Abstract
With the wide spread of multidrug-resistant bacteria, a variety of aminoglycosides have been used in clinical practice as one of the effective options for antimicrobial combinations. However, in recent years, the emergence of high-level resistance against pan-aminoglycosides has worsened the status of antimicrobial resistance, so the production of 16S rRNA methyltransferase (16S-RMTase) should not be ignored as one of the most important resistance mechanisms. What is more, on account of transferable plasmids, the horizontal transfer of resistance genes between pathogens becomes easier and more widespread, which brings challenges to the treatment of infectious diseases and infection control of drug-resistant bacteria. In this review, we will make a presentation on the prevalence and genetic environment of 16S-RMTase encoding genes that lead to high-level resistance to aminoglycosides.
1. Introduction
Aminoglycoside antibiotics were discovered and isolated from soil Actinobacteria in the 1940s, and streptomycin was the first aminoglycoside antibiotic used in the clinical treatment of tuberculosis and Gram-negative bacteria infection [1]. The mechanism of aminoglycosides is to bind the A site of 16S rRNA, which consists of the 30S ribosomal subunit, leading to the inhibition of protein synthesis and bacteria death [2]. Therefore, aminoglycosides have a broad spectrum of antimicrobial activity against both Gram-positive and Gram-negative bacteria and are often used in combination with β-lactam antibiotics, especially the third-generation cephalosporins [3].
Since the 1980s, due to the side effects of ototoxicity and nephrotoxicity, aminoglycosides have been used less and less in clinics and gradually replaced by β-lactams and quinolones, which are less toxic and have a wider antimicrobial spectrum [4]. However, with the increase of β-lactams and quinolone antimicrobial resistance and the prevalence of multidrug-resistant bacteria, the retained potency of aminoglycoside antibiotics has renewed interest in their use in clinical practice [5]. Additionally, aminoglycosides can be used as one of the effective antimicrobial combinations for the treatment of life-threatening infections caused by multidrug-resistant bacteria. Because aminoglycosides can be used in combination with other antibiotics, the toxicity can be decreased by adjusting the dosage [6]. However, in recent years, reports about high-level resistance to aminoglycosides have increased and spread widely around the world. In this review, we will make a presentation on the prevalence and genetic environment of 16S-RMTases, which lead to high-level resistance to pan-aminoglycosides.
2. Antimicrobial Resistance Mechanism of Aminoglycosides
According to the difference in chemical structures, aminoglycosides can be divided into 4,5-disubstituted 2-deoxystreptamine (DOS), such as neomycin and paromomycin; 4,6-disubstituted 2-DOS, such as gentamicin, amikacin, kanamycin, arbekacin, and tobramycin; monosubstituted DOS, such as apramycin; and no DOS ring, such as streptomycin [1,4]. The mechanisms of aminoglycosides resistance include: (1) modification or inactivation of aminoglycosides modifying enzymes; (2) increased expression of efflux pump; (3) decreased drug permeability; and (4) modification of the drug target, such as generating 16S-RMTases that interfere with binding of the aminoglycosides [7]. Numerous studies have shown that aminoglycosides modifying enzymes and 16S-RMTases can lead to high-level resistance to aminoglycosides in bacteria. Aminoglycosides modifying enzymes are the most common mechanism of aminoglycosides resistance in bacteria, consisting of aminoglycoside acetyltransferases, aminoglycoside phosphotransferases, and aminoglycoside nucleotidyltransferases [5]. The above three aminoglycoside modification enzymes can catalyze the modification of different OH and NH2 groups of 2-DOS molecules. Because the encoding genes of aminoglycoside modifying enzymes often occur with mutations, many subclasses of the enzymes are produced, making more aminoglycosides become the substrates of the modifying enzymes and increasing the antimicrobial resistance [8].
Although aminoglycoside modification enzymes are the most common resistance mechanism, due to the specificity of substrates, they cannot mediate high-level resistance to multiple aminoglycosides [9]. However, another mechanism of bacterial resistance to aminoglycosides, modification of drug target, leads to high-level resistance against pan-aminoglycoside, with minimal inhibitory concentration (MIC) > 256 μg/mL. There are two ways to modify the drug target: post-transcriptional methylation of 16S rRNA to block aminoglycosides binding to the target [7]; gene point mutation (nucleotide substitution), for example, Mycobacterium tuberculosis rpsL gene encodes S12 protein to mediate streptomycin resistance [10].
3. Plasmid-Mediated 16S rRNA Methylase Resistance Gene and Its Transfer Mechanism
Acquired 16S rRNA methylase is the most clinically significant aminoglycoside resistance mechanism. The concrete resistance mechanism is that with the catalysis of 16S-RMTase, adding a CH3 group provided by S-adenosine methionine (SAM) to specific residues at the A site of 16S rRNA, the binding ability of methylated 16S rRNA to aminoglycosides is significantly reduced, resulting in extensive and high-level resistance to various aminoglycosides [4]. The post-transcriptional modification of 16S rRNA is intrinsic to Actinomycetes (e.g., Streptomyces and Micromonospora) species that produce aminoglycosides to protect themselves from the damage of endogenous aminoglycosides [11,12].
Since the first acquired 16S-RMTase gene was identified in 2003, new genotypes and subtypes have emerged continuously, mediating high-level resistance to a variety of aminoglycosides. So far, a total of eleven acquired 16S-RMTases have been identified and reported, which include ArmA, NpmA, NpmB, and RmtA through RmtH [4,12]. In order to figure out the evolutionary relationships of 16S-RMTases encoding genes and their subtypes, a phylogenetic tree is reconstructed and shown in Figure 1. About the enzymatic function of 16S-RMTases, different types of methyltransferases have different action sites of 16S rRNA: ArmA, RmtA through RmtH methylate N7-G1405 of 16S rRNA and confer high-level resistance to 4,6-disubstituted 2-DOS; NpmA and NpmB methylate N1-A1408 of 16S rRNA and are resistant to 4,5-disubstituted 2-DOS, 4,6-disubstituted 2-DOS, and monosubstituted DOS but only susceptible to streptomycin [1,12,13]. Although the action site is kind of different, the whole types of methyltransferases are able to mediate high-level resistance to pan-aminoglycosides.
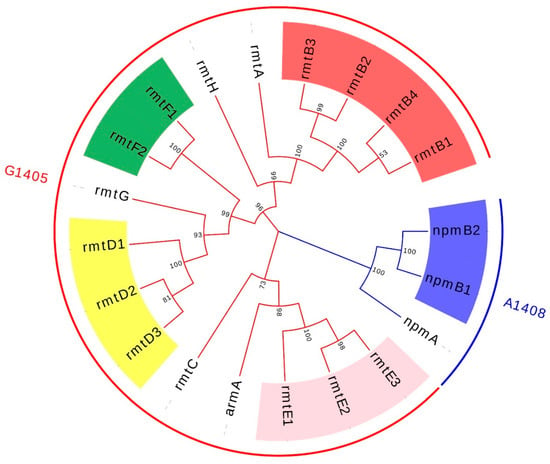
Figure 1.
The phylogenetic reconstruction of 16S-RMTases encoding genes and their subtypes. The colors of the branch and external circle are typed by the action sites of 16S-RMTase. The subtypes of rmtB, rmtD, rmtE, rmtF, and npmA are highlighted with red, yellow, pink, green, and blue. GenBank: rmtA, NG_048057.1; rmtB1, NG_048058.1; rmtB2, NG_048059.1; rmtB3, NG_051535.1; rmtB4, NG_051536.1; rmtC, NG_048060.1; rmtD1, NG_048061.1; rmtD2, NG_050557.1; rmtD3, LC229698.1; rmtE1, NG_050558.1; rmtE2, NG_050559.1; rmtE3, MH572011.1; rmtF1, NG_048062.1; rmtF2, NG_051537.1; rmtG, NG_048064.1; rmtH, NG_048065.1; npmA, NG_048018.1; npmB1, NG_077965.1; npmB2, NG_077964.1.
Except for the high-level antimicrobial resistance, the 16S-RMTase encoding genes found so far are mostly located within transferable plasmid and/or associated with mobile genetic elements such as transposons, integrons, and insertion sequence [14]. Furthermore, with the support of plasmid and mobile genetic elements, 16S-RMTase encoding genes are often associated with other resistance genes, leading multi-drug resistance even pan-drug resistance. Because of the high-level antimicrobial resistance and the mobility conferred by plasmids as well as other mobile genetic elements, we need pay more attention to the pan-aminoglycoside resistance caused by 16S-RMTase. Here are detailed descriptions of each 16S-RMTase encoding gene and its transfer mechanism; epidemic strains carrying 16S-RMTase encoding genes and their distribution are shown in Table 1, and the genetic context of the 16S-RMTase encoding genes are shown in Figure 2.

Table 1.
The strains with 16S rRNA methylase resistance gene and their distribution.
