog. 2016, 99, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Coraça-Huber, D.C.; Steixner, S.; Wurm, A.; Nogler, M. Antibacterial and Anti-Biofilm Activity of Omega-3 Polyunsaturated Fatty Acids against Periprosthetic Joint Infections-Isolated Multi-Drug Resistant Strains. Biomedicines 2021, 9, 334. [Google Scholar] [CrossRef] [PubMed]
- Herbst, E.A.; Paglialunga, S.; Gerling, C.; Whitfield, J.; Mukai, K.; Chabowski, A.; Holloway, G.P. Omega-3 supplementation alters mitochondrial membrane composition and respiration kinetics in human skeletal muscle. J. Physiol. 2014, 592, 1341–1352. [Google Scholar] [CrossRef]
- Pastagia, M.; Kleinman, L.C.; de la Lacerda Cruz, E.G.; Jenkins, S.G. Predicting risk for death from MRSA bacteremia. Emerg. Infect. Dis. 2012, 18, 1072–1080. [Google Scholar] [CrossRef]
- Weterings, V.; Bosch, T.; Witteveen, S.; Landman, F.; Schouls, L.; Kluytmans, J. Next-Generation Sequence Analysis Reveals Transfer of Methicillin Resistance to a Methicillin-Susceptible Staphylococcus aureus Strain That Subsequently Caused a Methicillin-Resistant Staphylococcus aureus Outbreak: A Descriptive Study. J. Clin. Microbiol. 2017, 55, 2808–2816. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Rupp, M.E.; Ulphani, J.S.; Fey, P.D.; Bartscht, K.; Mack, D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 1999, 67, 2627–2632. [Google Scholar] [CrossRef] [Green Version]
- Gerke, C.; Kraft, A.; Süssmuth, R.; Schweitzer, O.; Götz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998, 273, 18586–18593. [Google Scholar] [CrossRef] [Green Version]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [Green Version]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lasa, I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef] [Green Version]
- Cucarella, C.; Tormo, M.A.; Ubeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Penadés, J.R. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef] [Green Version]
- Geoghegan, J.A.; Corrigan, R.M.; Gruszka, D.T.; Speziale, P.; O’Gara, J.P.; Potts, J.R.; Foster, T.J. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2010, 192, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 2011, 79, 1153–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knobloch, J.K.; Bartscht, K.; Sabottke, A.; Rohde, H.; Feucht, H.H.; Mack, D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: Differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001, 183, 2624–2633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ica A | 5′-CGC ACT CAA TCA AGG CAT TA-3′ | 5′-CCA GCA AGT GTC TGA CTT CG-3′ |
| ica B | 5′-CAC ATA CCC ACG ATT TGC AT-3′ | 5′-TCG GAG TGA CTG CTT TTT CC-3′ |
| ica C | 5′-CTT GGG TAT TTG CAC GCA TT-3′ | 5′-GCA ATA TCA TGC CGA CAC CT-3′ |
| ica D | 5′-ACC CAA CGC TAA AAT CAT CG-3′ | 5′-GCG AAA ATG CCC ATA GTT TC-3′ |
| SigB | 5′-AGA AGC AAT GGA AAT GGG AC-3′ | 5′-CTT AAA CCG ATA CGC TCA CC-3′ |
| SarA | 5′-AAG GAC AAT CAC ATC ACG AAG-3′ | 5′-GAA CGC TCT AAT TCA GCG G-3′ |
| agrA | 5′-GCC CTC GCA ACT GAT AAT CC-3′ | 5′-CAT CGC TGC AAC TTT GTA GAC-3′ |
| agrC | 5′-GCA TCA ACT GAA ATT GAT GAC C-3′ | 5′-ACC TAA ACC ACG ACC TTC AC-3′ |
| 16s | 5′-GAA AGC CAC GGC TAA CTA CG-3′ | 5′-CAT TTC ACC GCT ACA CAT GG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spiegel, C.; Steixner, S.J.M.; Coraça-Huber, D.C. Antibiofilm Activity of Omega-3 Fatty Acids and Its Influence on the Expression of Biofilm Formation Genes on Staphylococcus aureus. Antibiotics 2022, 11, 932. https://doi.org/10.3390/antibiotics11070932
Spiegel C, Steixner SJM, Coraça-Huber DC. Antibiofilm Activity of Omega-3 Fatty Acids and Its Influence on the Expression of Biofilm Formation Genes on Staphylococcus aureus. Antibiotics. 2022; 11(7):932. https://doi.org/10.3390/antibiotics11070932
Chicago/Turabian StyleSpiegel, Christopher, Stephan Josef Maria Steixner, and Débora C. Coraça-Huber. 2022. "Antibiofilm Activity of Omega-3 Fatty Acids and Its Influence on the Expression of Biofilm Formation Genes on Staphylococcus aureus" Antibiotics 11, no. 7: 932. https://doi.org/10.3390/antibiotics11070932
APA StyleSpiegel, C., Steixner, S. J. M., & Coraça-Huber, D. C. (2022). Antibiofilm Activity of Omega-3 Fatty Acids and Its Influence on the Expression of Biofilm Formation Genes on Staphylococcus aureus. Antibiotics, 11(7), 932. https://doi.org/10.3390/antibiotics11070932








