Abstract
Biofilm has garnered a lot of interest due to concerns in various sectors such as public health, medicine, and the pharmaceutical industry. Biofilm-producing bacteria show a remarkable drug resistance capability, leading to an increase in morbidity and mortality. This results in enormous economic pressure on the healthcare sector. The development of biofilms is a complex phenomenon governed by multiple factors. Several attempts have been made to unravel the events of biofilm formation; and, such efforts have provided insights into the mechanisms to target for the therapy. Owing to the fact that the biofilm-state makes the bacterial pathogens significantly resistant to antibiotics, targeting pathogens within biofilm is indeed a lucrative prospect. The available drugs can be repurposed to eradicate the pathogen, and as a result, ease the antimicrobial treatment burden. Biofilm formers and their infections have also been found in plants, livestock, and humans. The advent of novel strategies such as bioinformatics tools in treating, as well as preventing, biofilm formation has gained a great deal of attention. Development of newfangled anti-biofilm agents, such as silver nanoparticles, may be accomplished through omics approaches such as transcriptomics, metabolomics, and proteomics. Nanoparticles’ anti-biofilm properties could help to reduce antimicrobial resistance (AMR). This approach may also be integrated for a better understanding of biofilm biology, guided by mechanistic understanding, virtual screening, and machine learning in silico techniques for discovering small molecules in order to inhibit key biofilm regulators. This stimulated research is a rapidly growing field for applicable control measures to prevent biofilm formation. Therefore, the current article discusses the current understanding of biofilm formation, antibiotic resistance mechanisms in bacterial biofilm, and the novel therapeutic strategies to combat biofilm-mediated infections.
1. Introduction
The existence of microbial pathogens has been recognized for many years, and since then, researchers have continuously tried to eliminate already existing and emerging pathogens causing infectious diseases, and develop antimicrobial agents to treat and eliminate the infectious disease. Antimicrobials represent a broad spectrum of compounds that may act against a wide repertoire of disease-causing microorganisms, such as bacteria, viruses, parasites, fungi, and protozoa [1]. These compounds have been used since the early 20th century to treat infected patients and have helped significantly in lowering the morbidity as well as mortality rates of most infectious diseases. In 1928, Alexander Fleming discovered penicillin, and in the 1940s, it came into clinical use just in time for World War II [2]. After only four years of use, the first penicillin-resistant strains of bacteria emerged; this resulted in the evolution of antimicrobial resistance (AMR). Subsequently, AMR has accelerated rapidly and expanded to different pathogenic species due to the persistent exposure and non-targeted application of antimicrobials in clinical and agricultural settings. Many current antimicrobials have become ineffective due to the advancement of AMR; it is believed that the non-judicious use and overdosing of antimicrobials are one of the foremost factors for the rise in drug-resistant bacteria [3]. Over last few decades, researchers have been trying to understand the mechanism of AMR and to develop new antimicrobials.
The ability to form biofilms is a universal attribute of bacteria. Biofilms are multicellular communities held together by a self-produced extracellular matrix. The mechanisms that a number of bacteria employ to form biofilms vary, frequently depending on environmental conditions and specific strain attributes. In the 17th century, Antonie van Leeuwenhoek first observed “animalcules” on his own teeth. In 1940, the “bottle effect” was observed in marine microorganisms [4]. This showed that the bacteria grow more often on the surface. Then, in 1943, biofilms were made by Zobell, and he found that bacteria on surfaces were greater in number compared with the surrounding seawater [5].
Today, we generally define such biofilms as microbial communities adhered to a substratum and encased within an extracellular polymeric substance (EPS) produced by the microbial cells themselves [6,7]. Microbial biofilms are found on the surfaces of medical devices, dental implants, suturing materials, catheters, and human and animal tissues; also in aquatic mediums, damp structures, natural and artificial environmental conditions, and plant roots in the disease-causing forms with the ability to release toxins to the surrounding matrix [4,7]. Apart from that, biofilms also occur in human or animal alimentary canals, wastewater filters, or aquatic bodies in symbiotic form [8]. Biofilms are most prevalent in natural environments; and are responsible for causing infections in humans and animals due to their resistance to antimicrobials [9,10]. Therefore, it is very much pertinent to fully understand biofilm-led survival against antibiotics [11]. In this review, we will discuss the AMR, mechanism of biofilm-led AMR, and small molecules and drug candidates for the potential anti-biofilm therapies.
2. Overview of Biofilm-Led Antibiotic Survival
Biofilm is a consortium of microorganisms in which cells adhere to each other and often to almost every surface; it can form one layer in direct contact with the substratum or form in flocs, is mobile, and can form complex communities (Figure 1).
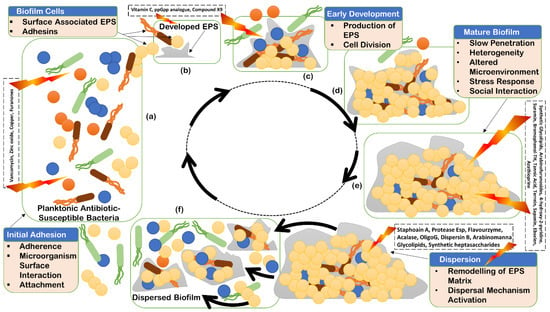
Figure 1.
Stages of biofilm life cycle: initial adhesion (a); initial developed extracellular polymeric substances (EPS) (b); early development of EPS production (c); further development of biofilm EPS for maturation (d); mature biofilm (e); and dispersion of biofilm (f).
These adherent cells grow in multicellular aggregates and are embedded in a matrix composed of extracellular polymeric substances (EPS). New properties usually emerge in the biofilm, which are clearly distinct from the bacterial life and could not be predicted from the bacterial life. Biofilms are most widely distributed in water, soil, sediment, and subsurface environment, and navigate the biogeochemical cycling processes of most elements. Microorganisms colonize all higher organisms, including humans, and form biofilms [12]. Biofilm can form on living as well as non-living surfaces such as prosthetic or artificial teeth (Figure 2e,f), and may be prevalent in nature, industries, and hospitals. For instance, biofilm can form as early soft dental plaque or more mature mineralized and calcified plaque (calculus) on human teeth (Figure 2), skin, and urinary tract; also on medical devices such as catheters and implants that can result in chronic infections [4,13]. Moreover, they are responsible for contamination of process water, as well as biofouling, and they worsen the hygienic quality of drinking water [12,14].
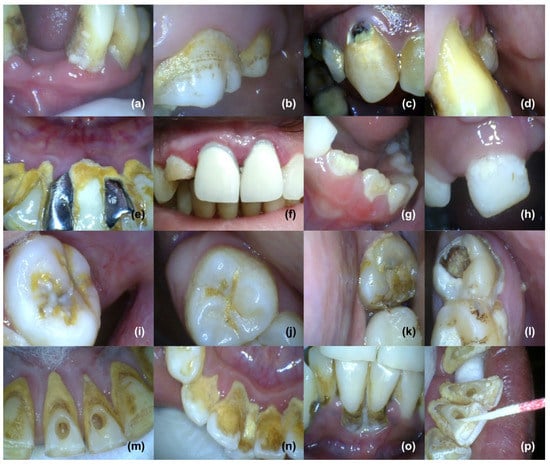
Figure 2.
Biofilm on human teeth in the form of plaque and/or calcified plaque (calculus): soft plaque at the neck (near gingiva), crown (more towards gingiva), and more permeable dentogingival junction area of human teeth (a–d). Dentogingival junction can be seen shifted towards cemento-enamel junction (a,d), providing more surface for biofilm deposition, exposure of dentine area, and serving as an easy route for passage of bacterial products from biofilm into deep tissues. Hard (calcified) plaque or calculus on artificial surfaces of prosthetic teeth (dental crown or fixed partial denture): more calcified plaque can be seen near the dentogingival junction area, as compared to the crown of prosthetic teeth, and gingival inflammation (gingivitis) of free gingiva may also be seen (e,f). Plaque on early secondary or permanent, as well as primary, teeth (g,h). Calcified or hard plaque on the occlusal area forms mainly in pits, fissures, fossae, and grooves of permanent human teeth (i,j). Change in color of plaque can also be seen, indicating entrapment of minerals from saliva into biofilm and hardening, and soft as well as hard plaque on occlusal, interdental, dentogingival, and cervical areas of permanent human molar teeth (k,l). Very hard plaque of dark yellow, light yellow, or brown in color near the dentogingival, interdental, and cervical areas of permanent human lower anterior teeth (m–p). V-shaped shifting of dentogingival junction and free gingiva away from the neck of the teeth, towards the cemento-enamel junction, can also be seen. (Intraoral and dental pictures were provided by Dr. Mamta of Mamta Dental Clinic, Faridabad, India).
Biofilm can accommodate several bacterial species (Figure 1), form complex systems, and can have high cell densities ranging from 108 to 1011 cells g−1 wet weight [15]. Biofilms form a heterogenous ecosystem as soon as their cells undergo differentiation and synchronize their life cycles. Novel structures, activities, patterns, and properties that arise during the process, and of self-organization, are the emergent properties of biofilm communities (Figure 1). The main component of the matrix is water (up to 97%). Structural and functional components of the matrix include soluble, gel-forming polysaccharides, proteins, and extracellular DNA (eDNA), as well as insoluble components, such as amyloids, cellulose, fimbriae, pili, and flagellae. As the biofilm formation is a dynamic process, intermolecular interactions between EPS components produce new EPS molecules in the matrix that determine the physiological activity and mechanical properties of the matrix of the organism, and the biofilm architecture is solely because of these EPS molecules [16]. Pores and channels between microcolonies form the voids in the matrix that ease the liquid transport. Extracellular DNA forms a stable filamentous network structure (Figure 3). Tolerance to desiccation is also an emergent property of biofilm conferred by the matrix [17].
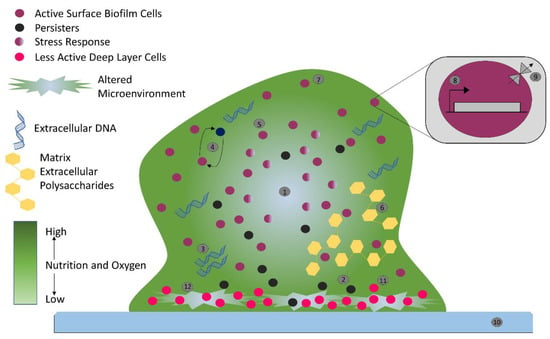
Figure 3.
Biofilm-mediated antimicrobial resistance in bacteria: nutrient gradient indicated by color gradient (1), persister cells (2), extracellular DNA (3), intercellular interactions (4), stress response (5), matrix extracellular polysaccharides (6), biofilm cells (7), genetic determinants (8), multidrug efflux pump (9), host surface (10), less active deep layer cells (11), and altered environment (12).
Bacterial biofilms are one of the key factors in chronic infections on the grounds of higher tolerance to antibiotics and disinfectants; they can combat phagocytosis and other components of the immune system [18]. Consequently, microorganisms in biofilms become less susceptible to multiple antimicrobial agents, which drives biofilms to an impending predicament in therapeutics [19]. Nonetheless, antibiotic tolerance is predominantly contingent on the formation of biofilm, the composition of ECM with proteins, lipids, water, glycolipids, polysaccharides, surfactants, extracellular DNA (eDNA), membrane vesicles, and extracellular RNA, and the architecture of biofilm, which refers to the biomass and space organization within the biofilm [16,20,21,22].
