Polymicrobial Biofilm Dynamics of Multidrug-Resistant Candida albicans and Ampicillin-Resistant Escherichia coli and Antimicrobial Inhibition by Aqueous Garlic Extract
Abstract
1. Introduction
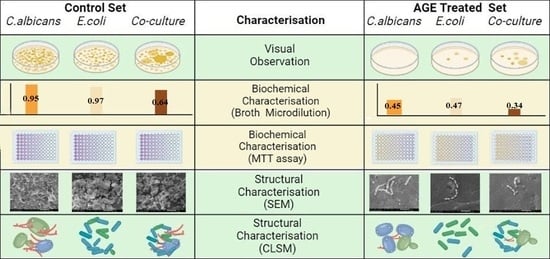
2. Results and Discussion
2.1. Microbial Culture and Identification
2.2. Point Inoculation
2.3. Agar Diffusion
2.4. VITEK Antimicrobial Susceptibility Test (AST)
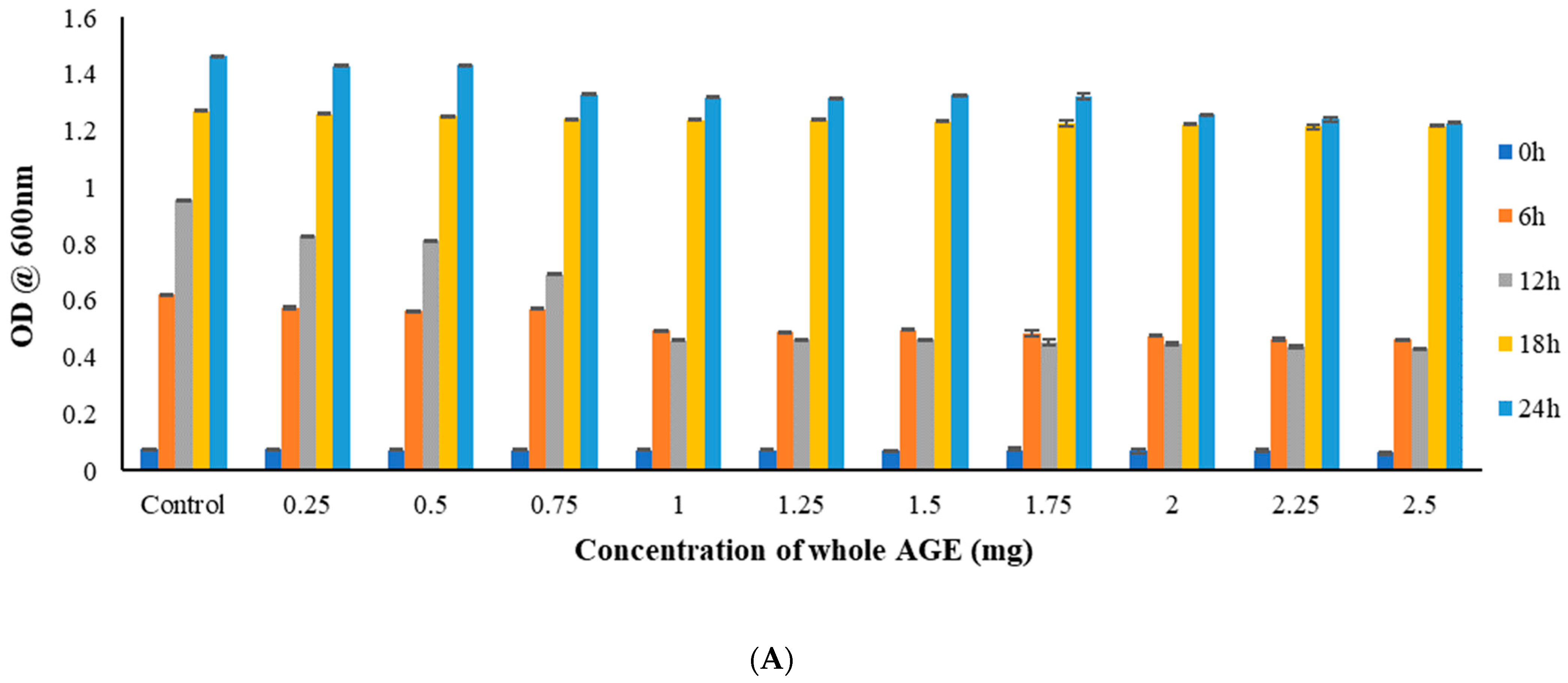
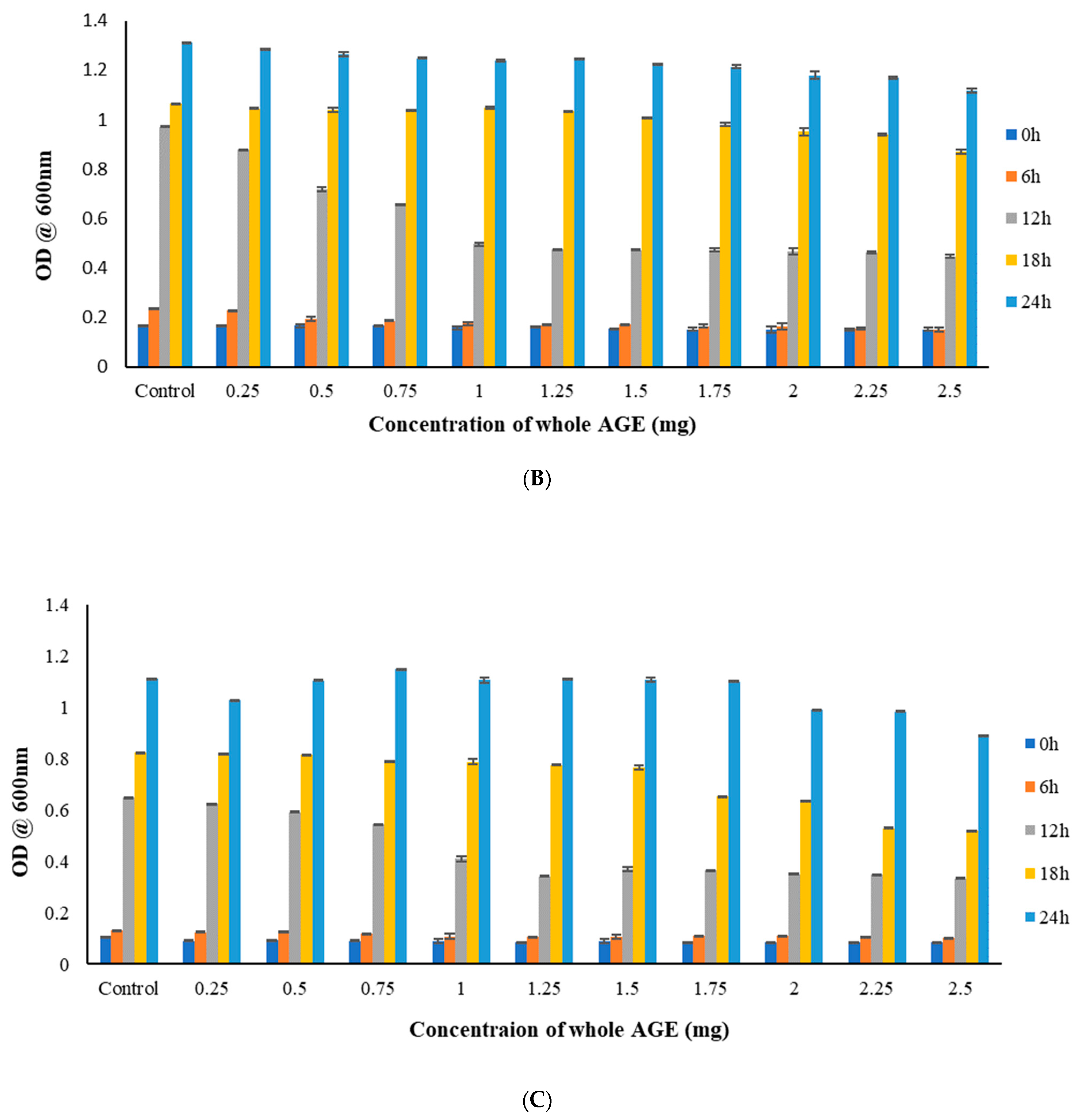
2.5. Broth Microdilution Method to Determine the Minimum Inhibitory Concentration (MIC)
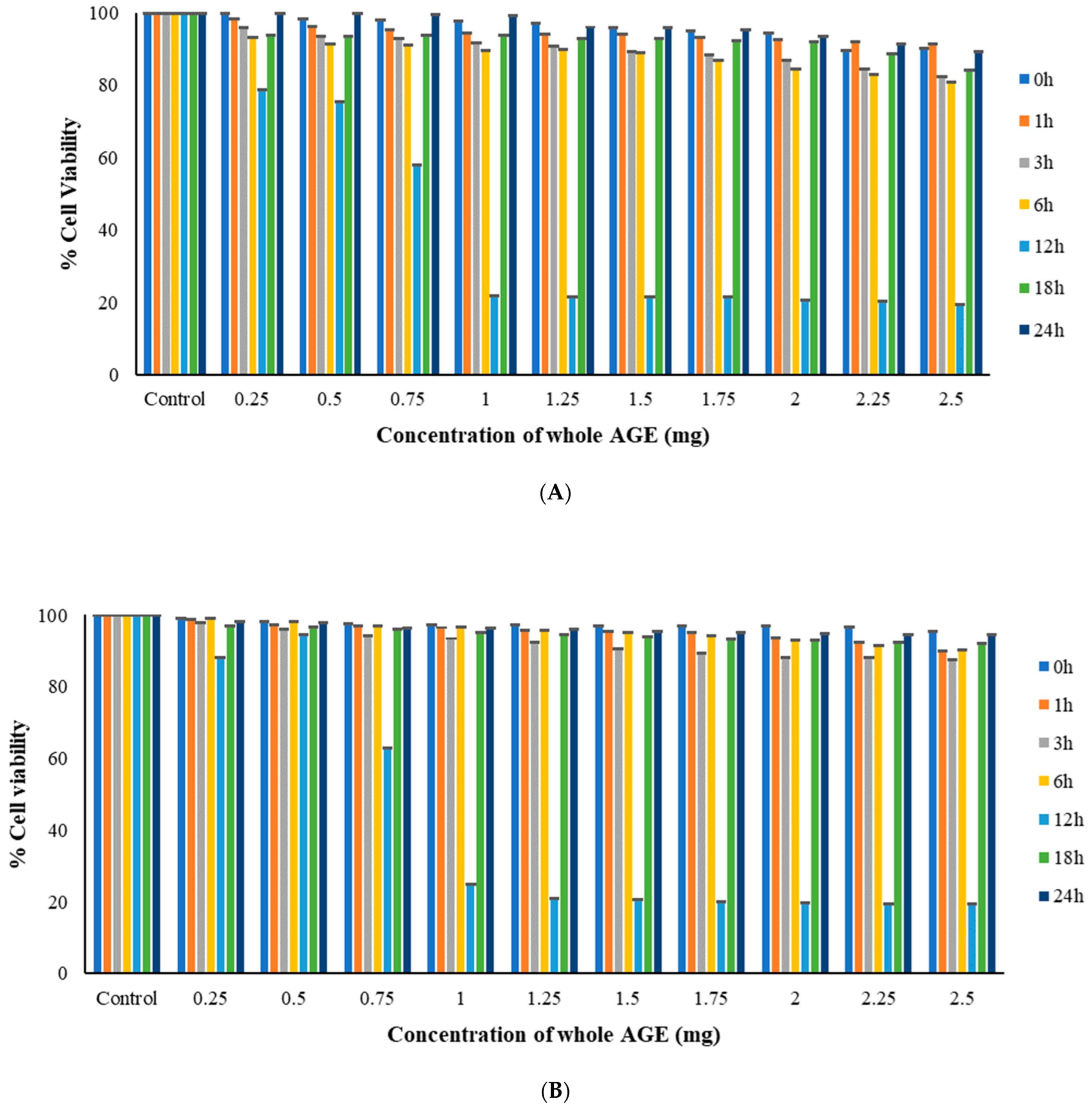
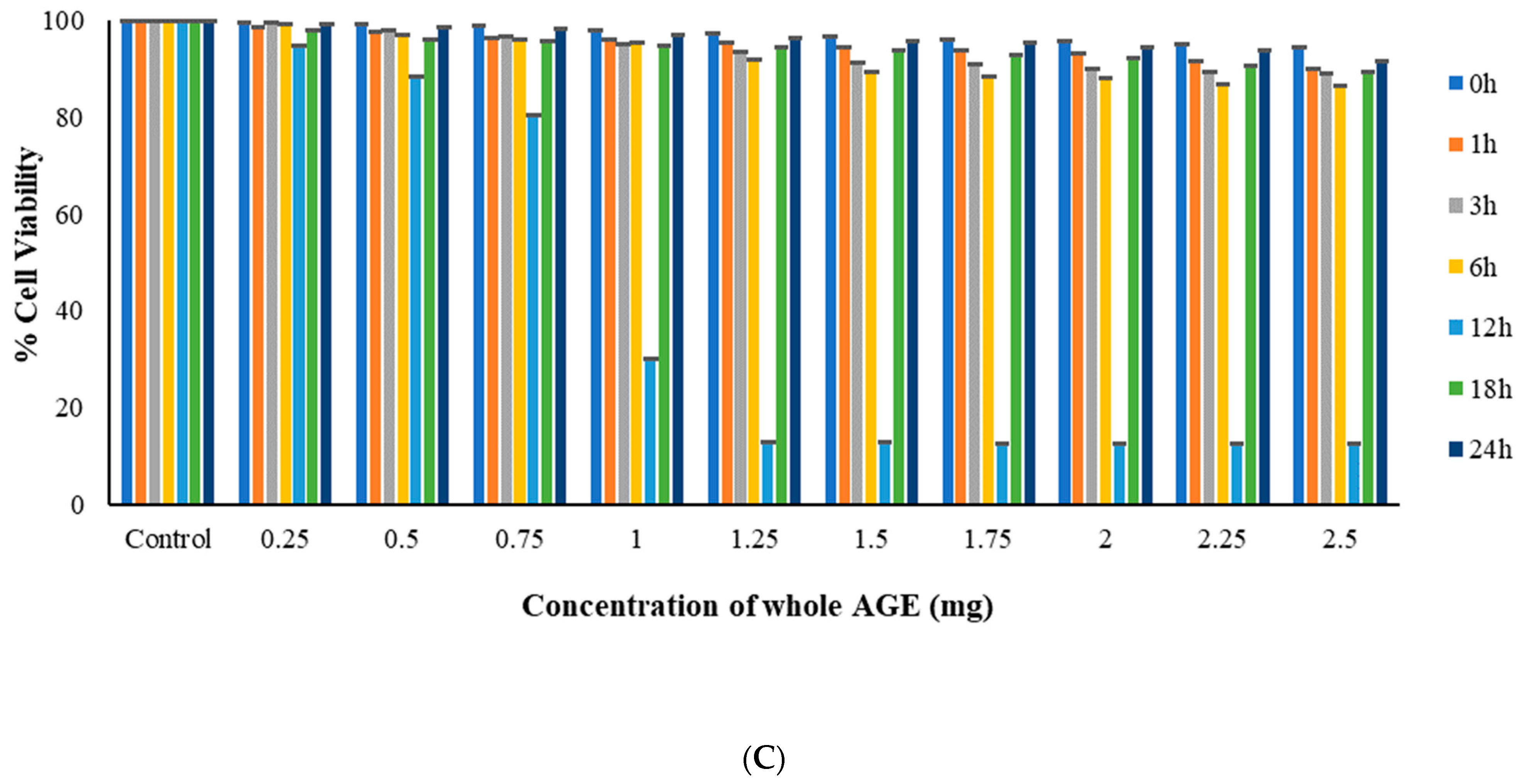
2.6. MTT Assay
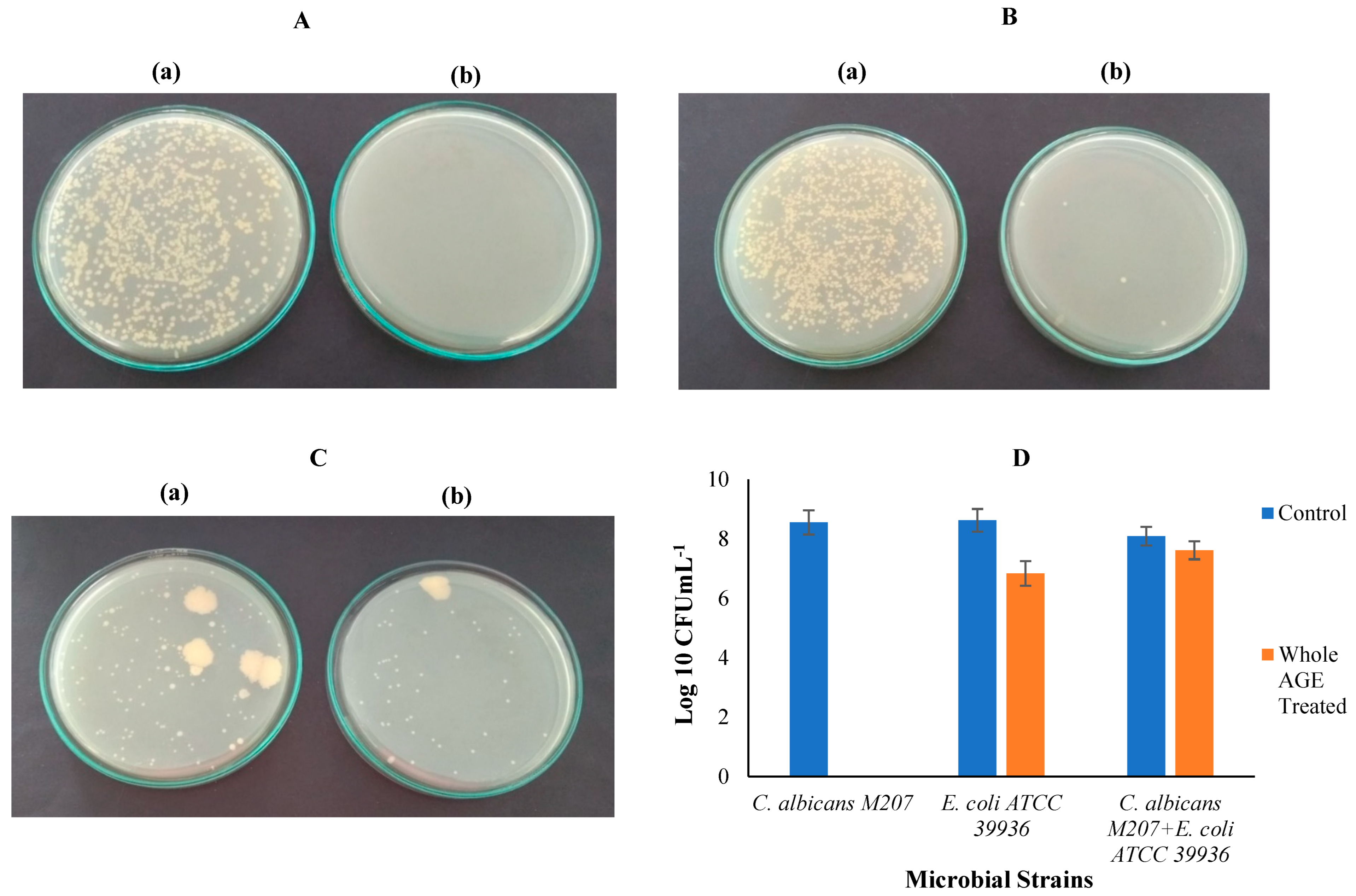
2.7. Colony-Forming Unit (CFU)
2.8. Biofilm Inhibition in Petri dish
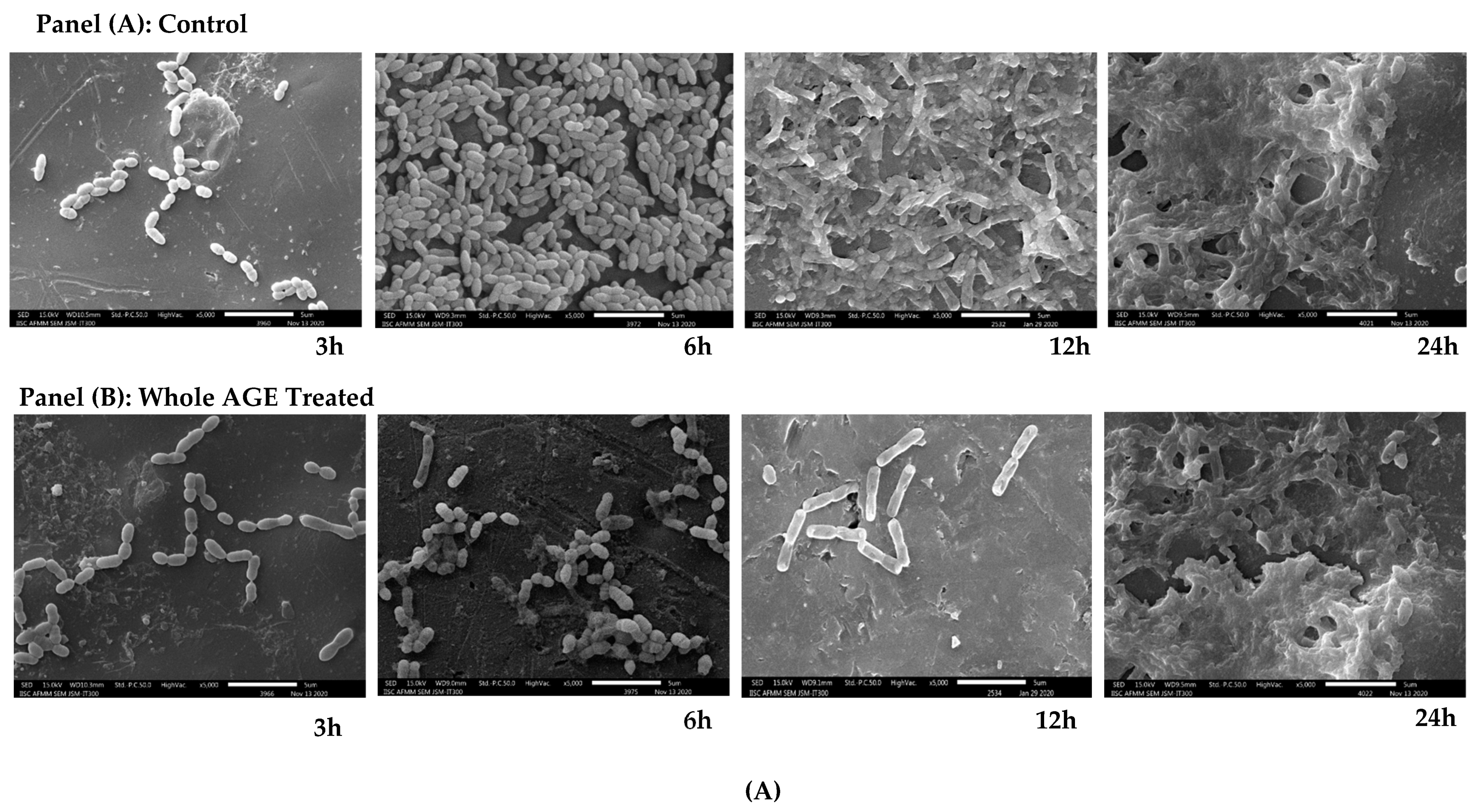
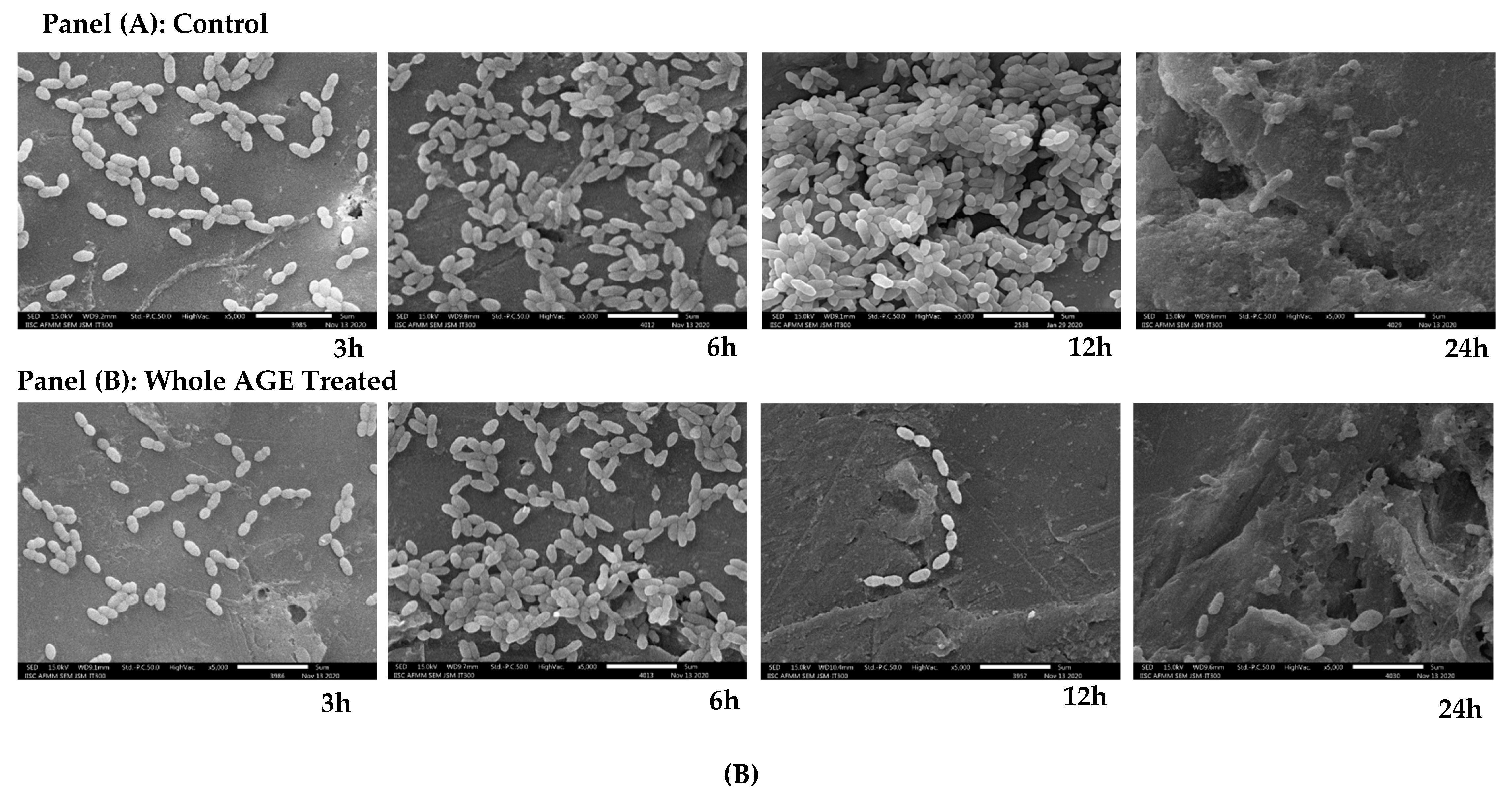
2.9. Scanning Electron Microscopy (SEM)
2.10. Confocal Laser Scanning Microscopy (CLSM)
3. Materials and Methods
3.1. Subculturing and Maintenance of Cultures
3.2. Microbial Cultures and Identification
3.3. VITEK-MALDI TOF Identification
3.4. Point Inoculation
3.5. Substrate Material
3.6. Biofilm Control Using Natural Antimicrobial Agents
Preparation of Aqueous Extracts
3.7. Agar Well Diffusion Method
3.8. VITEK Antimicrobial Susceptibility Test (AST)
3.9. Biofilm Formation on Polystyrene Material
3.9.1. Broth Microdilution Method to Determine the Minimum Inhibitory Concentration (MIC)
3.9.2. MTT Assay
3.10. Colony-Forming Unit (CFU)
3.11. Biofilm Inhibition in Petri dish
3.12. Scanning Electron Microscope (SEM)
3.13. Confocal Laser Scanning Microscopy (CLSM)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGE | Aqueous Garlic Extract |