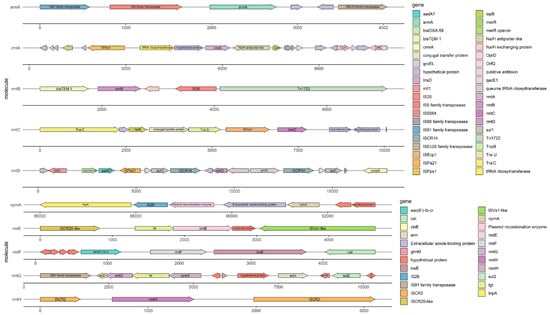
Figure 2.
The sequence of the 16S rRNA methylase resistance gene and its genetic environment. GenBank: rmtA, AB083212.2; rmtB, FJ556899.1; rmtC, AB194779.2; rmtD, DQ914960.2; rmtE, KT428293.1; rmtF, JQ808129.1; rmtG, VLNW01000106.1; rmtH, KC544262.1; armA, AY220558.1; npmA, AB261016.2.
3.1. ArmA
The 16S-RMTase gene armA was first identified on a transferable plasmid of K. pneumoniae in French [107]. Subsequently, armA has been widely spread throughout the world, primarily found in A. baumanii, P. rettgeri, and K. pneumoniae, especially prevalent in A. baumanii. What is more, armA often came along with the rmtB gene and led to high-level resistance to aminoglycosides with MIC ≥ 256 μg/mL [108,109,110,111]. Research studies showed that armA was often located on transferable plasmids belonging to different Inc types [19], and insertion sequences such as IS26 might be involved in the mobilization of resistant genes around pathogens. In addition, these 16S-RMTases producers also showed resistance to β-lactams and carbapenems through various antimicrobial resistance genetic determinants, such as blaTEM-1, blaCTX-M, blaNDM, and other resistance genes [112,113]. The example of armA and its genetic context is shown in Figure 2.
3.2. RmtA
The 16S-RMTase gene rmtA was first identified in P. aeruginosa isolated from Japan in 2003 [9]. Then, rmtA gene was also detected among P. aeruginosa, E. cloacae, and K. pneumoniae, which were isolated from Korea, China, and Switzerland, mediating high-level resistance to aminoglycosides with MIC ≥ 512 μg/mL [9,29,31,114]. Further research studies showed that the type of plasmids carrying rmtA was usually IncA/C [29]. According to the genetic environment around the rmtA, it was speculated that rmtA was located on mercury-resistant transposons Tn5041 and mobile genetic elements, such as κ-λ elements and IS6100, flank the rmtA gene and mediate horizontal transfer and homologous recombination between strains [114]. In Figure 2, you can acquire more details of the rmtA and its genetic context which is shown as an example. Several studies had also confirmed that the rmtA gene was usually associated with blaCTX-M-15, blaNDM-1, blaSHV, blaTEM-1, and other resistant genes, which largely limits the treatment of multidrug-resistant bacteria [9,114,115].
3.3. RmtB
The 16S-RMTase gene rmtB was first detected in S. marcescens isolated from French in 2004 [116]. rmtB is primarily found in E. coli and K. pneumoniae all over the world and results in resistance to aminoglycosides with MIC ≥ 512 μg/mL [7,51,54,117]. In China, rmtB was the main prevalent genotype of 16S-RMTase and was often combined with armA. Recently, with the development of molecular diagnostic techniques, several alleles of rmtB have been identified: rmtB2 was found in K. pneumoniae and P. rettgeri [42,103]; rmtB3 and rmtB4 were found in P. aeruginosa [92]. Compared with the sequence of rmtB1 (GenBank: NG_048058.1), rmtB2 (GenBank: NG_048059.1) showed 96.6% nucleotide identity (26 nucleotides of difference), rmtB3 (GenBank: NG_051535.1) showed 99.6% nucleotide identity (3 nucleotides of difference), and rmtB4 (GenBank: NG_051536.1) showed 97.0% nucleotide identity (23 nucleotides of difference). Compared with RmtB1, the amino acid sequence of RmtB2 had 6 amino acid substitutions: Ala41Thr, Val124Ile, Val132Ile, Thr166Ile, Ile194Leu, and Thr229Ala; RmtB3 had 1 amino acid substitution: Ala82Val; RmtB4 had 4 amino acid substitutions: Val124Ile, Val132Ile, Thr166Ala, and Ile194Leu. Further studies of its genetic environment show that the rmtB gene is usually located on transposons of transferable plasmids with insertion sequences such as ISCR1, ISCfr1, and IS26, leading to the transfer and transmission of rmtB much easier among different strains [118,119]. A typical example of rmtB and its genetic environment is displayed in Figure 2.
3.4. RmtC
The 16S-RMTase gene rmtC was firstly detected in P. mirabilis isolated from Japan in 2004 [63]. Since then, rmtC has been spread widely around the world and distributed in various species, such as K. pneumoniae, E. coli, P. aeruginosa, and A. baumanii [24,60,69], resulting in high-level resistance to aminoglycosides with MIC ≥ 1024 μg/mL [63]. According to the genetic sequences around the rmtC, there was an obvious pattern that ISEcp1 was located upstream of rmtC and played an important role in the expression of rmtC as well as the transfer among different Gram-negative strains [60,63]. This ISEcp1 element belonged to the IS1380 family which was located at the ends of rmtC and contains a transposase gene (tnpA) and provided a promoter activity for expression of the adjacent rmtC. This structure enabled the rmtC gene to be transposed onto another plasmid [120,121]. As you can see, in Figure 2, we show an instance of rmtC and its genetic environment to help you understand the above statement. Recently, RmtC methyltransferase has been reported in China and isolated from S. stanley and K. aerogenes respectively. Further studies showed that the multidrug resistance regions and their genetic environment of the two strains’ plasmid were homologous to some extent, associated with carbapenemase and β-lactamase resistance genes such as blaNDM-1 and blaCMY-6 to mediate multidrug resistance [68,73].
3.5. RmtD
The 16S-RMTase gene rmtD was first identified in P. aeruginosa isolated from Brazil in 2007, mainly distributed in South America (such as Brazil, Chile, Argentina, etc.), commonly found in P. aeruginosa and K. pneumoniae [77,78,79]. Consistent with the characteristics of 16S-RMTase, the strains carrying rmtD show a high level of resistance to aminoglycosides with MIC ≥ 256 μg/mL, even rmtD3 mediates MIC of aminoglycosides up to 1024 μg/mL [75,81].
What is more, several alleles of rmtD have continually been reported in recent years. rmtD2 was isolated from the plasmid of E. aerogenes and C. freundii in Argentina [79] and displayed 97.3% nucleotide identity (20 nucleotides of difference) and 96.4% amino acid identity (9 residues of difference) with RmtD1 [77,79]. The rmtD3 genes were found in two P. aeruginosa strains from Myanmar and Poland, respectively, and both of them were located on chromosomes [81,82]. Compared with RmtD3, there were 9 amino acid substitutions in RmtD and 4 amino acid substitutions in RmtD2 [81]. Recent studies have found that different gene subtypes have different transfer mechanisms. By the means of IS26-mediated recombinational events, rmtD with other genetic elements form complex transposons and may facilitate the future spread of the gene within Enterobacteriaceae [122]. rmtD2 was located on transposon Tn21, and sequence analysis showed that the antimicrobial resistance region of rmtD1 and rmtD2 is formed through transposition or homologous recombination by ISCR3 and ISCR14 [79]. When it comes to rmtD3 identified so far, it often resided in chromosomal mosaic regions, which comprise integrative and conjugative elements (ICEs) with variable cargo regions, carrying IS- or transposon-associated resistance genes [82]. Therefore, with the help of so many transfer elements, rmtD often occurred together with blaSPM-1, blaKPC-2, blaTEM-1, and blaCTX-M-2 [75,77,79]. We draw the Figure 2 to help you understand the genetic context of rmtD more comprehensively.
3.6. RmtE
The 16S-RMTase gene rmtE was first identified in cattle origin E. coli isolated from America in 2010 [84]. In 2014, a case of human infection by RmtE-producing E. coli was firstly reported in America and mediated resistance to aminoglycosides with MIC ≥ 256 μg/mL [85]. Obviously, the distribution of strains carrying rmtE was relatively simple and mainly exists in E. coli. The rmtE2 was identified in swine-origin E. coli isolated from China, which existed in the IncI1 plasmid. More details about rmtE2 were that a single base mutation of T→C is detected at nucleotide 20 of rmtE, which caused a replacement of Val (6) by Ala in the gene product [74]. rmtE was detected among A. baumanii from the UK. Compared with the sequences of rmtE1 and rmtE2 indicated that rmtE3 had two SNPs: one at nucleotide 20 (T→C, Val 7 Ala) and another at nucleotide 141 (T→A, Asn 47 Lys) [87].