3. Mechanism of Biofilm Resistance
Antimicrobials (antibiotics, antivirals, antifungals, and antiparasitics) are compounds that kill microbes, stop their growth, and prevent or treat the infections in humans, animals, and plants. Antimicrobial resistance is the ability of microorganisms to survive against an antimicrobial drug at a higher concentration for a longer period, and is measured as minimum inhibitory concentration (MIC) [23]. Biofilm resistance can be antibiotic resistance or antibiotic tolerance. Microorganisms develop mechanisms against antimicrobials either through acquisition of foreign genetic material coding for resistant determinants by horizontal gene transfer (HGT) in biofilm EPS, or through genetic mutation (Figure 3). Mechanisms of AMR are: prevention of access or reduced permeability of antimicrobials, mutational changes in antibiotic targets, modification of targets, and enzymatic degradation of the antimicrobials by hydrolysis or chemical change (Figure 4). Antibiotic resistance (ABR) is a subdivision of AMR, as antibiotics are effective against the bacteria, but the bacteria become resistant to antibiotics.
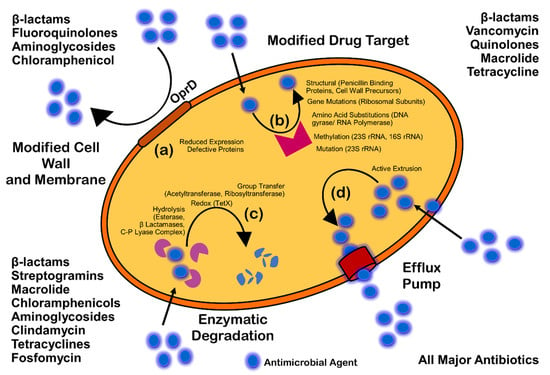
Figure 4.
Mechanism of antimicrobial resistance: cell wall modification (a); modification of drug target (b); enzymatic degradation or destruction of antimicrobials (c); and efflux pump (d).
Antimicrobial resistance can be intrinsic (naturally acquired) by genetic mutation, genes encoding inherent structural and functional traits of the molecular target, acquired extrinsically, or can be adaptive. Intrinsically, antibiotics such as vancomycin and daptomycin, which are active against Gram-positive bacteria, might not be effective against the Gram-negative bacteria due to distinct cell wall composition. On the other hand, acquired resistance involves genetic modification either through HGT or mutation. The bacteria can also adapt the capacity of resistance via gene expression and protein production rapidly in response to stress or other environmental conditions, and also in the presence of specific antibiotics. Thioredoxin A (Ttrx A), thioredoxin reductase (trxB), D-Ala-D-Ala carboxypeptidase, DacA, FabI, and SapC are a few resistance genes which are responsible for the innate resistance to antibiotics such as fluoroquinolones, β-lactams, and aminoglycosides [24].
However, tolerance is the ability of microorganisms to withstand or survive antibiotics higher than the inhibitory concentration for a period of time [25]. Tolerance is an adaptive mechanism that reflects the change in cellular behavior from an active state to a dormant or inactive state (Figure 3) for a transient period [26]. A major rearrangement of energy production or few miscellaneous functions are witnessed during the tolerant state, and can be seen during poor growth or in the presence of few antibiotics or stress. Entrapment of antibiotics to the ECM without attachment to the target can also trigger tolerance, and results in dormancy or non-growth of bacterial cells. Persistence is a phenomenal form of tolerance, and persisters (Figure 3) are the tolerant form of cells in the population that are capable of surviving antibiotics but can be killed with longer exposure [27]. They can be either triggered persisters, induced in the presence of environmental stress or condition, or spontaneous persisters, converted to the form after a stochastic process. Persistence is also called heterotolerance [28].
Biofilm-mediated resistance (Figure 3) is a complex form of resistance that requires both the mechanisms of antibiotic resistance, as well as tolerance. Additionally, bacterial strains and species, antimicrobial agents, condition and developmental state of biofilm, and the growth condition of biofilm can highly affect the overall process [25].
3.1. Prevention of Access or Reduced Penetration
The architecture, as well as composition, of ECM through gradients of dispersion can severely affect the penetration of antibiotics, the access to cells, and, finally, affect the efficacy of antibiotics. Diffusion of antibiotics also varies due to interaction with the ECM components [29]. For example, extracellular DNA enhances the resistance of Pseudomonas biofilm against aminoglycosides, but not against beta-lactams and fluoroquinolones [30,31,32]. In the same way, eDNA enhances the resistance of Staphylococcus epidermidis biofilm against glycopeptides. It has been seen that negatively charged eDNA binds to negatively charged glycopeptides (vancomycin) and aminoglycosides (tobramycin). It has also been demonstrated that the binding of vancomycin and eDNA is 100-fold higher than the vancomycin and D-Ala-D-Ala peptides in peptidoglycan precursors; this may result in the accumulation of eDNA in the ECM [32]. The retention of eDNA in the ECM may result in a cation-limited environment through the chelation of Mg2+ cations. The chelation of Mg2+ can also initiate the AMR-linked two-component system of PhoPQ and PmrAB for Psuedomonas aeruginosa and S. enterica serovar Typhimurium [31,33].
Additionally, the antibiotic modifying enzymes can be released and located in the ECM; these can also be used by other sensitive species of bacteria within a mixed species biofilm. For instance, beta lactamases released by Moraxella catarrhalis protect the S. pneumoniae and H. influenza against amoxicillin and ampicillin, respectively [34,35]. Therefore, the biofilm architecture can alter the diffusion of antibiotics, and also the exposure of cells.
3.2. Stress Responses and Nutritional Limitation
Physiological heterogeneity is characterized by the genetic and phenotypic expression, metabolic activity, and antibiotic tolerance between the cells within different areas of biofilm [36,37]. The organization and architecture of biofilm generates gradients (Figure 3) of dispersion of water, nutrients, pH, signaling molecules, and waste products. It is believed that cells near the surface of biofilm microcolony utilize most of the nutrient supplies and create a deprived area further down [38,39,40,41]. Development of oxygen and nutritional depletion (Figure 3) in microcolonies and lower niches can contribute to the development of diverse physiological states of aerobic, anaerobic, microaerobic, and fermentative conditions; also the development of persisters, slow growth, fast growth, and dormant cells [17,36]. One such unique feature was observed by Yogesh and Anjali in 2021 (unpublished data), when MDR Enterococcus faecalis strains were re-cultured directly from the biofilm stage, which were stored for an extended period of 16 to 18 months at −70 °C. They found that the re-cultured colonies could grow only after 60–72 h of incubation at 37 °C on nutrient or chromogenic UTI agar without any supplement. When examined by the investigators of this study, the re-cultured bacterial strains were found to be resistant for the additional antibiotics. Cells in the oxygen deprived area (Figure 3) can show reduced metabolic activities and may undergo dormancy; this is believed to be the reason for tolerance against antibiotics such as tobramycin and ciprofloxacin that target protein synthesis and DNA gyrase [42].
It is well established that slow glowing cells are susceptible to colistin, which acts on the cell membrane [43]. However, the presence of colistin-tolerant cells within oxygen rich areas was observed, suggesting disagreement with the connection of slow growth rate and development of antibiotic tolerance within biofilm [44,45]. This fact was examined by Yogesh and Anjali in 2021 (unpublished data) when they found biofilm-producing Enterococcus faecalis resistant to colistin. Despite the full thickness antibiotic penetration, visible cellular activities and protein synthesis have also been seen in oxygen rich areas [46,47]. Through denitrification and fermentation, P. aeruginosa can sustain the anaerobic conditions, and supplementation of nitrate or L-arginine can increase the metabolic activity within nutrient deprived areas, thereby increasing the susceptibility to ciprofloxacin and tobramycin [42].
Stringent response (SR) and SOS response are adaptive mechanisms in response to stress and starvation of amino acids, iron, and carbon [48]. The tolerance of E. coli for cell wall inhibitor antibiotics such as carbapenems, penicillin, and cephalosporins is believed to be due to SR; this is also thought to be the case for cell division inhibitors such as norfloxacin and ofloxacin [49,50,51,52]. DNA damage may induce the SOS response and can initiate antibiotic tolerance. DNA damage leads to the generation of single-stranded DNA; that may activate RecA, stimulate self-cleavage of the repressor LexA and result in the de-repression of SOS genes [53]. The SOS response may lead to tolerance to antibiotics such as fluoroquinolones that can cause damage to DNA [54]. The tolerance of E. coli biofilm to fluoroquinolones has been observed due to the SOS response [52].
3.3. Enzymatic Cell Wall Modification
The dlt genes are crucial for Enterococcus faecalis and Staphylococcus aureus to form biofilm [55,56]. This has been observed with the deletion of S. aureus dltA and the subsequent reduction of resistance to vancomycin, and also the reduction of planktonic resistance of E. faecalis to colistin and polymyxin B [57]. The dltABCD operon was a positive hit in a screening for biofilm-specific gentamicin tolerance genes in Streptococcus mutans, a dental pathogen that can also cause infective endocarditis [58]. The dltABCD homologues are important for D-alanylation of teichoic acid in many Gram-positive species [59]. Due to its inability to add D-alanine to the anionic teichoic acid molecules in the cell wall, the ΔdltA mutant was more negatively charged than the wild-type [58]. It is thought that the increased negative charge of the ΔdltA strain promotes uptake of gentamicin, a positively charged aminoglycoside [58].
3.4. Multispecies Interaction
The interaction between different species (Figure 3) in biofilm can initiate antibiotic tolerance. For example, polymicrobial biofilms of E. faecalis, Finogoldia magna, and S. aureus were observed to be two-fold more resistant than the mono-species biofilm of P. aeruginosa [60]. In the same way, in a dual species biofilm model, M. catarhhalis released beta-lactamase, which protected S. pneumoniae from amoxicillin [34,61]. In the research by Ryan et al. (2008) on P. aeruginosa and Stenotrophomonas maltophilia dual-species biofilms, it was observed that the diffusible signal factor is an intercellular signaling molecule produced by S. maltophilia, and is sensed by the two-component sensor BptS in P. aeruginosa, leading to upregulation of the PmrA-regulated PA3552-3559 and PA4773-4775 genes [62]. Furthermore, the interaction between fungi and bacteria in a multispecies biofilm has also been studied. The resistance of Staphylococcus to vancomycin was increased in C. albicans and S. aureus biofilm due to the fungal matrix component beta-1,3-glucan, which is believed to act as a barrier against vancomycin [63,64]. It was also noted that C. albicans can increase the biofilm production of P. aeruginosa through alcohol production [65].
3.5. Mutation
Genomic mutation, even without any strong spontaneous selective stress or pressure, may lead to AMR. Mutation usually occurs at the rate of 10−10 to 10−9 per nucleotide per generation in most of the bacteria [66,67]. Oxidative stress-causing agents that damage DNA can also accelerate the mutation rate. Although with a sublethal dose of a bactericidal, the build-up of ROS might be low but would be enough to induce synthesis of multidrug efflux pumps, mutagenesis, and promote resistance [68]. The defect in mutS, mutL, and uvrD genes can further promote the mutation frequency up to 100-fold due to failure of the DNA repair mechanism [69,70]. An example of evolutionary mutation in microorganisms can be seen with hypermutators with the capability to acquire favorable mutations under selective stress that can lead to AMR [71]. This particular phenotype has also been observed with Pseudomonas biofilm, with resistance against rifampicin and ciprofloxacin [72]. Apart from that the abovementioned phenotypic characteristics, hypermutations have also been reported in S. aureus and H. influenza isolated from cystic fibrosis infection but not in Enterobacteriaceae isolated from acute urinary tract infection; which indicates hypermutability is favored in certain environments [73,74,75].