| ECM | Extracellular Matrix |
| MDR | Multi Drug Resistant |
| Ampicillin | Amp |
References
- Harriott, M.M.; Noverr, M.C. Importance of Candida–Bacterial Polymicrobial Biofilms in Disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, Y.Y.; Wang, Y.; Wang, X.; Hamblin, M.R. Antimicrobial Photodynamic Therapy to Control Clinically Relevant Biofilm Infections. Front. Microbiol. 2018, 9, 1299. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Chen, M.; Ma, K.; Wang, T.; Wu, D.; Yan, G.; Wang, C.; Shao, J. Abundance Interaction in Candida albicans and Candida glabrata Mixed Biofilms under Diverse Conditions. Med. Mycol. 2021, 59, 158–167. [Google Scholar] [CrossRef]
- Liu, X.; Ma, Z.; Zhang, J.; Yang, L. Antifungal Compounds against Candida Infections from Traditional Chinese Medicine. Biomed Res. Int. 2017, 2017, 4614183. [Google Scholar] [CrossRef]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. Adhesion, Biofilm Formation, Cell Surface Hydrophobicity, and Antifungal Planktonic Susceptibility: Relationship among Candida spp. Front. Microbiol. 2015, 6, 205. [Google Scholar] [CrossRef] [PubMed]
- El-Azizi, M.; Farag, N.; Khardori, N. Antifungal Activity of Amphotericin B and Voriconazole against the Biofilms and Biofilm-Dispersed Cells of Candida albicans Employing a Newly Developed in Vitro Pharmacokinetic Model. Ann. Clin. Microbiol. Antibact. 2012, 14, 1–9. [Google Scholar] [CrossRef][Green Version]
- Ramage, G.; Vande Walle, K.; Wickes, B.L.; López-Ribot, J.L. Standardized Method for in Vitro Antifungal Susceptibility Testing of Candida albicans Biofilms. Antimicrob. Agents Chemother. 2001, 45, 2475–2479. [Google Scholar] [CrossRef]