According to the genetic environment of rmtE and its alleles, there were several mobile genetic elements that probably mediated the transfer of rmtE between plasmids or between plasmids and chromosomes. rmtE1 was identified on a blaCMY-2-carrying IncA/C plasmid called pYDC637 of E. coli in 2015. Within this unit, rmtE1 was bound by an ISCR20-like element and an IS1294-like insertion sequence [123]. Interestingly, in 2017, rmtE1 was identified on the chromosome of E. coli with a similar structure to the former. It had shown that the subunit containing rmtE1 and its surrounding insertion sequences were similar to that of pYDC637 [86]. As to the genetic environment of rmtE2, an ISCR20-like transposase was located upstream of rmtE2 and an ISVs1-like transposase was located downstream [74]. As reported, there is an ISVs1-like transposase located downstream of rmtE3 [87]. In comparing the genetic context of rmtE1 and rmtE2, ISCR20-like transposase was located upstream of rmtE1 and rmtE2. However, the transposase genes located downstream of the 16S RMTase genes were distinct. ISVs1-like transposase was located downstream of rmtE2 and rmtE3. A general instance of the genetic environment surrounding rmtE is shown in Figure 2. To sum up, whether insertion sequences or broad-host-range, self-conjugative plasmids all played an important role in the initial mobilization of rmtE and the recombination with other resistance genes, such as blaTEM-1, blaCMY-2, blaTEM-1 [15,86,123].
3.7. RmtF
The 16S-RMTase gene rmtF was first identified in K. pneumoniae isolated from French in 2011, mediating resistance to aminoglycosides with MIC ≥ 256 μg/mL [89]. Subsequently, rmtF has been spread widely around the world, especially in K. pneumoniae, as well as can be found in P. aeruginosa, E. coli, and C. freundii [56,59,60,89]. In 2017, a new rmtF variant, rmtF2, was identified in P. aeruginosa in Nepal mediating resistance to aminoglycosides with MIC up to 1024 μg/mL [92]. Analysis of its predicted amino acid sequence reveals a substitution (Lys65Glu) compared with the sequence of RmtF [92].
Through whole-genome sequencing and analysis of antimicrobial resistance gene sequences, it was found that ISCR5 was often located on both sides of rmtF and its allele rmtF2 whether on plasmids or chromosomes, which used to be called insE as shown Figure 2 [89,92]; In recent years, transposase family genes and insertion sequence elements such as Tn3, Tn1721, IS91, and IS6100 also have been found on both sides of rmtF or rmtF2 [62,95]. rmtF is usually located on various conjugative plasmids, which belong to broad-host-range incompatibility groups such as IncA/C, IncR, IncFII, and IncFIB [58,90,94,95]. With the help of transferable plasmids and insertion sequences, rmtF was often associated with β-lactam and carbapenem resistance genes such as blaNDM, blaCTX-M, blaOXA-232, and blaTEM-1, especially in K. pneumoniae belongs to several high-risk clones ST231, ST147 [90,93,124,125]. The identification of RmtF coresident in strains harboring ESBLs, acquired AmpC enzymes, the NDM-type carbapenemases, and fluoroquinolone-resistance mechanisms not only leads to the potential for coselection and maintenance of resistance by the use of other antibiotics but also seriously compromises the treatment of life-threatening infections caused by Gram-negative organisms [56].
3.8. RmtG
The 16S-RMTase gene rmtG was first identified in K. pneumoniae isolated from Brazil in 2011, conferring resistance to aminoglycosides with MIC ≥ 256 μg/mL [80]. At present, the rmtG gene was still mainly prevalent in South America, especially in K. pneumoniae isolated from Brazil. It also could be found in K. aerogenes, P. aeruginosa, and E. coli isolated in America, India, Switzerland [3,59,96,97,98,100,101]. However, no strain carrying rmtG has been found in China so far.
Further analyses of the mobile genetic elements around the rmtG gene showed that rmtG was frequently located on the Tn3 transposon of conjugative plasmids belonging to IncN, IncA/C types [80,96,102,126]. What is more, there was another rule that rmtG is part of an operon that includes genes related to rRNA and tRNA modification such as rsmH, tgt, and rsmL [101]. Around the multidrug resistance region, rmtG was flanked by ISCR2 and IS91-like elements, which were responsible for the mobilization of the array [101,102]. Furthermore, the association of rmtG with ISCR2 in a 2-fold tandem repeat suggested a gene amplification process [101]. An example of rmtG and its genetic context is shown in Figure 2. In addition, it was important to emphasize that the RmtG-producing strains such as K. pneumoniae were predominantly clonal complex 258 (CC258), which included sequence types such as ST11, ST258, ST437, and ST340 [35,80,96,97,127]. The coproduction of RmtG, ESBLs of the CTX-M type (eg., CTX-M-2, CTX-M-15, CTX-M-59) and carbapenemases (e.g., KPC-2) could further limit the treatment options for multidrug-resistant bacteria [35,96,101,127].
3.9. RmtH
The reports about rmtH were relatively bare. In 2013, the rmtH gene was first identified on a conjugative plasmid isolated from K. pneumoniae in Iraq, mediating high-level resistance to various aminoglycosides with MIC ≥ 256 μg/mL [103]. In 2017, a strain of K. pneumoniae carrying the rmtH gene isolated from Lebanon was reported and a complete sequence analysis was carried out. It was found that rmtH and blaSHV-12 were both located on the same IncFII plasmid, and integrated on the Tn6329 transposon, with IS26 and ISCR2 insertion sequences on both sides involved in the formation of the multidrug resistance region [103,104]. You can find out the surrounding genetic elements of rmtH in Figure 2.
3.10. NpmA
The aminoglycoside-resistance 16S-RMTases were functionally divided into two subfamilies that modify the ribosome at either the N7 position of 16S rRNA nucleotide G1405 (m7G1405) or the N1 position of A1408 (m1A1408). Although enzymes from both subfamilies were found in aminoglycoside-producing bacteria, the m7G1405 methyltransferases were far more clinically prevalent than their m1A1408 methyltransferase counterparts [128].
NpmA was clinically isolated from E. coli strain ARS3 in Japan in 2007 [13]. Due to the posttranslational methylation of the A site of 16S rRNA at position A1408, NpmA leads to pan-aminoglycoside resistance encompassing both 4,5- and 4,6-disubstituted 2-DOS aminoglycosides with MIC ≥ 256 μg/mL, including neomycin and apramycin [12]. So, apramycin resistance seemed to be a good indicator for the detection of an A1408 16S-RMTase producer [13]. Later, the allele of npmA was identified in C. difficile and named npmA2. The study suggested that hospital-acquired C. difficile might be a reservoir for uncommon antibiotic resistance determinants such as npmA [105].
In 2021, NpmB was discovered in the process of screening for NpmA-like enzymes in the NCBI sequence databases, whose sequences were identified in E. coli genomes registered from the United Kingdom. NpmB1 and NpmB2 consisted of 217 amino acids, and only one amino acid substitution was identified in their sequences at position 21 (arginine for NpmB1 and cysteine for NpmB2). NpmB1 possesses 40% amino acid identity with NpmA1 and conferred resistance to all clinically relevant aminoglycosides, including 4,5-DOS agents. Phylogenetic analysis of NpmB1 and NpmB2, its single-amino-acid variant, revealed that the encoding gene was likely acquired by E. coli from a soil bacterium. The structure of NpmB1 suggested that it required a structural change of the β6/7 linker to bind to 16S rRNA. These findings established NpmB1 and NpmB2 as the second group of acquired pan-aminoglycoside resistance 16S-RMTases [12]. In the end, about the genetic context, an example of npmA is shown in Figure 2. However, we cannot find the sequence of npmB’s genetic context in the NCBI database, so the figure does not contain the npmB gene for the moment.
4. Conclusions
With the wide spread of multidrug-resistant bacteria and pan-drug resistant bacteria, monotherapy such as β-lactams or carbapenem for serious infections has become ineffective and unsuccessful. At this moment, the drug combination therapy containing kinds of antibiotics plays an irreplaceable role in the treatment of infectious diseases. As one of the first identified and used clinically, aminoglycoside antibiotics have a broad antimicrobial spectrum and can inhibit both Gram-negative bacteria and part of Gram-positive bacteria [5]. Aminoglycoside antibiotics regain the focus on the clinic as one of the effective options for the drug combination therapy [129].
However, a variety of resistance mechanisms lead to aminoglycosides resistance in bacteria. Among the mechanisms, the production of 16S-RMTases poses a potential threat. The reason for this statement is that they can confer not only complete resistance to a broad range of aminoglycosides but also high-level resistance that cannot be corrected by upregulating the dosage. In addition, several studies have shown that many subtypes of corresponding genotypes often lead to higher levels of resistance to aminoglycosides [81,92]; for example, rmtF2 is able to mediate resistance to amikacin and arbekacin with MIC > 1024 μg/mL.