Mutation in the rspL gene, the gene that codes 16S rDNA and S12 ribosomal protein, affects the antibiotic targeting for aminoglycosides, whereas mutation in the mexZ gene results in overproduction of the MexXY-OprM efflux system [76,77]. Additionally, the mutations in the genes coding for the PmrAB two-component regulatory system, which regulates the addition of aminoarabinose to lipid A, has been associated with colistin resistance [78]. Conversely, mutations in the promoter of the chromosomal ampC gene that increase the plasmid copy number may result in increased production of β-lactamases [79,80]. A large number of β-lactamase variants with point mutations in the gene, resulting in changes in the amino-acid sequence, may lead to the development of extended-spectrum β-lactamases (ESBLs) that also degrade first-, second-, and third-generation cephalosporins and/or became resistant to β-lactamase inhibitors [81].
3.6. Efflux Pump
This is the movement of a drug from the intracellular to extracellular matrix without attachment to an intracellular target (Figure 3 and Figure 4); therefore, this mechanism confers resistance to bacterial cells [82]. Planktonic resistance in P. aeruginosa to low concentration ofloxacin has been said to be due to the multiple multidrug efflux pumps, such asMaxAB-OprM [83]. Another major multidrug efflux pump PA1875-1877 is believed to be a contributor to resistance in P. aeruginosa biofilm [84]. A two-fold to four-fold increase in susceptibility of biofilm to tobramycin, gentamicin, and ciprofloxacin was observed after the deletion of PA1875, PA1876, and PA1876; on the other hand, planktonic susceptibility was not affected much [84]. In addition, the resistance of P. aeruginosa biofilm to azithromycin was said to be due to the MexAB-OprM or MexCD-OprJ efflux pumps, whereas these efflux pumps are said to be required for the resistance against colistin in a metabolically active state [44,85].
3.7. Quorum Sensing (QS)
QS is a population-density-dependent regulatory mechanism for interbacterial communication, which acts through signaling molecules named autoinducers. These autoinducers are recognized by the cell-surface receptors in order to induce gene transcription for virulence factors, surface proteins, transcriptional factors, and biofilm production [86,87]. A biofilm formed by P. aeruginosa lacking lasR and rhlR was observed as more susceptible to tobramycin than the wild-type biofilms [88]. In the same way, S. aureus deficient with QS-specific agrD was observed as less resistant as compared to the wild-type [89]. Moreover, fsrA and gelE mutants of E. faecalis for QS and QS-controlled protease were seen with the impairment of biofilm formation in the presence of daptomycin, gentamicin, or linezolid [90].
4. Mechanism of AMR
The four major mechanisms of AMR are illustrated in Figure 4.
4.1. Cell Wall or Cell Membrane Modification
Enterobacteriaceae show resistance to carbapenems because of reduced permeability of the bacterial membrane to this antibiotic. In most Enterobacteriaceae, OmpC and OmpF of E. coli are the major porins. The downregulation in the expression of porin or the substitution of major porins with more-selective membrane channels has been engaged in this resistance mechanism [91]. Gram-negative bacteria such as Pseudomonas aeruginosa, V. cholerae, and S. enteric show resistance due to the reduced permeability (Figure 4) of antibiotics like azithromycin, clarithromycin, roxithromycin, and erythromycin. Bacteria can also prevent the access of antibiotic to the target by pumping the antibiotics out of the bacterial cell through efflux pumps. Thus, in Gram-negative bacteria, intrinsic resistance is achieved [92].
4.2. Modification or Protection of Targets
Antibiotics are specifically designed to bind to their specific targets with high affinity, and therefore disrupt the normal activity of the target. Antibiotics are less efficient at binding to their targets if the targets alter their structure (Figure 4). A single nucleotide mutation (SNP) in the gene, encoding an antibiotic target, results in resistance towards the given antibiotic. For example, an amino acid substitution in the rpoB gene develops resistance towards rifampin. This mutation reduces the affinity of rifampin to its target, and the transcription continues [93].
Streptococcus pneumoniae produces the penicillin binding proteins (PBPs), which reduce the affinity to beta-lactam antimicrobial agents. S. pneumonia is also resistant to cephalosporin (third-generation drug) due to altered PBP1a and 2x alterations. In PBPs, alteration occurs by the recombination of the PBP gene of S. pneumonia and related PBP genes of other closely related streptococcal species by transformation [94]. Without any mutation, antibiotic resistance can be achieved by post-translational modification of targets. In the erythromycin ribosome methylase (erm) family, the drug-binding sites alter due to the methylation of 16S rRNA, which therefore prevents the binding of lincosamides, macrolides, and streptogramin to the 16S rRNA [95]. Antibiotics such as lincosamides, phenicols, oxazolidinones, streptogramins, and pleuromutilins may not interact with the target 23S rRNA due to methylation of the A2503 residue by chloramphenicol florfenicol resistance gene (cfr) product [96].
4.3. Enzymatic Degradation of Antimicrobials
Bacteria can also modify the structure of antibiotics, prevent their entry into the cell, and render them inactive (Figure 4). Bacteria inactivate antibiotics via hydrolysis reactions. Antibiotics such as aminoglycosides, β-lactams, phenicols, and macrolides can be degraded by enzymes such as carbapenemases and chloramphenicol acetyltransferase. The active enzymes against β-lactams include both the early and the extended-spectrum β-lactamases (ESBLs), which are also active against oxyimino-cephalosporins [97]. TEM-1 β-lactamase and SHV-1 (sulphydril variable active site) enzyme, found on the plasmid in a strain of Escherichia coli, are the major ESBLs and hydrolyze a broad range of extended spectrum cephalosporins. Changes within antimicrobial functional groups such as thiol, phosphoryl, acyl, or ribosyl due to degrading enzymes may lead to failure of binding of lincomycin, macrolides, and chloramphenicol to targets [98].
4.4. Ribosome Protection
Ribosome protection is a resistance mechanism which is developed by some bacteria. Tetracycline is a bacterial protein synthesis inhibitor, and the bacteria produce ribosome protection proteins that bind to the ribosomal target and prevent the binding of tetracycline to the ribosome. In such cases, bacteria can grow even in the presence of tetracycline due to synthesized ribosome protection proteins [99].
5. Impact of AMR
The most persistent multidrug-resistant bacteria implicated in high mortality and morbidity globally are S. aureus, Escherichia coli, Enterococcus faecium, Streptococcus pneumoniae, Klebsiella pneumonia, and Pseudomonas aeruginosa. Apart from that, cancer-related neutropenia has been observed with high antimicrobial resistance. Moreover, AMR has complicated the management and treatment of neonatal sepsis due to biofilm formation. Failure of first- and second-generation antibiotics inevitably forces the development of next generation antimicrobials, requiring huge amounts of resources. Resistance of biofilm-producing MDR Enterococcus faecalis for a second-line drug (vancomycin) and a last-resort drug (linezolid) was observed by Yogesh and Anjali in 2021 (unpublished data). Morbidity and mortality rates are severely affected by MDR bacterial strains. Without effective antibiotics, organ transplantation, intensive care for pre-term newborns, hip replacement surgery, and chemotherapy for cancer treatment are not usually performed [100].
In the 21st century, biofilm-led AMR has become a global threat to the public health system. Resistant microbes are more difficult to treat, and substitute medications or even higher doses are required to treat them, which may be toxic and more expensive. Microbes resistant to multiple antimicrobials are called multidrug resistant (MDR). All classes of microbes, including bacteria, viruses, fungi, and protozoa, can evolve resistance. Bacteria which are totally drug-resistant (TDR) or extensively drug-resistant (XDR) are called “superbugs” [101]. Bacterial resistance occurs naturally, by genetic mutation, or by procuring genetic material from other bacteria. However, extensive use of antimicrobials appears to encourage selection for mutations that can render antimicrobials ineffective.
6. Current Therapeutic Practices against Biofilm Producing Bacteria
Biofilm can be understood by the fact that it is a mechanism of self-motivation in the process of pathogenesis. Numerous techniques and methods have been worked on in order to understand biofilm, target biofilm, and biofilm-producing microorganisms through the medium of natural products, surface coating, lasers, texturing and patterning, nanostructures, and peptides [102,103,104,105]. Figure 5 illustrates a few strategies for alleviating biofilm-related ailments through the means of inhibition, dispersal, or eradication of the biofilm.
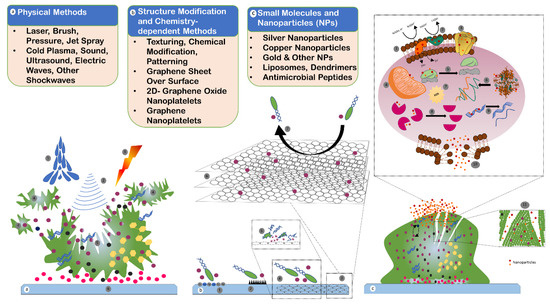
Figure 5.
Strategies against biofilm: (a) physical methods of biofilm removal include water jet (1), sound, ultrasonic, electric and other shockwaves (2), and laser and photodynamic therapy (3) to dislodge and disrupt the biofilm in small parts (4) and to break the main biofilm region (5). The host surface or substrate (6) is not usually disturbed or modified in this method. (b) Structural modification and chemistry-dependent methods include techniques to change the texture, pattern, surface and material properties, surface charge, and chemical coating to repel or avoid bacterial cells attaching to the surface (1,2). Additionally, graphene sheets on substrate (3), graphene nanoplatelets (GNP) (6), and chemistry-dependent polyamine-functionalized quantum dots (QDs) are some advanced methods that can be used to inhibit the cell adhesion to substrate (7), and to damage the bacterial cells (4,5) that would provide antimicrobial properties. (c) Small molecules and nanoparticles developed and used for biocompatibility and antimicrobial properties include organic nanoparticles (liposomes and polymers), inorganic nanoparticles (silver, copper, gold, and iron oxide nanoparticles), and specifically designed antimicrobial peptides (AMPs) of 5 to 90 amino acids and with a molecular mass of 1 to 5 kDa. AMPs are capable of disrupting bacterial cell membranes, in addition to having enzymatic and protein activities. Nanoparticles can cause damage to the electron transport chain (1,2), inhibit membrane protein (3), dysfunction of the mitochondria (4), disassembly of ribosome (5), denaturation of proteins (6), oxidative stress by producing ROS (7), inactivation of enzymes (8), damage to DNA (9), damage to cell membrane (10), and degradation of EPS (11).
6.1. Biofilm Inhibition Strategy
The material matrix of implanted medical devices, prosthetic surfaces such as dental filling materials and artificial teeth, and biomaterials provide an ideal site for bacterial adhesion, promoting mature biofilm (Figure 2e,f) formation [106]. Consequently, bacterial adhesion or attachment methods can be worked upon with the aim of preventing biofilm. In doing so, modification of the attachment surface, for example coating the external surface of devices, is an ideal method [107]. Various biomaterials and coating materials which alter the surface of target, making it unfavorable for bacteria, are being developed [106,107]. This has led to the development of coated medical devices for the prevention of biofilm-related infections and/or complications associated with orthopedic implants. Aiming to prevent biofilm, application of therapeutic agents or inhibitors is usually done with orthopedic, dental implants, and dental filling material, such as incorporation of anti-biofilm nanoparticles in dental restorative composite materials; this has resulted in a significant reduction of biofilm, as well as it’s associated infections [106]. Small molecule biofilm inhibitors (Figure 1 and Table 1) are applied to biomaterials and devices as another approach for preventing biofilm formation, as well as to make the surfaces non-reactive or inert. In a clinical trial, 5-fluorouracil was found to be as effective as chlorhexidine/silver sulfadiazine in controlling central venous catheter colonization with no bloodstream infection [108]. Conversely, azithromycin prevented P. aeruginosa ventilator-associated pneumonia by targeting quorum sensing in high risk patients [109]. Similarly, azithromycin was seen to improve quality of life without any adverse events in patients with cystic fibrosis infected with P. aeruginosa [110]. Natural products such as garlic extracts have also been examined as an inhibitor of quorum sensing; however, no significant effects were detected in patients with garlic therapy as compared with placebo [111]. Already used or utilized inhibitors are therefore becoming the focus of anti-biofilm strategies and research [112].