- Cabral, D.J.; Penumutchu, S.; Norris, C.; Morones-Ramirez, J.R.; Belenky, P. Microbial Competition between Escherichia coli and Candida albicans Reveals a Soluble Fungicidal Factor. Microb. Cell 2018, 5, 249–255. [Google Scholar] [CrossRef]
- Rossi, E.; Cimdins, A.; Luthje, P.; Brauner, A.; Sjoling, A.; Landini, P.; Romling, U. “It’s a Gut Feeling”—Escherichia coli Biofilm Formation in the Gastrointestinal Tract Environment. Crit. Rev. Microbiol. 2018, 44, 1–30. [Google Scholar] [CrossRef]
- Bandara, H.M.H.N.; Yau, J.Y.Y.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Escherichia coli and Its Lipopolysaccharide Modulate in Vitro Candida Biofilm Formation. J. Med. Microbiol. 2009, 58, 1623–1631. [Google Scholar] [CrossRef]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-Kingdom Interactions: Candida albicans and Bacteria. FEMS Microbiol. Lett. 2009, 299, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Corte, L.; Pierantoni, D.C.; Tascini, C.; Roscini, L.; Cardinali, G. Biofilm Specific Activity: A Measure to Quantify Microbial Biofilm. Microorganisms 2019, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, A.; Kordbacheh, P.; Nazari, A.; Ghazvini, R.D.; Mirhendi, H.; Safara, M.; Bakhshi, H.; Yaghoubi, R. Candiduria in Hospitalized Patients and Identification of Isolated Candida Species by Morphological and Molecular Methods in Ilam. Iran. J. Public Health 2019, 48, 156. [Google Scholar]
- Gajdacs, M.; Doczi, I.; Abrok, M.; Lazar, A.; Burian, K. Epidemiology of Candiduria and Candida Urinary Tract Infections in Inpatients and Outpatients: Results from a 10-Year Retrospective Survey. Cent. Eur. J. Urol. 2019, 72, 209–214. [Google Scholar] [CrossRef]