What is more, most of the 16S-RMTases encoding genes are often located on transferable plasmids which have a broad host range. With the development of whole-genome sequencing, it is abundantly clear that almost each 16S-RMTases gene is surrounded by some kinds of mobile genetic elements including transposons, integrons, and insertion sequences. Mobile genetic elements and conjugative plasmids not only promote resistance genes to transfer horizontally among all the members of Enterobacteriaceae but also upregulate the expression of the resistance genes resulting in higher MIC [102,120]. From another point of view, there is no doubt that mobile genetic elements also facilitate 16S-RMTases genes to associate with other resistance genes so that forming multidrug resistance regions. The coproduction of 16S-RMTases with ESBLs, AmpC enzymes, and carbapenemases largely limits the drug combination therapy for life-threatening infections caused by multidrug-resistant pathogens [121].
Up to now, there are 2 methods to screen the strain expressing 16S-RMTases: disk diffusion method shows that there is no inhibition zone around multiple aminoglycosides disk or the diameter of inhibition zone shrinking; microbroth dilution method shows that the high-level MIC of multiple aminoglycosides exceeds 128 μg/mL [14]. If the above results occur, it can be preliminarily determined that the strain carries 16S-RMTase genes. What is more, because of the difference in methylation site, acquired G1405 16S-RMTases and A1408 16S-RMTases confer resistance to different aminoglycosides [12]. For example, apramycin which belongs to 4,5-disubstituted 2-DOS can be used to screen A1408 16S-RMTase [13]. However, the above methods are still not routinely as well as widely applied to the clinical laboratory to detect the resistance phenotypes, enzyme types even corresponding subtypes of 16S-RMTase gene [121]. In the future, it is important to appeal to the clinician and researchers to place particular emphasis on the current situation of aminoglycosides resistance and explore optimized screening procedures and methods to detect the strains expressing 16S-RMTase as early as possible. Additionally, to control the prevalence and global dissemination of multidrug-resistant bacteria, it is highly recommended to evaluate the molecular epidemiology of aminoglycosides resistance genes and perform in-depth analysis of related mobile genetic elements.
Author Contributions
Methodology, W.Y. and F.H.; writing—original draft preparation, W.Y.; writing—review and editing, F.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2701800, 2021YFC2701803), the National Natural Science Foundation of China (grant no. 82172311, 32141002, and 81861138052), and China Antimicrobial Surveillance Network (Independent Medical Grants from Pfizer, 2018QD100). The funders had no role in study design, data collection, analysis, publishing decisions, or manuscript preparation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/ (accessed on 17 June 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Arakawa, Y. 16S ribosomal RNA methylation: Emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 2007, 45, 88–94. [Google Scholar] [CrossRef]
- Wangkheimayum, J.; Bhattacharjee, M.; Das, B.J.; Singha, K.M.; Chanda, D.D.; Bhattacharjee, A. Expansion of acquired 16S rRNA methytransferases along with CTX-M-15, NDM and OXA-48 within three sequence types of Escherichia coli from northeast India. BMC Infect. Dis. 2020, 20, 544. [Google Scholar] [CrossRef]
- Wachino, J.-I.; Doi, Y.; Arakawa, Y. Aminoglycoside Resistance: Updates with a Focus on Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. N. Am. 2020, 34, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.; Cooper, M.A. Aminoglycoside antibiotics in the 21st century. ACS Chem. Biol. 2013, 8, 105–115. [Google Scholar] [CrossRef]
- Serio, A.W.; Keepers, T.; Andrews, L.; Krause, K.M. Aminoglycoside Revival: Review of a Historically Important Class of Antimicrobials Undergoing Rejuvenation. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef]
- Caméléna, F.; Morel, F.; Merimèche, M.; Decousser, J.-W.; Jacquier, H.; Clermont, O.; Darty, M.; Mainardis, M.; Cambau, E.; Tenaillon, O.; et al. Genomic characterization of 16S rRNA methyltransferase-producing Escherichia coli isolates from the Parisian area, France. J. Antimicrob. Chemother. 2020, 75, 1726–1735. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Yokoyama, K.; Doi, Y.; Yamane, K.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Yagi, T.; Kato, H.; Arakawa, Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 2003, 362, 1888–1893. [Google Scholar] [CrossRef]
- Cooksey, R.C.; Morlock, G.P.; McQueen, A.; Glickman, S.E.; Crawford, J.T. Characterization of streptomycin resistance mechanisms among Mycobacterium tuberculosis isolates from patients in New York City. Antimicrob. Agents Chemother. 1996, 40, 1186–1188. [Google Scholar] [CrossRef]
- Tang, M.; He, Z.; Pu, W. The research progress of acquired 16S rRNA methyltransferases. Acta Vet. Zootech. Sin. 2021, 52, 2369–2383. [Google Scholar]
- Kawai, A.; Suzuki, M.; Tsukamoto, K.; Minato, Y.; Doi, Y. Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance. Antimicrob. Agents Chemother. 2021, 65, e0100921. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.; Shibayama, K.; Kurokawa, H.; Kimura, K.; Yamane, K.; Suzuki, S.; Shibata, N.; Ike, Y.; Arakawa, Y. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob. Agents Chemother. 2007, 51, 4401–4409. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.-i.; Arakawa, Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: An update. Drug Resist. Updat. 2012, 15, 133–148. [Google Scholar] [CrossRef]
- Oshiro, S.; Tada, T.; Watanabe, S.; Tohya, M.; Hishinuma, T.; Uchida, H.; Kuwahara-Arai, K.; Mya, S.; Zan, K.N.; Kirikae, T.; et al. Emergence and Spread of Carbapenem-Resistant and Aminoglycoside-Panresistant Complex Isolates Coproducing NDM-Type Metallo-β-Lactamase and 16S rRNA Methylase in Myanmar. mSphere 2020, 5, e00054-20. [Google Scholar] [CrossRef]
- Zhou, Y.; Ai, W.; Guo, Y.; Wu, X.; Wang, B.; Xu, Y.; Rao, L.; Zhao, H.; Wang, X.; Yu, F. Co-Occurrence of Rare ArmA-, RmtB-, and KPC-2-Encoding Multidrug-Resistant Plasmids and Hypervirulence iuc Operon in ST11-KL47 Klebsiella pneumoniae. Microbiol. Spectr. 2022, 10, e0237121. [Google Scholar] [CrossRef]
- Usui, M.; Kajino, A.; Kon, M.; Fukuda, A.; Sato, T.; Shirakawa, T.; Kawanishi, M.; Harada, K.; Nakajima, C.; Suzuki, Y.; et al. Prevalence of 16S rRNA methylases in Gram-negative bacteria derived from companion animals and livestock in Japan. J. Vet. Med. Sci. 2019, 81, 874–878. [Google Scholar] [CrossRef]
- Taylor, E.; Bal, A.M.; Balakrishnan, I.; Brown, N.M.; Burns, P.; Clark, M.; Diggle, M.; Donaldson, H.; Eltringham, I.; Folb, J.; et al. A prospective surveillance study to determine the prevalence of 16S rRNA methyltransferase-producing Gram-negative bacteria in the UK. J. Antimicrob. Chemother. 2021, 76, 2428–2436. [Google Scholar] [CrossRef]
- Lu, X.; Zeng, M.; Zhang, N.; Wang, M.; Gu, B.; Li, J.; Jin, H.; Xiao, W.; Li, Z.; Zhao, H.; et al. Prevalence of 16S rRNA Methylation Enzyme Gene armA in Salmonella from Outpatients and Food. Front. Microbiol. 2021, 12, 663210. [Google Scholar] [CrossRef]