Table 1.
Potential small molecules and drug candidates for biofilm inhibition.
6.2. Dispersal of Biofilm for Treatment
Bacteria in biofilms are inherently more tolerant to antimicrobial treatment when compared directly to planktonic cells (Figure 1) of the same strain [169,170]. The mechanisms of antibiotic resistance in planktonic bacteria, such as mutations, efflux pumps, and antibiotic modifying enzymes, are well understood [93]. These do not hold true for resistance in biofilm formers, as there are many examples where drug-susceptible bacterial strains often exhibit significant antibiotic tolerance in the biofilm; however, they become susceptible once the integrity of biofilm is compromised [39]. Thus, biofilm antibiotic tolerance is thought to involve alternative mechanisms to bacterial antimicrobial resistance.
Quorum sensing, as a principal chemical pathway, enables biofilm-mediated bacterial maintenance and survival [171]. As dispersed cells are generally more susceptible to antimicrobial treatment than biofilm-residing cells, the above strategy has emerged as an intense area of study, resulting in the review, as well as development, of chemical agents capable of effective biofilm dispersal [172,173]. Usually, dispersal agents are employed in parallel with antimicrobials in the case of untreated dispersed cell translocation to different areas and seeding infection [174,175]. Co-treatment generally involves administering a combination of drugs concurrently—in this case, a biofilm dispersal agent and an antibiotic—to exert a synergistic effect [156,174]. Limitation of this treatment strategy can be the availability of both agents at the target site in the correct concentration [176].
6.3. Agents to Eradicate Biofilm
A variety of promising candidates, such as antimicrobial peptides (illustrated in Figure 5), capable of targeting and eradicating biofilm, have already been developed; their activity, design, and potential uses have become an emerging field in biofilm research.
Antimicrobial peptides (AMPs) are one of the most well-studied classes of biofilm-eradicating agents, and are often considered an attractive alternative to antibiotics [177,178]. They are composed of 5 to 90 amino acids with a molecular mass of 1 to 5 kDa, and are ubiquitous compounds, produced in a variety of plant, invertebrate, and animal species. While they are mostly cationic in nature, anionic forms have also been reported [80,81]. Despite the intensive research on biofilms, their mode of action is yet to be recognized, except their capabilities of disrupting the cell membrane and their enzymatic or protein activities. While their actions have been observed as a substitute to antimicrobials, AMPs are yet to be recognized as effective agents against biofilms.
Nanoparticles (NPs) are one of the leading drug carriers and are widely researched among all drug delivery systems. They can act as an important tool to breach the biofilm shield and enhance the availability of drugs to the bacterial population (illustrated in Figure 5). Innumerable synthetic polymers are used, along with natural polymers for manufacturing nanoparticles. Inorganic compounds, as well as synthetic and natural polymers, have been used to produce nanoparticles [179]. The most accepted mechanism of action is the ability of NPs to enter the cell in order to induce endocytosis. The authors of this study, in 2021, synthesized silver nanoparticles (AgNPs) (illustrated in Figure 6) of 118–157.7 nm diameter (Figure 7), derived from Azadirachta indica (neem), and tested them in various concentrations against multidrug resistant oral pathogen Enterococcus faecalis-formed biofilm through a human dentine-block-based biofilm assay (illustrated in Figure 6) and a crystal violet assay. They observed a significant reduction in colony forming units (CFU) (107) upon treatment with AgNPs. Additionally, a significant reduction in CFUs (Figure 8) upon treatment with AgNPs in combination with clove or eugenol was also observed by the authors of this study.
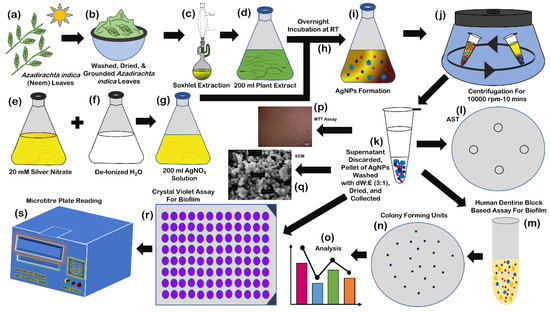
Figure 6.
Synthesis of silver nanoparticles: collection of Azadirachta indica (neem) leaves (a); washing, drying, and grounding of leaves (b); Soxhlet extraction (c); collection of extract (d); preparation of 200 mL 20 mM silver nitrate solution (e); 200 mL deionized water (f); mixing of silver nitrate solution and deionized water (at the time of mixing) (g); incubation at room temperature for 12–18 h (h); change of color of mixture after 12–18 h (i); centrifugation at 1000 rpm for 10 min (j); collection of pellet after discarding the supernatant, washing of pallet with solution of deionized water and ethanol (3:1) for three times, drying, and collection of nanoparticles (k); human dentine-block-based biofilm assay (l–o); cytotoxicity assay (p); SEM analysis (q); and crystal violet-based biofilm assay (r,s).
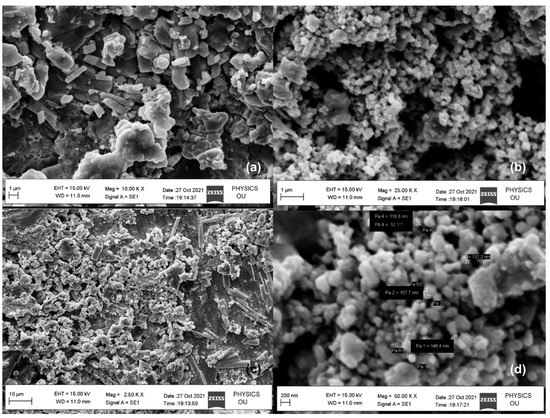
Figure 7.
SEM analyses of biosynthesized silver nanoparticles (a–d).
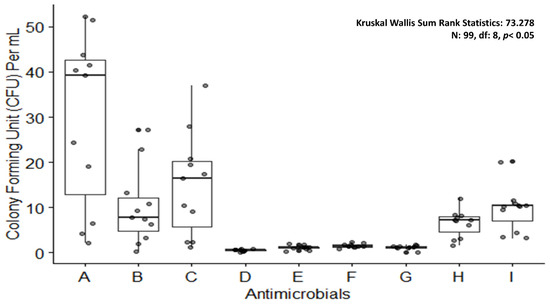
Figure 8.
Boxplot showing colony forming units upon treating Enterococcus faecalis biofilm with antimicrobials. A: control; B: silver nanoparticles 20 µg/mL; C: silver nanoparticles 10 µg/mL; D: silver nanoparticles 20 µg/mL and clove 2500 µg/mL combination; E: silver nanoparticles 10 µg/mL and clove 1700 µg/mL combination; F: silver nanoparticles 5 µg/mL and eugenol 850 µg/mL combination; G: sodium hypochlorite 5%; H: sodium hypochlorite 3; I: chlorhexidine 2%.
Other proven agents for anti-biofilm properties are chlorhexidine and sodium hypochlorite, which have exhibited a notable reduction in CFU/mL (Figure 8) as compared to AgNPs alone or in combinations with other agents when tested by the authors of this study. It was observed that sodium hypochlorite was as potent as the combined dose of AgNPs and clove. In order to eradicate biofilm-producing pathogens, nanoparticles could be an answer due to their low toxicity, high efficacy power, high and efficient penetration ability into the host cells, and site-specific drug release. Additionally, these properties of NPs can be considered in order to develop an effective method against biofilm as a means to keep AMR at bay.
6.4. Bioinformatic Approach to Identify Anti-Biofilm Agent
Accelerated discovery of novel anti-biofilm agents needs new sequencing and computational technologies which are biologically relevant models to better understand biofilm formation. As of now, most studies exploring biofilm mechanisms rely on omics studies, such as transcriptomics and proteomics, to uncover new genetic and protein targets for novel anti-biofilm agents to modulate. Apart from the omics studies, in silico screening can be used to screen for molecules from large databases. Another approach is machine learning, which can predict the anti-biofilm activity of a molecule and can also play a vital role. Candidate molecules thus identified can then be synthesized and validated in several biological models, including biofilms grown in microtiter plates, flow cells, animal models, and human organoids. Therefore, integration of both lab and computational science can provide a better chance of developing a successful anti-biofilm agent.
RNA-Seq, which is a high-throughput technology, is employed to measure gene regulation and expression. It is able to identify transcriptomic signatures highly distinct to biofilms or biofilm-mediated bacterial dispersion [173]. RNA-Seq can also be used to study the effects of antibiotics and potential anti-biofilm agents on biofilm formation [180,181]. Transposon insertion sequencing (Tn-Seq) is a recent technology which offers a high-throughput approach to help in identifying genes required to survive in a specific condition, including in a biofilm. Novel techniques have been identified in Tn-Seq, including sorting mutant cells and probing essential gene functions using inducible promotors [182].
Additionally, for supplementary information, proteomics can be useful, especially meta-proteomics, which can identify proteins essential for multi-species aggregation. Meta-proteomics can also identify the abundance of proteins for each species in key metabolic and energy pathways, such as amino acid metabolism and fermentation absence among communities of single species [183]. Community of four co-cultivated soil bacteria (Stenotrophomonas rhizophila, Xanthomonas retroflexus, Microbacterium oxydans, and Paenibacillus amylolyticus) has been seen with increased biofilm formation as compared to single species communities; that suggests a cooperative as well as competitive mechanism of bacterial survival. Metabolomics analyzes differential production of small molecule (Table 1) metabolites and metabolism intermediates (e.g., carbohydrates, nucleotides, and amino acids). Not only that, smaller molecules and intermediates such as amino acids and nucleotides can be analyzed through metabolomics, along with their production. Metabolomics offers a snapshot of the functional changes that result from the transcriptomic and proteomic changes measured by the above methods. Biofilm activity can also be predicted by analyzing metabolites [184,185,186].
7. Conclusions
It is a well-established fact that biofilm further complicates the infection associated with communicable, as well as non-communicable, diseases. In addition, biofilm has also been implicated in post-operative infections and digestive disorders such as recurrent sialadenitis. Furthermore, widely studied cystic fibrosis has been identified as a biofilm-associated genetic disorder. Bacteria involved in infective endocarditis are able to form biofilms; that can be associated with the failure of anti-microbials and compromised heart valves. Bacteria capable of producing biofilm have also been identified from atherosclerotic arteries [187]. Surface adherence is one of the prevailing modes for bacterial growth, while the biofilm environment supports the development of antimicrobial resistance [18,188,189]. Our understanding of how this lifestyle influences the evolution of AMR, whether by different population genetic dynamics or molecular mechanisms, is limited. One example is that the close proximity of cells in biofilms may facilitate the horizontal transfer and persistence of resistance genes in bacterial populations [190,191]. There could be many contributing factors assisting the surge in AMR, limiting the use of many antimicrobial agents. Since the role of biofilm in AMR is evident, this consequently opens up avenues for repurposing the existing drugs targeting the biofilm. Forecasting the AMR by analyzing the population of biofilm formers would go a long way to tackle AMR. Secondly, the use of nanoparticles in biomaterials or as a drug delivery mode can be of great use. On the grounds of this, there is compelling requisite for the development of bioinformatics tools with the aim of analyzing, and also predicting, AMR emanating from biofilm.