- He, Z.; Huo, X.; Lei, D.; Zhao, H.; Jia, K.; Wang, F. Management of Candiduria in Hospitalized Patients: A Single-Center Study on the Implementation of IDSA Guidelines and Factors Affecting Clinical Decisions. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 59–65. [Google Scholar] [CrossRef]
- Dias, V. Candida Species in the Urinary Tract: Is It a Fungal Infection or Not? Future Microbiol. 2020, 15, 81–83. [Google Scholar] [CrossRef]
- Willems, H.M.; Xu, Z.; Peters, B.M. Polymicrobial Biofilm Studies: From Basic Science to Biofilm Control. Curr. Oral Health Rep. 2016, 3, 36–44. [Google Scholar] [CrossRef]
- Liu, W.; Roder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sorensen, S.J.; Burmølle, M. Interspecific Bacterial Interactions Are Reflected in Multispecies Biofilm Spatial Organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef]
- Gabrilska, R.A.; Rumbaugh, K.P. Biofilm Models of Polymicrobial Infection. Future Microbiol. 2015, 10, 1997–2015. [Google Scholar] [CrossRef]
- Rodrigues, M.E.; Gomes, F.; Rodrigues, C.F. Candida spp./Bacteria Mixed Biofilms. J. Fungi 2019, 6, 5. [Google Scholar] [CrossRef]
- Haque, F.; Alfatah, M.; Ganesan, K.; Shankar Bhattacharyya, M. Inhibitory Effect of Sophorolipid on Candida albicans Biofilm Formation and Hyphal Growth. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Shao, J.; Da, W.; Shi, G.; Wang, T.; Wu, D.; Wang, C. Decreasing Cell Population of Individual Candida Species Does Not Impair the Virulence of Candida albicans and Candida glabrata Mixed Biofilms. Front. Microbiol. 