- Shen, X.; Liu, L.; Yu, J.; Ai, W.; Cao, X.; Zhan, Q.; Guo, Y.; Wang, L.; Yu, F. High Prevalence of 16S rRNA Methyltransferase Genes in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates Associated with Bloodstream Infections in 11 Chinese Teaching Hospitals. Infect. Drug Resist. 2020, 13, 2189–2197. [Google Scholar] [CrossRef]
- Chen, F.; Wang, L.; Wang, M.; Xie, Y.; Xia, X.; Li, X.; Liu, Y.; Cao, W.; Zhang, T.; Li, P.; et al. Genetic characterization and in vitro activity of antimicrobial combinations of multidrug-resistant Acinetobacter baumannii from a general hospital in China. Oncol. Lett. 2018, 15, 2305–2315. [Google Scholar] [CrossRef] [PubMed]
- Nafplioti, K.; Galani, I.; Angelidis, E.; Adamou, P.; Moraitou, E.; Giannopoulou, P.; Chra, P.; Damala, M.; Vogiatzakis, E.; Trikka-Graphakos, E.; et al. Dissemination of International Clone II Acinetobacter baumannii Strains Coproducing OXA-23 Carbapenemase and 16S rRNA Methylase ArmA in Athens, Greece. Microb. Drug Resist. 2020, 26, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Gur, D.; Hasdemir, U.; Cakar, A.; Cavusoglu, I.; Celik, T.; Aksu, B.; Sancak, B.; Altun, B.; Soyletir, G.; Ulger, N.; et al. Comparative in vitro activity of plazomicin and older aminoglyosides against Enterobacterales isolates; prevalence of aminoglycoside modifying enzymes and 16S rRNA methyltransferases. Diagn. Microbiol. Infect. Dis. 2020, 97, 115092. [Google Scholar] [CrossRef] [PubMed]
- da Paz Pereira, J.N.; das Neves de Andrade, C.A.; da Costa Lima, J.L.; de Lima Neto, R.G.; Ramos de Araujo, P.S.; Vieira Maciel, M.A. Clonal Dissemination of Clinical Isolates of Acinetobacter baumannii Carriers of 16S rRNA Methylase Genes in an Oncological Hospital in Recife, Brazil. Curr. Microbiol. 2020, 77, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Longo, L.G.A.; de Sousa, V.S.; Kraychete, G.B.; Justo-da-Silva, L.H.; Rocha, J.A.; Superti, S.V.; Bonelli, R.R.; Martins, I.S.; Moreira, B.M. Colistin resistance emerges in pandrug-resistant Klebsiella pneumoniae epidemic clones in Rio de Janeiro, Brazil. Int. J. Antimicrob. Agents 2019, 54, 579–586. [Google Scholar] [CrossRef]
- Galani, I.; Nafplioti, K.; Adamou, P.; Karaiskos, I.; Giamarellou, H.; Souli, M.; Maraki, S.; Mauromanolaki, V.E.; Papaioannou, V.; Tsiplakou, S.; et al. Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect. Dis. 2019, 19, 167. [Google Scholar] [CrossRef]
- Aishwarya, K.V.L.; Geetha, P.V.; Shanthi, M.; Uma, S. Co occurrence of two 16S rRNA methyltrasferases along with NDM and OXA 48 like carbapenamases on a single plasmid in Klebsiella pneumoniae. J. Lab. Physicians 2019, 11, 305–311. [Google Scholar] [CrossRef]
- Jin, J.S.; Kwon, K.T.; Moon, D.C.; Lee, J.C. Emergence of 16S rRNA methylase rmtA in colistin-only-sensitive Pseudomonas aeruginosa in South Korea. Int. J. Antimicrob. Agents 2009, 33, 490–491. [Google Scholar] [CrossRef]
- Poirel, L.; Schrenzel, J.; Cherkaoui, A.; Bernabeu, S.; Renzi, G.; Nordmann, P. Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J. Antimicrob. Chemother. 2011, 66, 1730–1733. [Google Scholar] [CrossRef]
- Nagasawa, M.; Kaku, M.; Kamachi, K.; Shibayama, K.; Arakawa, Y.; Yamaguchi, K.; Ishii, Y. Loop-mediated isothermal amplification assay for 16S rRNA methylase genes in Gram-negative bacteria. J. Infect. Chemother. 2014, 20, 635–638. [Google Scholar] [CrossRef]
- Zhao, Z.; Lan, F.; Liu, M.; Chen, W.; Huang, L.; Lin, Q.; Li, B. Evaluation of automated systems for aminoglycosides and fluoroquinolones susceptibility testing for Carbapenem-resistant. Antimicrob. Resist. Infect. Control 2017, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Wang, L.D.; Li, D.; Du, F.-l.; Long, D.; Liu, Y.; Ng, O.; Zhang, W. High Prevalence of 16s rRNA Methylase Genes among Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Isolates in a Chinese Tertiary Hospital. Microbial. Drug Resist. 2021, 27, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Sefidan, F.Y.; Mohammadzadeh-Asl, Y.; Ghotaslou, R. High-Level Resistance to Aminoglycosides due to 16S rRNA Methylation in Enterobacteriaceae Isolates. Microbial. Drug Resist. 2019, 25, 1261–1265. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Tao, J.; Li, G.; Jia, W. Aminoglycoside resistance and the prevalence of 16S rRNA methylase genes in Enterobacteriaceae strains. Chin. J. Infect. Chemother. 2021, 21, 593–598. [Google Scholar] [CrossRef]
- Fournier, C.; Poirel, L.; Despont, S.; Kessler, J.; Nordmann, P. Increasing Trends of Association of 16S rRNA Methylases and Carbapenemases in Enterobacterales Clinical Isolates from Switzerland, 2017–2020. Microorganisms 2022, 10, 615. [Google Scholar] [CrossRef] [PubMed]
- El-Far, A.; Samir, S.; El-Gebaly, E.; Omar, M.; Dahroug, H.; El-Shenawy, A.; Soliman, N.S.; Gamal, D. High Rates of Aminoglycoside Methyltransferases Associated with Metallo-Beta-Lactamases in Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Clinical Isolates from a Tertiary Care Hospital in Egypt. Infect. Drug Resist. 2021, 14, 4849–4858. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, L.-X.; Jiang, Y.-W.; Wu, D.-S.; Jiang, Q.; Sun, R.-Y.; Wang, M.-G.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Comparison of the prevalence and molecular characteristics of fosA3 and fosA7 among Salmonella isolates from food animals in China. J. Antimicrob. Chemother. 2022, 77, 1286–1295. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.-Y.; Wang, Y.; Sun, F.; Li, W.; Wu, H.; Shen, P.-C.; Pan, Z.-M.; Jiao, X. Emergence of 16S rRNA Methylase Gene rmtB in Salmonella Enterica Serovar London and Evolution of RmtB-Producing Plasmid Mediated by IS26. Front. Microbiol. 2020, 11, 604278. [Google Scholar] [CrossRef]
- Roch, M.; Sierra, R.; Sands, K.; Martins, W.M.B.S.; Schrenzel, J.; Walsh, T.R.; Gales, A.C.; Andrey, D.O. Vertical and horizontal dissemination of an IncC plasmid harbouring rmtB 16S rRNA methylase gene, conferring resistance to plazomicin, among invasive ST258 and ST16 KPC-producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 24, 183–189. [Google Scholar] [CrossRef]
- Nafplioti, K.; Souli, M.; Adamou, P.; Moraitou, E.; Giannopoulou, P.; Chra, P.; Damala, M.; Vogiatzakis, E.; Trikka-Graphakos, E.; Baka, V.; et al. Characterization of 16S rRNA methylase genes in Enterobacterales and Pseudomonas aeruginosa in Athens Metropolitan area, 2015–2016. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 111–121. [Google Scholar] [CrossRef]
- Musila, L.; Kyany’a, C.; Maybank, R.; Stam, J.; Oundo, V.; Sang, W. Detection of diverse carbapenem and multidrug resistance genes and high-risk strain types among carbapenem non-susceptible clinical isolates of target gram-negative bacteria in Kenya. PLoS ONE 2021, 16, e0246937. [Google Scholar] [CrossRef] [PubMed]
- Mc Gann, P.; Geringer, M.R.; Hall, L.R.; Lebreton, F.; Markelz, E.; Kwak, Y.I.; Johnson, S.; Ong, A.C.; Powell, A.; Tekle, T.; et al. Pan-drug resistant contributing to a fatal case of COVID-19. J. Med. Microbiol. 2021, 70, 001406. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Wang, X.; Lei, C.; Tang, Y.; He, J.; Gao, Y.; Zhang, Y.; Wang, H. Identification of Three Novel PmGRI1 Genomic Resistance Islands and One Multidrug Resistant Hybrid Structure of Tn7-like Transposon and PmGRI1 in Proteus mirabilis. Antibiotics 2021, 10, 1268. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Willcox, M.D.P.; Rice, S.A.; Sharma, S.; Stapleton, F. Development of antibiotic resistance in the ocular Pseudomonas aeruginosa clone ST308 over twenty years. Exp. Eye Res. 2021, 205, 108504. [Google Scholar] [CrossRef]