Furthermore, drug co-administration modalities, as suggested by a number of studies, can precipitate challenges such as complex treatment schedules, increased risk of adverse effects, increased treatment costs, and antagonism [192,193]. Accordingly, in order to predict AMR, in addition to formulating viable therapeutic strategies for biofilm associated ailments, aforementioned facts need to be addressed robustly. Identification of potential protein targets, biofilm formation pathways, and modulators associated with protein targets through in-vitro, and high-throughput virtual screening should be considered. Small molecules, such as 5-fluorouracil, which targets quorum sensing, and synthetic molecules, such as terrain, which targets both quorum sensing as well as c-di-GMP, can be considered for anti-biofilm strategies. Novel molecules, such as SYG-180-2-2, with the ability to reduce the bacterial adhesion and polysaccharide intercellular adhesin (PIA) in methicillin-resistant Staphylococcus aureus should be seen as potential antibacterial agents [194,195]. Although the exact anti-biofilm mechanism of a few small molecules or drug candidates such as TAGE and CAGE are not entirely understood, their effectiveness against biofilm-causing microorganisms can still pave the road to combat biofilm-led AMR. Coating of medical devices including implants with silver and zinc oxide nanoparticles is one of the most effective strategies against biofilm-led infection, which can be explored further. Another novel method, machine learning modeling of anti-biofilm agents from a database can also be considered before in-vitro anti-biofilm assays and biological models.
Author Contributions
Conceptualization, Y.D. and R.P.P.; methodology, Y.D. and A.P.; software, Y.D.; validation, Y.D., R.P.P. and R.D.; formal analysis, Y.D. and R.P.P.; investigation, A.G.; resources, A.V.; data curation, A.P.; writing—original draft preparation, Y.D., R.D. and T.S.; writing—review and editing, Y.D., R.P.P., T.S. and C.-M.C.; visualization, T.S.; supervision, A.P. and V.S.R.; project administration, V.S.R.; funding acquisition, C.-M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by two industry–academia collaboration projects, VtR Inc-CGU, R.O.C. grant number SCRPD1L0221, DOXABIO-CGU, R.O.C. grant number SCRPD1K0131, and also the CGU project grant number UZRPD1L0011 and the APC was funded by VtR Inc-CGU, R.O.C., DOXABIO-CGU, R.O.C., and CGU project.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available they have not been added to any repository yet.
Acknowledgments
The authors of this study acknowledge the support of Mamta from Mamta Dental Clinic, Faridabad, India for intraoral and dental pictures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Burnett-Boothroyd, S.C.; McCarthy, B.J. 13-Antimicrobial treatments of textiles for hygiene and infection control applications: An Industrial perspective. In Textiles for Hygiene and Infection Control; McCarthy, B.J., Ed.; Woodhead Publishing Series in Textiles; Woodhead Publishing: Sawston, UK, 2011; pp. 196–209. ISBN 978-1-84569-636-8. [Google Scholar]
- Saga, T.; Yamaguchi, K. History of antimicrobial agents and resistant. Jpn. Med. Assoc. J. 2009, 52, 103–108. [Google Scholar]
- Ellis, A. Overcoming Resistance: A Rational Emotive Behavior Therapy Integrated Approach, 2nd ed.; Springer Publishing Company: Cham, Switzerland, 2002; ISBN 978-0-8261-4913-8. [Google Scholar]
- Percival, S.L.; Malic, S.; Cruz, H.; Williams, D.W. Introduction to biofilms. In Biofilms and Veterinary Medicine; Percival, S., Knottenbelt, D., Cochrane, C., Eds.; Springer Series on Biofilms; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–68. ISBN 978-3-642-21289-5. [Google Scholar]
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Irvin, R.T.; Cheng, K.J. The bacterial glycocalyx in nature and disease. Annu. Rev. Microbiol. 1981, 35, 299–324. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef]
- Mah, T.-F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Sinclair, J. Importance of a one health approach in advancing global health security and the sustainable development goals. Rev. Sci. Tech. Int. Off. Epizoot. 2019, 38, 145. [Google Scholar] [CrossRef]
- Flemming, H.-C. Microbial biofouling: Unsolved problems, insufficient approaches, and possible solutions. In Biofilm Highlights; Flemming, H.-C., Wingender, J., Szewzyk, U., Eds.; Springer Series on Biofilms; Springer: Berlin/Heidelberg, Germany, 2011; pp. 81–109. ISBN 978-3-642-19940-0. [Google Scholar]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef]
- Wingender, J.; Flemming, H.-C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health 2011, 214, 417–423. [Google Scholar] [CrossRef]
- Morgan-Sagastume, F.; Larsen, P.; Nielsen, J.L.; Nielsen, P.H. Characterization of the loosely attached fraction of activated sludge bacteria. Water Res. 2008, 42, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.; Fazen, C.; Henry, T.; Mok, W.; Orman, M.; Sandvik, E.; Volzing, K.; Brynildsen, M. The role of metabolism in bacterial persistence. Front. Microbiol. 2014, 5, 70. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Neu, D.T.R.; Wingender, D.J. The Perfect Slime: Microbial Extracellular Polymeric Substances (EPS); IWA Publishing: London, UK, 2016; ISBN 978-1-78040-741-8. [Google Scholar]
- Ch’ng, J.-H.; Chong, K.K.L.; Lam, L.N.; Wong, J.J.; Kline, K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019, 17, 82–94. [Google Scholar] [CrossRef]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 2021, 10, 3. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in gram-negative bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General mechanisms leading to persister formation and awakening. Trends Genet. TIG 2019, 35, 401–411. [Google Scholar] [CrossRef] [PubMed]
- De Beer, D.; Stoodley, P.; Lewandowski, Z. Liquid flow in heterogeneous biofilms. Biotechnol. Bioeng. 1994, 44, 636–641. [Google Scholar] [CrossRef]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLOS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef]
- Wilton, M.; Charron-Mazenod, L.; Moore, R.; Lewenza, S. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 60, 544–553. [Google Scholar] [CrossRef]
- Doroshenko, N.; Tseng, B.S.; Howlin, R.P.; Deacon, J.; Wharton, J.A.; Thurner, P.J.; Gilmore, B.F.; Parsek, M.R.; Stoodley, P. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob. Agents Chemother. 2014, 58, 7273–7282. [Google Scholar] [CrossRef]
- Johnson, L.; Horsman, S.R.; Charron-Mazenod, L.; Turnbull, A.L.; Mulcahy, H.; Surette, M.G.; Lewenza, S. Extracellular DNA-induced antimicrobial peptide resistance in Salmonella Enterica serovar typhimurium. BMC Microbiol. 2013, 13, 115. [Google Scholar] [CrossRef]
- Perez, A.C.; Pang, B.; King, L.B.; Tan, L.; Murrah, K.A.; Reimche, J.L.; Wren, J.T.; Richardson, S.H.; Ghandi, U.; Swords, W.E. Residence of Streptococcus Pneumoniae and Moraxella Catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 2014, 70, 280–288. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Hong, W.; Pang, B.; Weimer, K.E.D.; Juneau, R.A.; Turner, J.; Swords, W.E. Indirect pathogenicity of haemophilus influenzae and Moraxella Catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 2010, 1, e00102-10. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Williamson, K.S.; Richards, L.A.; Perez-Osorio, A.C.; Pitts, B.; McInnerney, K.; Stewart, P.S.; Franklin, M.J. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 2012, 194, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- De Beer, D.; Stoodley, P.; Roe, F.; Lewandowski, Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 1994, 43, 1131–1138. [Google Scholar] [CrossRef]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella Pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Zhang, T.; Xu, R.; Pitts, B.; Walters, M.C.; Roe, F.; Kikhney, J.; Moter, A. Reaction–diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. npj Biofilms Microbiom. 2016, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Borriello, G.; Richards, L.; Ehrlich, G.D.; Stewart, P.S. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2006, 50, 382–384. [Google Scholar] [CrossRef]
- Haagensen, J.A.J.; Klausen, M.; Ernst, R.K.; Miller, S.I.; Folkesson, A.; Tolker-Nielsen, T.; Molin, S. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2007, 189, 28–37. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the Pmr and MexAB-OprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Chiang, W.-C.; Pamp, S.J.; Nilsson, M.; Givskov, M.; Tolker-Nielsen, T. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol. Med. Microbiol. 2012, 65, 245–256. [Google Scholar] [CrossRef]
- Werner, E.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Heydorn, A.; Molin, S.; Pitts, B.; Stewart, P.S. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2004, 70, 6188–6196. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.C., III; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, G.S.; Oyeyemi, B.F.; Bhavesh, N.S. Stringent response protein as a potential target to intervene persistent bacterial infection. Biochimie 2019, 165, 67–75. [Google Scholar] [CrossRef]
- Kusser, W.; Ishiguro, E.E. Suppression of mutations conferring penicillin tolerance by interference with the stringent control mechanism of Escherichia coli. J. Bacteriol. 1987, 169, 4396–4398. [Google Scholar] [CrossRef] [PubMed]
- Tuomanen, E.; Tomasz, A. Induction of autolysis in nongrowing Escherichia coli. J. Bacteriol. 1986, 167, 1077–1080. [Google Scholar] [CrossRef]
- Wu, N.; He, L.; Cui, P.; Wang, W.; Yuan, Y.; Liu, S.; Xu, T.; Zhang, S.; Wu, J.; Zhang, W.; et al. Ranking of persister genes in the same Escherichia coli genetic background demonstrates varying importance of individual persister genes in tolerance to different antibiotics. Front. Microbiol. 2015, 6, 3. [Google Scholar] [CrossRef]
- Bernier, S.P.; Lebeaux, D.; DeFrancesco, A.S.; Valomon, A.; Soubigou, G.; Coppée, J.-Y.; Ghigo, J.-M.; Beloin, C. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 2013, 9, e1003144. [Google Scholar] [CrossRef] [PubMed]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef]
- Dörr, T.; Lewis, K.; Vulić, M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLOS Genet. 2009, 5, e1000760. [Google Scholar] [CrossRef]
- Gross, M.; Cramton, S.E.; Götz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef]