2019, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- De Brucker, K.; Tan, Y.; Vints, K.; De Cremer, K.; Braem, A.; Verstraeten, N.; Michiels, J.; Vleugels, J.; Cammue, B.P.; Thevissen, K. Fungal β-1,3-Glucan Increases Ofloxacin Tolerance of Escherichia coli in a Polymicrobial E. Coli/Candida albicans Biofilm. Antimicrob. Agents Chemother. 2022, 59, 3052–3058. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.M.; Felicio, M.R.; Boas, E.V.; Gonçalves, S.; Costa, F.F.; Samy, R.P.; Santos, N.C.; Franco, O.L. New Frontiers for Anti-Biofilm Drug Development. Pharmacol. Ther. 2016, 160, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Sadanandan, B.; Ashrit, P.; Nataraj, L.K.; Shetty, K.; Jogalekar, A.P.; Vaniyamparambath, V.; Hemanth, B. High Throughput Comparative Assessment of Biofilm Formation of Candida glabrata on Polystyrene Material. Korean J. Chem. Eng. 2022, 1–10. [Google Scholar] [CrossRef]
- La Cruz-Claure, D.; Maria, L.; Cespedes-Llave, A.A.; Ulloa, M.T.; Benito-Lama, M.; Dominguez-Alvarez, E.; Bastida, A. Inhibition-Disruption of Candida glabrata Biofilms: Symmetrical Selenoesters as Potential Anti-Biofilm Agents. Microorganisms 2019, 7, 664. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerson, J.W.; Shirtliff, M.E. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival Strategies of Infectious Biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef]
- Harding, J.L.; Reynolds, M.M. Combating Medical Device Fouling. Trends Biotechnol. 2014, 32, 140–146. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.