- Huang, L.; Fu, L.; Hu, X.; Liang, X.; Gong, G.; Xie, C.; Zhang, F.; Wang, Y.; Zhou, Y. Co-occurrence of Klebsiela varicola and Klebsiela pneumoniae Both Carrying bla(KPC) from a Respiratory Intensive Care Unit Patient. Infect. Drug Resist. 2021, 14, 4503–4510. [Google Scholar] [CrossRef]
- He, D.-D.; Cui, M.-M.; Zhang, T.-L.; Hu, G.-Z.; Liu, J.-H.; Pan, Y.-S. Characterization of blaCMY-2-carrying IncC and rmtB-carrying IncI1/ST136 plasmids in an avian Escherichia coli ST224 strain. Plasmid 2021, 114, 102555. [Google Scholar] [CrossRef]
- Cheng, K.; Fang, L.-X.; Ge, Q.-W.; Wang, D.; He, B.; Lu, J.-Q.; Zhong, Z.-X.; Wang, X.-R.; Yu, Y.; Lian, X.-L.; et al. Emergence of fosA3 and bla(CTX-M-14) in Multidrug-Resistant Citrobacter freundii Isolates from Flowers and the Retail Environment in China. Front. Microbiol. 2021, 12, 586504. [Google Scholar] [CrossRef]
- Zhu, X.; Li, P.; Qian, C.; Liu, H.; Lin, H.; Zhang, X.; Li, Q.; Lu, J.; Lin, X.; Xu, T.; et al. Prevalence of Aminoglycoside Resistance Genes and Molecular Characterization of a Novel Gene, aac(3)-IIg, among Clinical Isolates of the Enterobacter cloacae Complex from a Chinese Teaching Hospital. Antimicrob. Agents Chemother. 2020, 64, e00852-20. [Google Scholar] [CrossRef]
- Nishida, S.; Ono, Y. Genomic analysis of a pan-resistant Klebsiella pneumoniae sequence type 11 identified in Japan in 2016. Int. J. Antimicrob. Agents 2020, 55, 105854. [Google Scholar] [CrossRef]
- Wu, X.; Han, H.; Chen, C.; Zheng, B. Genomic characterisation of a colistin-resistant Klebsiella pneumoniae ST11 strain co-producing KPC-2, FloR, CTX-M-55, SHV-12, FosA and RmtB causing a lethal infection. J. Glob. Antimicrob. Resist. 2019, 19, 78–80. [Google Scholar] [CrossRef]
- Uchida, H.; Tada, T.; Tohya, M.; Sugahara, Y.; Kato, A.; Miyairi, I.; Kirikae, T. Emergence in Japan of an isolate of Klebsiella pneumoniae co-harbouring blaKPC-2 and rmtB. J. Glob. Antimicrob. Resist. 2019, 17, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Feng, Y.; Ma, K.; Liu, L.; McNally, A.; Zong, Z. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene bla(NDM-5) and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 2019, 9, 696. [Google Scholar] [CrossRef] [PubMed]
- Belaynehe, K.M.; Won, H.G.; Yoon, I.J.; Yoo, H.S. Prevalence and molecular characteristics of 16s rRNA methylase gene rmtB in amikacin resistant Escherichia coli isolated from South Korea. Korean J. Vet. Res. 2019, 59, 157–160. [Google Scholar] [CrossRef]
- Amin, M.; Mehdipour, G.; Navidifar, T. High distribution of 16S rRNA methylase genes and among strains isolated from an Ahvaz teaching hospital, Iran. Acta Microbiol. Immunol. Hung. 2019, 66, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Hishinuma, T.; Watanabe, S.; Uchida, H.; Tohya, M.; Kuwahara-Arai, K.; Mya, S.; Zan, K.N.; Kirikae, T.; Tin, H.H. Molecular Characterization of Multidrug-Resistant Isolates in Hospitals in Myanmar. Antimicrob. Agents Chemother. 2019, 63, e02397-18. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, L.; Hopkins, K.L.; Gutierrez, B.; Ovejero, C.M.; Shukla, S.; Douthwaite, S.; Prasad, K.N.; Woodford, N.; Gonzalez-Zorn, B. Association of the novel aminoglycoside resistance determinant RmtF with NDM carbapenemase in Enterobacteriaceae isolated in India and the UK. J. Antimicrob. Chemother. 2013, 68, 1543–1550. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Mishra, S.K.; Ohara, H.; Shimada, K.; Kirikae, T.; Pokhrel, B.M. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int. J. Antimicrob. Agents 2013, 42, 372–374. [Google Scholar] [CrossRef]
- Rubin, J.E.; Peirano, G.; Peer, A.K.; Govind, C.N.; Pitout, J.D.D. NDM-1-producing Enterobacteriaceae from South Africa: Moving towards endemicity? Diagn. Microbiol. Infect. Dis. 2014, 79, 378–380. [Google Scholar] [CrossRef]
- Filgona, J.; Banerjee, T.; Anupurba, S. Incidence of the novel rmtF and rmtG methyltransferases in carbapenem-resistant Enterobacteriaceae from a hospital in India. J. Infect. Dev. Ctries. 2015, 9, 1036–1039. [Google Scholar] [CrossRef][Green Version]
- Rahman, M.; Prasad, K.N.; Pathak, A.; Pati, B.K.; Singh, A.; Ovejero, C.M.; Ahmad, S.; Gonzalez-Zorn, B. RmtC and RmtF 16S rRNA Methyltransferase in NDM-1-Producing Pseudomonas aeruginosa. Emerg. Infect. Dis. 2015, 21, 2059–2062. [Google Scholar] [CrossRef]
- Erdem, F.; Abulaila, A.; Aktas, Z.; Oncul, O. In vitro evaluation of double carbapenem and colistin combinations against OXA-48, NDM carbapenemase-producing colistin-resistant Klebsiella pneumoniae strains. Antimicrob. Resist. Infect. Control 2020, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Jauneikaite, E.; Sriskandan, S.; Woodford, N.; Hopkins, K.L. Detection and characterisation of 16S rRNA methyltransferase-producing Pseudomonas aeruginosa from the UK and Republic of Ireland from 2003–2015. Int. J. Antimicrob. Agents 2022, 59, 106550. [Google Scholar] [CrossRef] [PubMed]
- Wachino, J.-I.; Yamane, K.; Shibayama, K.; Kurokawa, H.; Shibata, N.; Suzuki, S.; Doi, Y.; Kimura, K.; Ike, Y.; Arakawa, Y. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 2006, 50, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Partridge, S.R.; Iredell, J.R. RmtC 16S rRNA methyltransferase in Australia. Antimicrob. Agents Chemother. 2008, 52, 794–795. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Escudero, J.A.; Hidalgo, L.; Gonzalez-Zorn, B. 16S rRNA methyltransferase RmtC in Salmonella enterica serovar Virchow. Emerg. Infect. Dis. 2010, 16, 712–715. [Google Scholar] [CrossRef]
- Huang, J.; Wang, M.; Ding, H.; Ye, M.; Hu, F.; Guo, Q.; Xu, X.; Wang, M. New Delhi metallo-β-lactamase-1 in carbapenem-resistant Salmonella strain, China. Emerg. Infect. Dis. 2013, 19, 2049–2051. [Google Scholar] [CrossRef]
- Kapmaz, M.; Erdem, F.; Abulaila, A.; Yeniaras, E.; Oncul, O.; Aktas, Z. First detection of NDM-1 with CTX-M-9, TEM, SHV and rmtC in Escherichia coli ST471 carrying IncI2, A/C and Y plasmids from clinical isolates in Turkey. J. Glob. Antimicrob. Resist. 2016, 7, 152–153. [Google Scholar] [CrossRef]
- Huang, J.; Deng, S.; Ren, J.; Tu, J.; Ye, M.; Wang, M. Characterization of a blaNDM-1-harboring plasmid from a Salmonella enterica clinical isolate in China. Mol. Med. Rep. 2017, 16, 1087–1092. [Google Scholar] [CrossRef]
- Tada, T.; Tsuchiya, M.; Shimada, K.; Nga, T.T.T.; Thu, L.T.A.; Phu, T.T.; Ohmagari, N.; Kirikae, T. Dissemination of Carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of Carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA Methylases (RmtB and RmtC) in Vietnam. BMC Infect. Dis. 2017, 17, 467. [Google Scholar] [CrossRef]
- Bado, I.; Papa-Ezdra, R.; Delgado-Blas, J.F.; Gaudio, M.; Gutiérrez, C.; Cordeiro, N.F.; García-Fulgueiras, V.; Araújo Pirez, L.; Seija, V.; Medina, J.C.; et al. Molecular Characterization of Carbapenem-Resistant Acinetobacter baumannii in the Intensive Care Unit of Uruguay’s University Hospital Identifies the First rmtC Gene in the Species. Microb. Drug Resist. 2018, 24, 1012–1019. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; El-Mahdy, T.S.; Radwan, H.H.; Poirel, L. Cooccurrence of NDM-1, ESBL, RmtC, AAC(6′)-Ib, and QnrB in Clonally Related Isolates Together with Coexistence of CMY-4 and AAC(6′)-Ib in Isolates from Saudi Arabia. Biomed. Res. Int. 2019, 2019, 6736897. [Google Scholar] [CrossRef] [PubMed]