- Fabretti, F.; Theilacker, C.; Baldassarri, L.; Kaczynski, Z.; Kropec, A.; Holst, O.; Huebner, J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 2006, 74, 4164–4171. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Vuong, C.; Otto, M.; Götz, F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 2000, 44, 2845–2847. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Rybtke, M.; Givskov, M.; Høiby, N.; Twetman, S.; Tolker-Nielsen, T. The Dlt genes play a role in antimicrobial tolerance of Streptococcus mutans biofilms. Int. J. Antimicrob. Agents 2016, 48, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, F.C.; Baddiley, J. A Continuum of anionic charge: Structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 2011, 6, e27317. [Google Scholar] [CrossRef]
- Budhani, R.K.; Struthers, J.K. Interaction of Streptococcus Pneumoniae and Moraxella Catarrhalis: Investigation of the indirect pathogenic role of β-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 1998, 42, 2521–2526. [Google Scholar] [CrossRef]
- Ryan, R.P.; Fouhy, Y.; Garcia, B.F.; Watt, S.A.; Niehaus, K.; Yang, L.; Tolker-Nielsen, T.; Dow, J.M. Interspecies signalling via the Stenotrophomonas Maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 2008, 68, 75–86. [Google Scholar] [CrossRef]
- Adam, B.; Baillie, G.S.; Douglas, L.J. Mixed species biofilms of Candida Albicans and Staphylococcus epidermidis. J. Med. Microbiol. 2002, 51, 344–349. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Candida Albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef]
- Chen, A.I.; Dolben, E.F.; Okegbe, C.; Harty, C.E.; Golub, Y.; Thao, S.; Ha, D.G.; Willger, S.D.; O’Toole, G.A.; Harwood, C.S.; et al. Candida Albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014, 10, e1004480. [Google Scholar] [CrossRef]
- Woodford, N.; Ellington, M.J. The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 2007, 13, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.W.; Yeesin, P.; Simmons, L.A.; Wang, J.D. Sources of spontaneous mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Schaaper, R.M.; Dunn, R.L. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: The nature of in vivo DNA replication errors. Proc. Natl. Acad. Sci. USA 1987, 84, 6220–6224. [Google Scholar] [CrossRef]
- Leong, P.M.; Hsia, H.C.; Miller, J.H. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J. Bacteriol. 1986, 168, 412–416. [Google Scholar] [CrossRef]
- Blázquez, J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 37, 1201–1209. [Google Scholar] [CrossRef]
- Driffield, K.; Miller, K.; Bostock, J.M.; O’Neill, A.J.; Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 2008, 61, 1053–1056. [Google Scholar] [CrossRef]
- Prunier, A.-L.; Malbruny, B.; Laurans, M.; Brouard, J.; Duhamel, J.-F.; Leclercq, R. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 2003, 187, 1709–1716. [Google Scholar] [CrossRef]
- Román, F.; Cantón, R.; Pérez-Vázquez, M.; Baquero, F.; Campos, J. Dynamics of long-term colonization of respiratory tract by haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 2004, 42, 1450–1459. [Google Scholar] [CrossRef]
- Kovacs, B.; Le Gall-David, S.; Vincent, P.; Le Bars, H.; Buffet-Bataillon, S.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Is biofilm formation related to the hypermutator phenotype in clinical enterobacteriaceae isolates? FEMS Microbiol. Lett. 2013, 347, 116–122. [Google Scholar] [CrossRef][Green Version]
- Guénard, S.; Muller, C.; Monlezun, L.; Benas, P.; Broutin, I.; Jeannot, K.; Plésiat, P. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 58, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Springer, B.; Kidan, Y.G.; Prammananan, T.; Ellrott, K.; Böttger, E.C.; Sander, P. Mechanisms of streptomycin resistance: Selection of mutations in the 16S RRNA gene conferring resistance. Antimicrob. Agents Chemother. 2001, 45, 2877–2884. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, S.M.; Ernst, R.K.; Miller, S.I. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 2004, 186, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Paltansing, S.; Kraakman, M.; van Boxtel, R.; Kors, I.; Wessels, E.; Goessens, W.; Tommassen, J.; Bernards, A. Increased expression levels of chromosomal AmpC β-lactamase in clinical Escherichia coli isolates and their effect on susceptibility to extended-spectrum cephalosporins. Microb. Drug Resist. 2015, 21, 7–16. [Google Scholar] [CrossRef]
- van Boxtel, R.; Wattel, A.A.; Arenas, J.; Goessens, W.H.F.; Tommassen, J. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob. Agents Chemother. 2016, 61, e01413-16. [Google Scholar] [CrossRef]
- Ghafourian, S.; Sadeghifard, N.; Soheili, S.; Sekawi, Z. Extended spectrum beta-lactamases: Definition, classification and epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–21. [Google Scholar]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Brooun, A.; Liu, S.; Lewis, K. A Dose-Response Study of Antibiotic Resistance in Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2000, 44, 640–646. [Google Scholar] [CrossRef]
- Zhang, L.; Mah, T.-F. Involvement of a Novel Efflux System in Biofilm-Specific Resistance to Antibiotics. J. Bacteriol. 2008, 190, 4447–4452. [Google Scholar] [CrossRef]
- Gillis, R.J.; White, K.G.; Choi, K.-H.; Wagner, V.E.; Schweizer, H.P.; Iglewski, B.H. Molecular Basis of Azithromycin-Resistant Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2005, 49, 3858–3867. [Google Scholar] [CrossRef]
- Di Cagno, R.; De Angelis, M.; Calasso, M.; Gobbetti, M. Proteomics of the bacterial cross-talk by quorum sensing. J. Proteom. 2011, 74, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Ferro, C.; Hentzer, M.; Reil, G.; Görg, A.; Kjelleberg, S.; Givskov, M.; Riedel, K.; Eberl, L. Identification of Quorum-Sensing Regulated Proteins in the Opportunistic Pathogen Pseudomonas aeruginosa by Proteomics. Environ. Microbiol. 2003, 5, 1350–1369. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Burmølle, M.; Hentzer, M.; Haagensen, J.A.J.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; et al. Pseudomonas aeruginosa Tolerance to Tobramycin, Hydrogen Peroxide and Polymorphonuclear Leukocytes Is Quorum-Sensing Dependent. Microbiology 2005, 151, 373–383. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple Roles for Enterococcus Faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef]
- Baroud, M.; Dandache, I.; Araj, G.F.; Wakim, R.; Kanj, S.; Kanafani, Z.; Khairallah, M.; Sabra, A.; Shehab, M.; Dbaibo, G.; et al. Underlying Mechanisms of Carbapenem Resistance in Extended-Spectrum β-Lactamase-Producing Klebsiella Pneumoniae and Escherichia coli Isolates at a Tertiary Care Centre in Lebanon: Role of OXA-48 and NDM-1 Carbapenemases. Int. J. Antimicrob. Agents 2013, 41, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Wiese, A.; Brandenburg, K.; Ulmer, A.J.; Seydel, U.; Müller-Loennies, S. The Dual Role of Lipopolysaccharide as Effector and Target Molecule. Biol. Chem. 1999, 380, 767–784. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 4.2.15. [Google Scholar] [CrossRef]
- Nagai, K.; Davies, T.A.; Jacobs, M.R.; Appelbaum, P.C. Effects of Amino Acid Alterations in Penicillin-Binding Proteins (PBPs) 1a, 2b, and 2x on PBP Affinities of Penicillin, Ampicillin, Amoxicillin, Cefditoren, Cefuroxime, Cefprozil, and Cefaclor in 18 Clinical Isolates of Penicillin-Susceptible, -Intermediate, and -Resistant Pneumococci. Antimicrob. Agents Chemother. 2002, 46, 1273–1280. [Google Scholar] [CrossRef]
- Kumar, N.; Radhakrishnan, A.; Wright, C.C.; Chou, T.-H.; Lei, H.-T.; Bolla, J.R.; Tringides, M.L.; Rajashankar, K.R.; Su, C.-C.; Purdy, G.E.; et al. Crystal Structure of the Transcriptional Regulator Rv1219c of Mycobacterium Tuberculosis. Protein Sci. Publ. Protein Soc. 2014, 23, 423–432. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr RRNA Methyltransferase Confers Resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin a Antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Clark, N.M.; Zhanel, G.G. Evolution of Antimicrobial Resistance among Enterobacteriaceae (Focus on Extended Spectrum β-Lactamases and Carbapenemases). Expert Opin. Pharmacother. 2013, 14, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Bacterial Resistance to Antibiotics: Enzymatic Degradation and Modification. Adv. Drug Deliv. Rev. 2005, 57, 1451–1470. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Update on Acquired Tetracycline Resistance Genes. FEMS Microbiol. Lett. 2005, 245, 195–203. [Google Scholar] [CrossRef]
- Nesher, L.; Rolston, K.V.I. The Current Spectrum of Infection in Cancer Patients with Chemotherapy Related Neutropenia. Infection 2014, 42, 5–13. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Pletzer, D.; Hancock, R.E.W. Antibiofilm Peptides: Potential as Broad-Spectrum Agents. J. Bacteriol. 2016, 198, 2572–2578. [Google Scholar] [CrossRef]
- Li, X.; Wu, B.; Chen, H.; Nan, K.; Jin, Y.; Sun, L.; Wang, B. Recent Developments in Smart Antibacterial Surfaces to Inhibit Biofilm Formation and Bacterial Infections. J. Mater. Chem. B 2018, 6, 4274–4292. [Google Scholar] [CrossRef]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Parages, M.L.; O Gara, F. Coumarin: A Novel Player in Microbial Quorum Sensing and Biofilm Formation Inhibition. Appl. Microbiol. Biotechnol. 2018, 102, 2063–2073. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Mansour, S.C.; de la Fuente-Núñez, C.; Hancock, R.E.W.; Fairfull-Smith, K.E. Synthesis and Evaluation of Ciprofloxacin-Nitroxide Conjugates as Anti-Biofilm Agents. Molecules 2016, 21, E841. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Speziale, P.; Montanaro, L.; Costerton, J.W. Biofilm Formation in Staphylococcus Implant Infections. A Review of Molecular Mechanisms and Implications for Biofilm-Resistant Materials. Biomaterials 2012, 33, 5967–5982. [Google Scholar] [CrossRef] [PubMed]
- Bazaka, K.; Jacob, M.V.; Crawford, R.J.; Ivanova, E.P. Efficient Surface Modification of Biomaterial to Prevent Biofilm Formation and the Attachment of Microorganisms. Appl. Microbiol. Biotechnol. 2012, 95, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Walz, J.M.; Avelar, R.L.; Longtine, K.J.; Carter, K.L.; Mermel, L.A.; Heard, S.O.; 5-FU Catheter Study Group. Anti-Infective External Coating of Central Venous Catheters: A Randomized, Noninferiority Trial Comparing 5-Fluorouracil with Chlorhexidine/Silver Sulfadiazine in Preventing Catheter Colonization. Crit. Care Med. 2010, 38, 2095–2102. [Google Scholar] [CrossRef] [PubMed]
- Van Delden, C.; Köhler, T.; Brunner-Ferber, F.; François, B.; Carlet, J.; Pechère, J.-C. Azithromycin to Prevent Pseudomonas aeruginosa Ventilator-Associated Pneumonia by Inhibition of Quorum Sensing: A Randomized Controlled Trial. Intensive Care Med. 2012, 38, 1118–1125. [Google Scholar] [CrossRef]