-F.; Alarcon, E.I. Bacterial Biofilm Formation on Implantable Devices and Approaches to Its Treatment and Prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Sadanandan, B.; Vaniyamparambath, V.; Lokesh, K.N.; Shetty, K.; Joglekar, A.P.; Ashrit, P.; Hemanth, B. Candida albicans Biofilm Formation and Growth Optimization for Functional Studies Using Response Surface Methodology. J. Appl. Microbiol. 2022, 132, 3277–3292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liang, E.; Cheng, Y.; Mahmood, T.; Ge, F.; Zhou, K.; Bao, M.; Lv, L.; Li, L.; Yi, J.; et al. Is Combined Medication with Natural Medicine a Promising Therapy for Bacterial Biofilm Infection? Biomed. Pharmacother. 2020, 128, 110184. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Xia, G.; Shi, C.; Wan, J.; Liu, L.; Chen, Y.; Wu, Y.; Zhang, W.; Zhou, M.; He, H.; et al. Therapeutic Strategies against Bacterial Biofilms. Fundam. Res. 2021, 1, 193–212. [Google Scholar] [CrossRef]
- Qu, Y.; Locock, K.; Verma-Gaur, J.; Hay, I.D.; Meagher, L.; Traven, A. Searching for New Strategies against Polymicrobial Biofilm Infections: Guanylated Polymethacrylates Kill Mixed Fungal/Bacterial Biofilms. J. Antimicrob. Chemother. 2016, 71, 413–421. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural Anti-Biofilm Agents: Strategies to Control Biofilm-Forming Pathogens. Front. Microbiol. 2020, 11, 2640. [Google Scholar] [CrossRef]
- Li, G.; Ma, X.; Deng, L.; Zhao, X.; Wei, Y.; Gao, Z.; Jia, J.; Xu, J.; Sun, C. Fresh Garlic Extract Enhances the Antimicrobial Activities of Antibiotics on Resistant Strains in Vitro. Jundishapur J. Microbiol. 2015, 8, 1–6. [Google Scholar] [CrossRef]
- Mortale, S.P.; Karuppayil, S.M. Review on Combinatorial Approach for Inhibiting Candida albicans Biofilm. Am. J. Clin. Microbiol. Antimicrob. 2018, 1, 1–10. [Google Scholar]