- Kiaei, S.; Moradi, M.; Hosseini Nave, H.; Hashemizadeh, Z.; Taati-Moghadam, M.; Kalantar-Neyestanaki, D. Emergence of co-existence of bla with rmtC and qnrB genes in clinical carbapenem-resistant Klebsiella pneumoniae isolates in burning center from southeast of Iran. Folia Microbiol. 2019, 64, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, L.; Yu, J.; Cao, X.; Zhan, Q.; Guo, Y.; Wang, L.; Yu, F. Coexistence of blaNDM-1 and rmtC on a Transferrable Plasmid of a Novel ST192 Klebsiella aerogenes Clinical Isolate. Infect. Drug Resist. 2019, 12, 3883–3891. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sun, J.; Li, L.; Fang, L.-X.; Deng, H.; Yang, R.-S.; Li, X.-P.; Liao, X.-P.; Liu, Y.-H. First Report of the IncI1/ST898 Conjugative Plasmid Carrying rmtE2 16S rRNA Methyltransferase Gene in Escherichia coli. Antimicrob. Agents Chemother. 2015, 59, 7921–7922. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doi, Y.; de Oliveira Garcia, D.; Adams, J.; Paterson, D.L. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 2007, 51, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, T.R.; Castanheira, M.; Miller, G.H.; Jones, R.N.; Armstrong, E.S. Detection of methyltransferases conferring high-level resistance to aminoglycosides in enterobacteriaceae from Europe, North America, and Latin America. Antimicrob. Agents Chemother. 2008, 52, 1843–1845. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Rossi, F.; Barberino, M.G.; Adams-Haduch, J.M.; Doi, Y.; Paterson, D.L. 16S ribosomal RNA methylase RmtD produced by Klebsiella pneumoniae in Brazil. J. Antimicrob. Chemother. 2008, 61, 746–747. [Google Scholar] [CrossRef][Green Version]
- Fontes, L.C.; Neves, P.R.; Oliveira, S.; Silva, K.C.; Hachich, E.M.; Sato, M.I.; Lincopan, N. Isolation of Pseudomonas aeruginosa coproducing metallo-beta-lactamase SPM-1 and 16S rRNA methylase RmtD1 in an urban river. Antimicrob. Agents Chemother. 2011, 55, 3063–3064. [Google Scholar] [CrossRef]
- Tijet, N.; Andres, P.; Chung, C.; Lucero, C.; Group, W.H.-A.; Low, D.E.; Galas, M.; Corso, A.; Petroni, A.; Melano, R.G. rmtD2, a new allele of a 16S rRNA methylase gene, has been present in Enterobacteriaceae isolates from Argentina for more than a decade. Antimicrob. Agents Chemother. 2011, 55, 904–909. [Google Scholar] [CrossRef]
- Bueno, M.F.C.; Francisco, G.R.; O’Hara, J.A.; de Oliveira Garcia, D.; Doi, Y. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013, 57, 2397–2400. [Google Scholar] [CrossRef]
- Tada, T.; Shimada, K.; Mya, S.; Zan, K.N.; Kuwahara, K.; Kirikae, T.; Tin, H.H. A New Variant of 16S rRNA Methylase, RmtD3, in a Clinical Isolate of Pseudomonas aeruginosa in Myanmar. Antimicrob. Agents Chemother. 2018, 62, e01806-17. [Google Scholar] [CrossRef] [PubMed]
- Urbanowicz, P.; Izdebski, R.; Baraniak, A.; Zabicka, D.; Ziolkowski, G.; Hryniewicz, W.; Gniadkowski, M. Pseudomonas aeruginosa with NDM-1, DIM-1 and PME-1 beta-lactamases, and RmtD3 16S rRNA methylase, encoded by new genomic islands. J. Antimicrob. Chemother. 2019, 74, 3117–3119. [Google Scholar] [CrossRef] [PubMed]
- Bail, L.; Ito, C.A.S.; Arend, L.N.V.S.; Pilonetto, M.; Nogueira, K.d.S.; Tuon, F.F. Distribution of genes encoding 16S rRNA methyltransferase in plazomicin-nonsusceptible carbapenemase-producing Enterobacterales in Brazil. Diagn. Microbiol. Infect. Dis. 2021, 99, 115239. [Google Scholar] [CrossRef]
- Davis, M.A.; Baker, K.N.K.; Orfe, L.H.; Shah, D.H.; Besser, T.E.; Call, D.R. Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-S.; Hu, F.; Rivera, J.I.; Doi, Y. Escherichia coli sequence type 354 coproducing CMY-2 cephalosporinase and RmtE 16S rRNA methyltransferase. Antimicrob. Agents Chemother. 2014, 58, 4246–4247. [Google Scholar] [CrossRef]
- Li, B.; Pacey, M.P.; Doi, Y. Chromosomal 16S Ribosomal RNA Methyltransferase RmtE1 in Escherichia coli Sequence Type 448. Emerg. Infect. Dis. 2017, 23, 876–878. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Jauneikaite, E.; Sriskandan, S.; Woodford, N.; Hopkins, K.L. Novel 16S rRNA methyltransferase RmtE3 in Acinetobacter baumannii ST79. J. Med. Microbiol. 2022, 71, 001531. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sun, J.; Cheng, K.; Li, L.; Fang, L.X.; Zou, M.T.; Liao, X.P.; Liu, Y.H. Persistent spread of the rmtB 16S rRNA methyltransferase gene among Escherichia coli isolates from diseased food-producing animals in China. Vet. Microbiol. 2016, 188, 41–46. [Google Scholar] [CrossRef]
- Galimand, M.; Courvalin, P.; Lambert, T. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob. Agents Chemother. 2012, 56, 3960–3962. [Google Scholar] [CrossRef]
- Lee, C.-S.; Vasoo, S.; Hu, F.; Patel, R.; Doi, Y. Klebsiella pneumoniae ST147 coproducing NDM-7 carbapenemase and RmtF 16S rRNA methyltransferase in Minnesota. J. Clin. Microbiol. 2014, 52, 4109–4110. [Google Scholar] [CrossRef]
- Gamal, D.; Fernández-Martínez, M.; Salem, D.; El-Defrawy, I.; Montes, L.Á.; Ocampo-Sosa, A.A.; Martínez-Martínez, L. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 2016, 43, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Shimada, K.; Satou, K.; Hirano, T.; Pokhrel, B.M.; Sherchand, J.B.; Kirikae, T. Pseudomonas aeruginosa Clinical Isolates in Nepal Coproducing Metallo-β-Lactamases and 16S rRNA Methyltransferases. Antimicrob. Agents Chemother. 2017, 61, e00694-17. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.; Sriskandan, S.; Woodford, N.; Hopkins, K.L. High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int. J. Antimicrob. Agents 2018, 52, 278–282. [Google Scholar] [CrossRef]
- Shankar, C.; Muthuirulandi Sethuvel, D.P.; Neeravi, A.R.; Venkatesan, M.; Devanga Ragupathi, N.K.; Anandan, S.; Veeraraghavan, B. Identification of plasmids by PCR based replicon typing in bacteremic Klebsiella pneumoniae. Microb. Pathog. 2020, 148, 104429. [Google Scholar] [CrossRef]
- Shi, Q.; Han, R.; Guo, Y.; Zheng, Y.; Yang, Y.; Yin, D.; Zhang, R.; Hu, F. Emergence of ST15 Clinical Isolates Producing Plasmids-Mediated RmtF and OXA-232 in China. Infect. Drug Resist. 2020, 13, 3125–3129. [Google Scholar] [CrossRef]
- Hu, F.; Munoz-Price, L.S.; DePascale, D.; Rivera, J.I.; Doi, Y. Klebsiella pneumoniae sequence type 11 isolate producing RmtG 16S rRNA methyltransferase from a patient in Miami, Florida. Antimicrob. Agents Chemother. 2014, 58, 4980–4981. [Google Scholar] [CrossRef]
- Poirel, L.; Labarca, J.; Bello, H.; Rioseco, M.L.; Bernabeu, S.; Nordmann, P. Emergence of the 16S rRNA methylase RmtG in an extended-spectrum-β-lactamase-producing and colistin-resistant Klebsiella pneumoniae isolate in Chile. Antimicrob. Agents Chemother. 2014, 58, 618–619. [Google Scholar] [CrossRef]
- Francisco, G.R.; Nora, S.T.R.; Bueno, M.F.C.; da Silva Filho, L.V.R.F.; de Oliveira Garcia, D. Identification of aminoglycoside-resistant Pseudomonas aeruginosa producing RmtG 16S rRNA methyltransferase in a cystic fibrosis patient. Antimicrob. Agents Chemother. 2015, 59, 2967–2968. [Google Scholar] [CrossRef]
- Cerdeira, L.; Fernandes, M.R.; Francisco, G.R.; Bueno, M.F.C.; Ienne, S.; Souza, T.A.; de Oliveira Garcia, D.; Lincopan, N. Draft Genome Sequence of a Hospital-Associated Clone of Klebsiella pneumoniae ST340/CC258 Coproducing RmtG and KPC-2 Isolated from a Pediatric Patient. Genome Announc. 2016, 4, e01130-16. [Google Scholar] [CrossRef]
- Mancini, S.; Poirel, L.; Corthesy, M.; Greub, G.; Nordmann, P. Klebsiella pneumoniae co-producing KPC and RmtG, finally targeting Switzerland. Diagn. Microbiol. Infect. Dis. 2018, 90, 151–152. [Google Scholar] [CrossRef]