- Saiman, L.; Marshall, B.C.; Mayer-Hamblett, N.; Burns, J.L.; Quittner, A.L.; Cibene, D.A.; Coquillette, S.; Fieberg, A.Y.; Accurso, F.J.; Campbell, P.W.; et al. Azithromycin in Patients with Cystic Fibrosis Chronically Infected with Pseudomonas aeruginosa: A Randomized Controlled Trial. JAMA 2003, 290, 1749–1756. [Google Scholar] [CrossRef]
- Smyth, A.R.; Cifelli, P.M.; Ortori, C.A.; Righetti, K.; Lewis, S.; Erskine, P.; Holland, E.D.; Givskov, M.; Williams, P.; Cámara, M.; et al. Garlic as an Inhibitor of Pseudomonas aeruginosa Quorum Sensing in Cystic Fibrosis-A Pilot Randomized Controlled Trial. Pediatr. Pulmonol. 2010, 45, 356–362. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Agents That Inhibit Bacterial Biofilm Formation. Future Med. Chem. 2015, 7, 647–671. [Google Scholar] [CrossRef]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef]
- Lee, J.-H.; Regmi, S.C.; Kim, J.-A.; Cho, M.H.; Yun, H.; Lee, C.-S.; Lee, J. Apple Flavonoid Phloretin Inhibits Escherichia coli O157:H7 Biofilm Formation and Ameliorates Colon Inflammation in Rats. Infect. Immun. 2011, 79, 4819–4827. [Google Scholar] [CrossRef]
- Kelly, S.R.; Jensen, P.R.; Henkel, T.P.; Fenical, W.; Pawlik, J.R. Effects of Caribbean Sponge Extracts on Bacterial Attachment. Aquat. Microb. Ecol. 2003, 31, 175–182. [Google Scholar] [CrossRef]
- Brackman, G.; Celen, S.; Baruah, K.; Bossier, P.; van Calenbergh, S.; Nelis, H.J.; Coenye, T. AI-2 Quorum-Sensing Inhibitors Affect the Starvation Response and Reduce Virulence in Several Vibrio Species, Most Likely by Interfering with LuxPQ. Microbiol. Read. Engl. 2009, 155, 4114–4122. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; Wezeman, R.J.; Siegel, A.P.; Blackwell, H.E. Small Molecule Inhibitors of Bacterial Quorum Sensing and Biofilm Formation. J. Am. Chem. Soc. 2005, 127, 12762–12763. [Google Scholar] [CrossRef]
- Kim, B.; Park, J.-S.; Choi, H.-Y.; Yoon, S.S.; Kim, W.-G. Terrein Is an Inhibitor of Quorum Sensing and C-Di-GMP in Pseudomonas aeruginosa: A Connection between Quorum Sensing and c-Di-GMP. Sci. Rep. 2018, 8, 8617. [Google Scholar] [CrossRef]
- Costas, C.; López-Puente, V.; Bodelón, G.; González-Bello, C.; Pérez-Juste, J.; Pastoriza-Santos, I.; Liz-Marzán, L.M. Using Surface Enhanced Raman Scattering to Analyze the Interactions of Protein Receptors with Bacterial Quorum Sensing Modulators. ACS Nano 2015, 9, 5567–5576. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E.; Greenberg, E.P. Acyl Homoserine-Lactone Quorum-Sensing Signal Generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.-H. MomL, a Novel Marine-Derived N-Acyl Homoserine Lactonase from Muricauda Olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef]
- Migiyama, Y.; Kaneko, Y.; Yanagihara, K.; Morohoshi, T.; Morinaga, Y.; Nakamura, S.; Miyazaki, T.; Hasegawa, H.; Izumikawa, K.; Kakeya, H.; et al. Efficacy of AiiM, an N-Acylhomoserine Lactonase, against Pseudomonas aeruginosa in a Mouse Model of Acute Pneumonia. Antimicrob. Agents Chemother. 2013, 57, 3653–3658. [Google Scholar] [CrossRef]
- Dong, Y.-H.; Xu, J.-L.; Li, X.-Z.; Zhang, L.-H. AiiA, an Enzyme That Inactivates the Acylhomoserine Lactone Quorum-Sensing Signal and Attenuates the Virulence of Erwinia Carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef]
- Chun, C.K.; Ozer, E.A.; Welsh, M.J.; Zabner, J.; Greenberg, E.P. Inactivation of a Pseudomonas aeruginosa Quorum-Sensing Signal by Human Airway Epithelia. Proc. Natl. Acad. Sci. USA 2004, 101, 3587–3590. [Google Scholar] [CrossRef]
- Yang, F.; Wang, L.-H.; Wang, J.; Dong, Y.-H.; Hu, J.Y.; Zhang, L.-H. Quorum Quenching Enzyme Activity Is Widely Conserved in the Sera of Mammalian Species. FEBS Lett. 2005, 579, 3713–3717. [Google Scholar] [CrossRef]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant Role of Paraoxonases in Inactivation of the Pseudomonas aeruginosa Quorum-Sensing Signal N-(3-Oxododecanoyl)-l-Homoserine Lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; O’Neill, J.C.; Miller, D.M.; Wezeman, R.J.; Mattmann, M.E.; Lin, Q.; Blackwell, H.E. Comparative Analyses of N-Acylated Homoserine Lactones Reveal Unique Structural Features That Dictate Their Ability to Activate or Inhibit Quorum Sensing. ChemBioChem 2008, 9, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Gerdt, J.P.; Wittenwyler, D.M.; Combs, J.B.; Boursier, M.E.; Brummond, J.W.; Xu, H.; Blackwell, H.E. Chemical Interrogation of LuxR-Type Quorum Sensing Receptors Reveals New Insights into Receptor Selectivity and the Potential for Interspecies Bacterial Signaling. ACS Chem. Biol. 2017, 12, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.; Yuan, M.; Jakobsen, T.H.; Mortensen, K.T.; Delos Santos, M.M.S.; Chua, S.L.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Disulfide Bond-Containing Ajoene Analogues as Novel Quorum Sensing Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2017, 60, 215–227. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Bragason, S.K.; Phipps, R.K.; Christensen, L.D.; van Gennip, M.; Alhede, M.; Skindersoe, M.; Larsen, T.O.; Høiby, N.; Bjarnsholt, T.; et al. Food as a Source for Quorum Sensing Inhibitors: Iberin from Horseradish Revealed as a Quorum Sensing Inhibitor of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 2410–2421. [Google Scholar] [CrossRef]
- Ganin, H.; Rayo, J.; Amara, N.; Levy, N.; Krief, P.; Meijler, M.M. Sulforaphane and Erucin, Natural Isothiocyanates from Broccoli, Inhibit Bacterial Quorum Sensing. MedChemComm 2012, 4, 175–179. [Google Scholar] [CrossRef]
- Ren, D.; Zuo, R.; González Barrios, A.F.; Bedzyk, L.A.; Eldridge, G.R.; Pasmore, M.E.; Wood, T.K. Differential Gene Expression for Investigation of Escherichia coli Biofilm Inhibition by Plant Extract Ursolic Acid. Appl. Environ. Microbiol. 2005, 71, 4022–4034. [Google Scholar] [CrossRef]
- Musk, D.J.; Banko, D.A.; Hergenrother, P.J. Iron Salts Perturb Biofilm Formation and Disrupt Existing Biofilms of Pseudomonas aeruginosa. Chem. Biol. 2005, 12, 789–796. [Google Scholar] [CrossRef]
- Junker, L.M.; Clardy, J. High-Throughput Screens for Small-Molecule Inhibitors of Pseudomonas aeruginosa Biofilm Development. Antimicrob. Agents Chemother. 2007, 51, 3582–3590. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, T.; Wang, K.; Hou, C.; Cai, S.; Huang, Y.; Du, Z.; Huang, H.; Kong, J.; Chen, Y. Baicalein Inhibits Staphylococcus aureus Biofilm Formation and the Quorum Sensing System In Vitro. PLoS ONE 2016, 11, e0153468. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Gokhale, A.A.; Lao, W.; Parashar, V.; Neiditch, M.B.; Semmelhack, M.F.; Lee, I.; Waters, C.M. Identification of a Novel Benzimidazole That Inhibits Bacterial Biofilm Formation in a Broad-Spectrum Manner. Antimicrob. Agents Chemother. 2011, 55, 4369–4378. [Google Scholar] [CrossRef] [PubMed]
- Wenderska, I.B.; Chong, M.; McNulty, J.; Wright, G.D.; Burrows, L.L. Palmitoyl-Dl-Carnitine Is a Multitarget Inhibitor of Pseudomonas aeruginosa Biofilm Development. ChemBioChem 2011, 12, 2759–2766. [Google Scholar] [CrossRef]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-Aided Identification of Recognized Drugs as Pseudomonas aeruginosa Quorum-Sensing Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Persson, T.; Hansen, T.H.; Rasmussen, T.B.; Skindersø, M.E.; Givskov, M.; Nielsen, J. Rational Design and Synthesis of New Quorum-Sensing Inhibitors Derived from Acylated Homoserine Lactones and Natural Products from Garlic. Org. Biomol. Chem. 2005, 3, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of Quorum Sensing in Pseudomonas aeruginosa Biofilm Bacteria by a Halogenated Furanone Compound. Microbiology 2022, 148, 87–102. [Google Scholar] [CrossRef]
- Yamada, A.; Kitamura, H.; Yamaguchi, K.; Fukuzawa, S.; Kamijima, C.; Yazawa, K.; Kuramoto, M.; Wang, G.-Y.-S.; Fujitani, Y.; Uemura, D. Development of Chemical Substances Regulating Biofilm Formation. Bull. Chem. Soc. Jpn. 1997, 70, 3061–3069. [Google Scholar] [CrossRef]
- Kosikowska, U.; Malm, A.; Pitucha, M.; Rajtar, B.; Polz-Dacewicz, M. Inhibitory Effect of N-Ethyl-3-Amino-5-Oxo-4-Phenyl-2,5-Dihydro-1H-Pyrazole-1-Carbothioamide on Haemophilus Spp. Planktonic or Biofilm-Forming Cells. Med. Chem. Res. 2014, 23, 1057–1066. [Google Scholar] [CrossRef]
- Slusarenko, A.J.; Patel, A.; Portz, D. Control of Plant Diseases by Natural Products: Allicin from Garlic as a Case Study. In Sustainable Disease Management in a European Context; Collinge, D.B., Munk, L., Cooke, B.M., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 313–322. ISBN 978-1-4020-8780-6. [Google Scholar]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa Virulence by Quorum Sensing Inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Han, Y.; Hou, S.; Simon, K.A.; Ren, D.; Luk, Y.-Y. Identifying the Important Structural Elements of Brominated Furanones for Inhibiting Biofilm Formation by Escherichia coli. Bioorg. Med. Chem. Lett. 2008, 18, 1006–1010. [Google Scholar] [CrossRef]
- Iii, R.W.H.; Ma, L.; Gambino, C.; Moeller, P.D.R.; Basso, A.; Cavanagh, J.; Wozniak, D.J.; Melander, C. Control of Bacterial Biofilms with Marine Alkaloid Derivatives. Mol. Biosyst. 2008, 4, 614–621. [Google Scholar] [CrossRef]
- Peach, K.C.; Bray, W.M.; Shikuma, N.J.; Gassner, N.C.; Lokey, R.S.; Yildiz, F.H.; Linington, R.G. An Image-Based 384-Well High-Throughput Screening Method for the Discovery of Biofilm Inhibitors in Vibrio Cholerae. Mol. Biosyst. 2011, 7, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamoorthy, K.; Luo, C.; Pattabiraman, N.; Feng, X.; Koestler, B.; Waters, C.M.; Palys, T.J. Identification of Small Molecules Inhibiting Diguanylate Cyclases to Control Bacterial Biofilm Development. Biofouling 2014, 30, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.K.; Klinkenberg, L.G.; Vazquez, M.J.; Segura-Carro, D.; Colmenarejo, G.; Ramon, F.; Rodriguez-Miquel, B.; Mata-Cantero, L.; Francisco, E.P.D.; Chuang, Y.M.; et al. Inhibiting the Stringent Response Blocks Mycobacterium Tuberculosis Entry into Quiescence and Reduces Persistence. Sci. Adv. 2019, 5, eaav2104. [Google Scholar] [CrossRef] [PubMed]
- Antoniani, D.; Rossi, E.; Rinaldo, S.; Bocci, P.; Lolicato, M.; Paiardini, A.; Raffaelli, N.; Cutruzzolà, F.; Landini, P. The Immunosuppressive Drug Azathioprine Inhibits Biosynthesis of the Bacterial Signal Molecule Cyclic-Di-GMP by Interfering with Intracellular Nucleotide Pool Availability. Appl. Microbiol. Biotechnol. 2013, 97, 7325–7336. [Google Scholar] [CrossRef]