- Patel, R. A Moldy Application of MALDI: MALDI-ToF Mass Spectrometry for Fungal Identification. J. Fungi 2019, 5, 4. [Google Scholar] [CrossRef]
- Guo, L.; Ye, L.; Zhao, Q.; Ma, Y.; Yang, J.; Luo, Y. Comparative Study of MALDI-TOF MS and VITEK 2 in Bacteria Identification. J. Thorac. Dis. 2014, 6, 534–538. [Google Scholar] [CrossRef]
- Gonçalves, B.; Fernandes, L.; Henriques, M.; Silva, S.; Gonc¸alves, B.; Onia Silva, S. Environmental PH Modulates Biofilm Formation and Matrix Composition in Candida albicans and Candida glabrata glabrata. Biofouling 2020, 36, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Cobrado, L.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. In Vitro Antifungal Activity and in Vivo Antibiofilm Activity of Cerium Nitrate against Candida Species. J. Antimicrob. Chemother. 2015, 70, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.; Kastora, S.L.; Gomes-Gonçalves, A.; Azevedo, N.; Rodrigues, C.F.; Silva, S.; Demuyser, L.; Van Dijck, P.; Casal, M.; Brown, A.J.P.; et al. ARTICLE Transcriptional Responses of Candida glabrata Biofilm Cells to Fluconazole Are Modulated by the Carbon Source. NPJ Biofilms Microbiomes 2020, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.S.; Looi, L.J.; Ong, B.S.; Chong, P.P. Antifungal and Anti-Biofilm Effects of Shallot (Allium ascalonicum) Aqueous Extract on Candida albicans. J. Herbmed Pharmacol. 2018, 7, 236–242. [Google Scholar] [CrossRef]
- Bouza, E.; Guinea, J.; Guembe, M.A. The Role of Antifungals against Candida Biofilm in Catheter-Related Candidemia. Antibiotics 2015, 4, 1–17. [Google Scholar] [CrossRef]