- Passarelli-Araujo, H.; Palmeiro, J.K.; Moharana, K.C.; Pedrosa-Silva, F.; Dalla-Costa, L.M.; Venancio, T.M. Molecular epidemiology of 16S rRNA methyltransferase in Brazil: RmtG in Klebsiella aerogenes ST93 (CC4). An. Acad. Bras. Cienc. 2019, 91, e20180762. [Google Scholar] [CrossRef]
- Martins, E.R.; Bueno, M.F.C.; Francisco, G.R.; Casella, T.; de Oliveira Garcia, D.; Cerdeira, L.T.; Gerber, A.L.; de Almeida, L.G.P.; Lincopan, N.; de Vasconcelos, A.T.R.; et al. Genome and plasmid context of two rmtG-carrying Enterobacter hormaechei isolated from urinary tract infections in Brazil. J. Glob. Antimicrob. Resist. 2020, 20, 36–40. [Google Scholar] [CrossRef]
- O’Hara, J.A.; McGann, P.; Snesrud, E.C.; Clifford, R.J.; Waterman, P.E.; Lesho, E.P.; Doi, Y. Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob. Agents Chemother. 2013, 57, 2413–2416. [Google Scholar] [CrossRef]
- Beyrouthy, R.; Robin, F.; Hamze, M.; Bonnet, R. IncFIIk plasmid harbouring an amplification of 16S rRNA methyltransferase-encoding gene rmtH associated with mobile element ISCR2. J. Antimicrob. Chemother. 2017, 72, 402–406. [Google Scholar] [CrossRef][Green Version]
- Marsh, J.W.; Pacey, M.P.; Ezeonwuka, C.; Ohm, S.L.; Snyder, D.; Cooper, V.S.; Harrison, L.H.; Doi, Y.; Mustapha, M.M. Clostridioides difficile: A potential source of NpmA in the clinical environment. J. Antimicrob. Chemother. 2019, 74, 521–523. [Google Scholar] [CrossRef]
- Ishizaki, Y.; Shibuya, Y.; Hayashi, C.; Inoue, K.; Kirikae, T.; Tada, T.; Miyoshi-Akiyama, T.; Igarashi, M. Instability of the 16S rRNA methyltransferase-encoding npmA gene: Why have bacterial cells possessing npmA not spread despite their high and broad resistance to aminoglycosides? J. Antibiot. 2018, 71, 798–807. [Google Scholar] [CrossRef]
- Galimand, M.; Courvalin, P.; Lambert, T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 2003, 47, 2565–2571. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, M.; Yang, J.; Dai, M.; Chang, Y.; Zhang, C.; Luan, G.; Ling, B.; Jia, X. Prevalence of carbapenemases among high-level aminoglycoside-resistant isolates in a university hospital in China. Exp. Ther. Med. 2016, 12, 3642–3652. [Google Scholar] [CrossRef][Green Version]
- Tada, T.; Miyoshi-Akiyama, T.; Shimada, K.; Shimojima, M.; Kirikae, T. Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob. Agents Chemother. 2014, 58, 2916–2920. [Google Scholar] [CrossRef]
- Tada, T.; Miyoshi-Akiyama, T.; Dahal, R.K.; Sah, M.K.; Ohara, H.; Shimada, K.; Kirikae, T.; Pokhrel, B.M. NDM-1 Metallo-β-Lactamase and ArmA 16S rRNA methylase producing Providencia rettgeri clinical isolates in Nepal. BMC Infect. Dis. 2014, 14, 56. [Google Scholar] [CrossRef]
- Bogaerts, P.; Galimand, M.; Bauraing, C.; Deplano, A.; Vanhoof, R.; De Mendonca, R.; Rodriguez-Villalobos, H.; Struelens, M.; Glupczynski, Y. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 2007, 59, 459–464. [Google Scholar] [CrossRef]
- Galimand, M.; Sabtcheva, S.; Courvalin, P.; Lambert, T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 2005, 49, 2949–2953. [Google Scholar] [CrossRef]
- Poirel, L.; Goutines, J.; Aires-de-Sousa, M.; Nordmann, P. High Rate of Association of 16S rRNA Methylases and Carbapenemases in Enterobacteriaceae Recovered from Hospitalized Children in Angola. Antimicrob. Agents Chemother. 2018, 62, e00021-18. [Google Scholar] [CrossRef]
- Yamane, K.; Doi, Y.; Yokoyama, K.; Yagi, T.; Kurokawa, H.; Shibata, N.; Shibayama, K.; Kato, H.; Arakawa, Y. Genetic environments of the rmtA gene in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 2004, 48, 2069–2074. [Google Scholar] [CrossRef]
- Yamane, K.; Wachino, J.-i.; Doi, Y.; Kurokawa, H.; Arakawa, Y. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 2005, 11, 951–953. [Google Scholar] [CrossRef]
- Doi, Y.; Yokoyama, K.; Yamane, K.; Wachino, J.-I.; Shibata, N.; Yagi, T.; Shibayama, K.; Kato, H.; Arakawa, Y. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 2004, 48, 491–496. [Google Scholar] [CrossRef]
- Doi, Y.; Adams-Haduch, J.M.; Paterson, D.L. Escherichia coli isolate coproducing 16S rRNA Methylase and CTX-M-type extended-spectrum beta-lactamase isolated from an outpatient in the United States. Antimicrob. Agents Chemother. 2008, 52, 1204–1205. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, J.-H.; Du, X.-D.; Zong, Z.-Y.; Chen, M.; Hu, G.-Z.; Pan, Y.-S. Comparative genomics of rmtB-carrying IncI1 ST136 plasmids in avian escherichia coli isolates from chickens in China. Int. J. Antimicrob. Agents 2018, 51, 659–662. [Google Scholar] [CrossRef]
- Du, X.D.; Wu, C.-M.; Liu, H.-B.; Li, X.-S.; Beier, R.C.; Xiao, F.; Qin, S.; Huang, S.-Y.; Shen, J.-Z. Plasmid-mediated ArmA and RmtB 16S rRNA methylases in Escherichia coli isolated from chickens. J. Antimicrob. Chemother. 2009, 64, 1328–1330. [Google Scholar] [CrossRef][Green Version]
- Wachino, J.-i.; Yamane, K.; Kimura, K.; Shibata, N.; Suzuki, S.; Ike, Y.; Arakawa, Y. Mode of transposition and expression of 16S rRNA methyltransferase gene rmtC accompanied by ISEcp1. Antimicrob. Agents Chemother. 2006, 50, 3212–3215. [Google Scholar] [CrossRef]
- Mohanam, L.; Menon, T. Emergence of rmtC and rmtF 16S rRNA Methyltransferase in Clinical Isolates of Pseudomonas aeruginosa. Indian J. Med. Microbiol. 2017, 35, 282–285. [Google Scholar] [CrossRef]
- Doi, Y.; Adams-Haduch, J.M.; Paterson, D.L. Genetic environment of 16S rRNA methylase gene rmtD. Antimicrob. Agents Chemother. 2008, 52, 2270–2272. [Google Scholar] [CrossRef]
- Lee, C.-S.; Li, J.-J.; Doi, Y. Complete sequence of conjugative IncA/C plasmid encoding CMY-2 β-lactamase and RmtE 16S rRNA methyltransferase. Antimicrob. Agents Chemother. 2015, 59, 4360–4361. [Google Scholar] [CrossRef][Green Version]
- Mancini, S.; Poirel, L.; Tritten, M.L.; Lienhard, R.; Bassi, C.; Nordmann, P. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J. Antimicrob. Chemother. 2018, 73, 821–823. [Google Scholar] [CrossRef]
- Sherchan, J.B.; Tada, T.; Shrestha, S.; Uchida, H.; Hishinuma, T.; Morioka, S.; Shahi, R.K.; Bhandari, S.; Twi, R.T.; Kirikae, T.; et al. Emergence of clinical isolates of highly carbapenem-resistant Klebsiella pneumoniae co-harboring bla(NDM-5) and bla(OXA-181) or-232 in Nepal. Int. J. Infect. Dis. 2020, 92, 247–252. [Google Scholar] [CrossRef]
- Ramos, P.I.P.; Picão, R.C.; Almeida, L.G.P.d.; Lima, N.C.B.; Girardello, R.; Vivan, A.C.P.; Xavier, D.E.; Barcellos, F.G.; Pelisson, M.; Vespero, E.C.; et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genom. 2014, 15, 54. [Google Scholar] [CrossRef]
- Bueno, M.F.C.; Francisco, G.R.; Cerdeira, L.; Ienne, S.; Souza, T.A.; Lincopan, N.; de Oliveira Garcia, D. Draft genome sequence of an aminoglycoside-resistant RmtG-producing Pseudomonas aeruginosa ST235 isolated from a cystic fibrosis patient. J. Glob. Antimicrob. Resist. 2017, 8, 106–107. [Google Scholar] [CrossRef]
- Nosrati, M.; Dey, D.; Mehrani, A.; Strassler, S.E.; Zelinskaya, N.; Hoffer, E.D.; Stagg, S.M.; Dunham, C.M.; Conn, G.L. Functionally critical residues in the aminoglycoside resistance-associated methyltransferase RmtC play distinct roles in 30S substrate recognition. J. Biol. Chem. 2019, 294, 17642–17653. [Google Scholar] [CrossRef]
- Turnidge, J. Pharmacodynamics and dosing of aminoglycosides. Infect. Dis. Clin. N. Am. 2003, 17, 503–528. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