- Antoniani, D.; Bocci, P.; Maciąg, A.; Raffaelli, N.; Landini, P. Monitoring of Diguanylate Cyclase Activity and of Cyclic-Di-GMP Biosynthesis by Whole-Cell Assays Suitable for High-Throughput Screening of Biofilm Inhibitors. Appl. Microbiol. Biotechnol. 2010, 85, 1095–1104. [Google Scholar] [CrossRef]
- Fernicola, S.; Paiardini, A.; Giardina, G.; Rampioni, G.; Leoni, L.; Cutruzzolà, F.; Rinaldo, S. In Silico Discovery and In Vitro Validation of Catechol-Containing Sulfonohydrazide Compounds as Potent Inhibitors of the Diguanylate Cyclase PleD. J. Bacteriol. 2016, 198, 147–156. [Google Scholar] [CrossRef]
- Lieberman, O.J.; Orr, M.W.; Wang, Y.; Lee, V.T. High-Throughput Screening Using the Differential Radial Capillary Action of Ligand Assay Identifies Ebselen as an Inhibitor of Diguanylate Cyclases. ACS Chem. Biol. 2014, 9, 183–192. [Google Scholar] [CrossRef]
- Zheng, Y.; Tsuji, G.; Opoku-Temeng, C.; Sintim, H.O. Inhibition of P. Aeruginosa c-Di-GMP Phosphodiesterase RocR and Swarming Motility by a Benzoisothiazolinone Derivative Electronic Supplementary Information (ESI) Available: All Experimental, Spectroscopic Details and Supplemental Figures. Chem. Sci. 2016, 7, 6238–6244. [Google Scholar] [CrossRef]
- Zhou, E.; Seminara, A.B.; Kim, S.-K.; Hall, C.L.; Wang, Y.; Lee, V.T. Thiol-Benzo-Triazolo-Quinazolinone Inhibits Alg44 Binding to c-Di-GMP and Reduces Alginate Production by Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 3076–3085. [Google Scholar] [CrossRef]
- Barraud, N.; Hassett, D.J.; Hwang, S.-H.; Rice, S.A.; Kjelleberg, S.; Webb, J.S. Involvement of Nitric Oxide in Biofilm Dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7344–7353. [Google Scholar] [CrossRef]
- Huigens, R.W.; Richards, J.J.; Parise, G.; Ballard, T.E.; Zeng, W.; Deora, R.; Melander, C. Inhibition of Pseudomonas aeruginosa Biofilm Formation with Bromoageliferin Analogues. J. Am. Chem. Soc. 2007, 129, 6966–6967. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Pinkner, J.S.; Ford, B.; Obermann, R.; Nolan, W.; Wildman, S.A.; Hobbs, D.; Ellenberger, T.; Cusumano, C.K.; Hultgren, S.J.; et al. Structure-Based Drug Design and Optimization of Mannoside Bacterial FimH Antagonists. J. Med. Chem. 2010, 53, 4779–4792. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Sangwan, P.L.; Koul, S.; Pandey, A.; Bani, S.; Abdullah, S.T.; Sharma, P.R.; Kitchlu, S.; Khan, I.A. 4-Epi-Pimaric Acid: A Phytomolecule as a Potent Antibacterial and Anti-Biofilm Agent for Oral Cavity Pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Chorell, E.; Pinkner, J.S.; Phan, G.; Edvinsson, S.; Buelens, F.; Remaut, H.; Waksman, G.; Hultgren, S.J.; Almqvist, F. Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones; Pilicides with Increased Antivirulence Activity. J. Med. Chem. 2010, 53, 5690–5695. [Google Scholar] [CrossRef] [PubMed]
- Qvortrup, K.; Hultqvist, L.D.; Nilsson, M.; Jakobsen, T.H.; Jansen, C.U.; Uhd, J.; Andersen, J.B.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Small Molecule Anti-Biofilm Agents Developed on the Basis of Mechanistic Understanding of Biofilm Formation. Front. Chem. 2019, 7, 742. [Google Scholar] [CrossRef] [PubMed]
- Svensson, A.; Larsson, A.; Emtenäs, H.; Hedenström, M.; Fex, T.; Hultgren, S.J.; Pinkner, J.S.; Almqvist, F.; Kihlberg, J. Design and Evaluation of Pilicides: Potential Novel Antibacterial Agents Directed Against Uropathogenic Escherichia coli. ChemBioChem 2001, 2, 915–918. [Google Scholar] [CrossRef]
- Lee, Y.M.; Almqvist, F.; Hultgren, S.J. Targeting Virulence for Antimicrobial Chemotherapy. Curr. Opin. Pharmacol. 2003, 3, 513–519. [Google Scholar] [CrossRef]
- Pemberton, N.; Pinkner, J.S.; Edvinsson, S.; Hultgren, S.J.; Almqvist, F. Synthesis and Evaluation of Dihydroimidazolo and Dihydrooxazolo Ring-Fused 2-Pyridones—Targeting Pilus Biogenesis in Uropathogenic Bacteria. Tetrahedron 2008, 64, 9368–9376. [Google Scholar] [CrossRef]
- Åberg, V.; Das, P.; Chorell, E.; Hedenström, M.; Pinkner, J.S.; Hultgren, S.J.; Almqvist, F. Carboxylic Acid Isosteres Improve the Activity of Ring-Fused 2-Pyridones That Inhibit Pilus Biogenesis in E. Coli. Bioorg. Med. Chem. Lett. 2008, 18, 3536–3540. [Google Scholar] [CrossRef]
- Rasmussen, L.; White, E.L.; Pathak, A.; Ayala, J.C.; Wang, H.; Wu, J.-H.; Benitez, J.A.; Silva, A.J. A High-Throughput Screening Assay for Inhibitors of Bacterial Motility Identifies a Novel Inhibitor of the Na+-Driven Flagellar Motor and Virulence Gene Expression in Vibrio Cholerae. Antimicrob. Agents Chemother. 2011, 55, 4134–4143. [Google Scholar] [CrossRef]
- Quave, C.L.; Estévez-Carmona, M.; Compadre, C.M.; Hobby, G.; Hendrickson, H.; Beenken, K.E.; Smeltzer, M.S. Ellagic Acid Derivatives from Rubus Ulmifolius Inhibit Staphylococcus aureus Biofilm Formation and Improve Response to Antibiotics. PLoS ONE 2012, 7, e28737. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Jayaprakasha, G.K.; Jesudhasan, P.R.; Pillai, S.D.; Patil, B.S. Suppression of Bacterial Cell–Cell Signalling, Biofilm Formation and Type III Secretion System by Citrus Flavonoids. J. Appl. Microbiol. 2010, 109, 515–527. [Google Scholar] [CrossRef]
- Luppens, S.B.I.; Reij, M.W.; van der Heijden, R.W.L.; Rombouts, F.M.; Abee, T. Development of a Standard Test to Assess the Resistance of Staphylococcus aureus Biofilm Cells to Disinfectants. Appl. Environ. Microbiol. 2002, 68, 4194–4200. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; William Costerton, J. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should We Stay or Should We Go: Mechanisms and Ecological Consequences for Biofilm Dispersal. Nat. Rev. Microbiol. 2012, 10, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for Combating Bacterial Biofilms: A Focus on Anti-Biofilm Agents and Their Mechanisms of Action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Guilhen, C.; Forestier, C.; Balestrino, D. Biofilm Dispersal: Multiple Elaborate Strategies for Dissemination of Bacteria with Unique Properties. Mol. Microbiol. 2017, 105, 188–210. [Google Scholar] [CrossRef]
- Reffuveille, F.; de la Fuente-Núñez, C.; Fairfull-Smith, K.E.; Hancock, R.E.W. Potentiation of Ciprofloxacin Action against Gram-Negative Bacterial Biofilms by a Nitroxide. Pathog. Dis. 2015, 73, ftv016. [Google Scholar] [CrossRef]
- Marvasi, M.; Chen, C.; Carrazana, M.; Durie, I.A.; Teplitski, M. Systematic Analysis of the Ability of Nitric Oxide Donors to Dislodge Biofilms Formed by Salmonella Enterica and Escherichia coli O157:H7. AMB Express 2014, 4, 42. [Google Scholar] [CrossRef]
- Roizman, D.; Vidaillac, C.; Givskov, M.; Yang, L. In Vitro Evaluation of Biofilm Dispersal as a Therapeutic Strategy to Restore Antimicrobial Efficacy. Antimicrob. Agents Chemother. 2017, 61, e01088-17. [Google Scholar] [CrossRef]
- Flemming, K.; Klingenberg, C.; Cavanagh, J.P.; Sletteng, M.; Stensen, W.; Svendsen, J.S.; Flægstad, T. High in Vitro Antimicrobial Activity of Synthetic Antimicrobial Peptidomimetics against Staphylococcal Biofilms. J. Antimicrob. Chemother. 2009, 63, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, S.A.; Brown, M.H. Antimicrobial Peptides–Promising Alternatives to Conventional Antibiotics. Microb. Physiol. 2011, 20, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Frederick, H.; Sarah, R.D.; David, A.P. Anionic Antimicrobial Peptides from Eukaryotic Organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar]
- Götz, F. Staphylococcus and Biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef]
- Tan, X.; Qin, N.; Wu, C.; Sheng, J.; Yang, R.; Zheng, B.; Ma, Z.; Liu, L.; Peng, X.; Jia, A. Transcriptome Analysis of the Biofilm Formed by Methicillin-Susceptible Staphylococcus aureus. Sci. Rep. 2015, 5, 11997. [Google Scholar] [CrossRef]
- Liu, W.; Yin, D.; Li, N.; Hou, X.; Wang, D.; Li, D.; Liu, J. Influence of Environmental Factors on the Active Substance Production and Antioxidant Activity in Potentilla Fruticosa L. and Its Quality Assessment. Sci. Rep. 2016, 6, 28591. [Google Scholar] [CrossRef]
- Cain, A.K.; Barquist, L.; Goodman, A.L.; Paulsen, I.T.; Parkhill, J.; van Opijnen, T. A Decade of Advances in Transposon-Insertion Sequencing. Nat. Rev. Genet. 2020, 21, 526–540. [Google Scholar] [CrossRef]
- Herschend, J.; Damholt, Z.B.V.; Marquard, A.M.; Svensson, B.; Sørensen, S.J.; Hägglund, P.; Burmølle, M. A Meta-Proteomics Approach to Study the Interspecies Interactions Affecting Microbial Biofilm Development in a Model Community. Sci. Rep. 2017, 7, 16483. [Google Scholar] [CrossRef]
- Beale, D.J.; Barratt, R.; Marlow, D.R.; Dunn, M.S.; Palombo, E.A.; Morrison, P.D.; Key, C. Application of Metabolomics to Understanding Biofilms in Water Distribution Systems: A Pilot Study. Biofouling 2013, 29, 283–294. [Google Scholar] [CrossRef]
- Yeom, J.; Shin, J.-H.; Yang, J.-Y.; Kim, J.; Hwang, G.-S. 1H NMR-Based Metabolite Profiling of Planktonic and Biofilm Cells in Acinetobacter Baumannii 1656-2. PLoS ONE 2013, 8, e57730. [Google Scholar] [CrossRef]
- Hasan, S.; Danishuddin, M.; Khan, A.U. Inhibitory Effect of Zingiber Officinale towards Streptococcus Mutans Virulence and Caries Development: In Vitro and in Vivo Studies. BMC Microbiol. 2015, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. Biofilm-Specific Antibiotic Tolerance and Resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef]
- Abozed, M.A. Chitosan, Chitosan Nanoparticles, and Chlorhexidine Gluconate, as Intra Canal Medicaments in Primary Teeth: An In-Vivo Study. 2018. Available online: clinicaltrials.gov (accessed on 8 July 2022).
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. npj Biofilms Microbiom. 2016, 2, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ridenhour, B.J.; Metzger, G.A.; France, M.; Gliniewicz, K.; Millstein, J.; Forney, L.J.; Top, E.M. Persistence of Antibiotic Resistance Plasmids in Bacterial Biofilms. Evol. Appl. 2017, 10, 640–647. [Google Scholar] [CrossRef]
- Rybak, M.J.; McGrath, B.J. Combination Antimicrobial Therapy for Bacterial Infections. Drugs 1996, 52, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Cosgrove, S.E.; Maragakis, L.L. Combination Therapy for Treatment of Infections with Gram-Negative Bacteria. Clin. Microbiol. Rev. 2012, 25, 450–470. [Google Scholar] [CrossRef]
- Rao, L.; Sheng, Y.; Zhang, J.; Xu, Y.; Yu, J.; Wang, B.; Zhao, H.; Wang, X.; Guo, Y.; Wu, X.; et al. Small-Molecule Compound SYG-180-2-2 to Effectively Prevent the Biofilm Formation of Methicillin-Resistant Staphylococcus aureus. Front. Microbiol. 2022, 12, 657. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).