- Yu, S.; Zhu, X.; Zhou, J.; Cai, Z. Biofilm Inhibition and Pathogenicity Attenuation in Bacteria by Proteus Mirabilis. R. Soc. Open Sci. 2018, 5, 170702. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, L.; Wakabayashi, H.; Myers, J.; Jiang, Y.; Cao, Y.; Jimenez-Ortigosa, C.; Perlin, D.S.; Rustchenko, E. Tolerance to Caspofungin in Candida albicans Is Associated with at Least Three Distinctive Mechanisms That Govern Expression of FKS Genes and Cell Wall Remodeling. Antimicrob. Agents Chemother. 2017, 61, e00071-17. [Google Scholar] [CrossRef]
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front. Microbiol. 2017, 7, 2173. [Google Scholar] [CrossRef]
- Perlin, D.S. Echinocandin Resistance in Candida. Clin. Infect. Dis. 2015, 61, 612–617. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of Minimum Inhibitory Concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.L.; Jayaraman, A.; Kao, K.C. Relative Abundances of Candida albicans and Candida glabrata in in Vitro Coculture Biofilms Impact Biofilm Structure and Formation. Appl. Environ. Microbiol. 2018, 84, e02769-17. [Google Scholar] [CrossRef] [PubMed]
- Kruger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The Polymicrobial Nature of Biofilm Infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE 2011, 6, 27317. [Google Scholar] [CrossRef]
- Orazi, G.; O’Toole, G.A. “It Takes a Village”: Mechanisms Underlying Antimicrobial Recalcitrance of Polymicrobial Biofilms. J. Bacteriol. 2020, 202, e00530-19. [Google Scholar] [CrossRef] [PubMed]
- Zago, C.E.; Silva, S.; Sanitá, P.V.; Barbugli, P.A.; Maria, C.; Dias, I.; Lordello, V.B.; Vergani, C.E. Dynamics of Biofilm Formation and the Interaction between Candida albicans and Methicillin-Susceptible (MSSA) and Resistant Staphylococcus aureus (MRSA). PLoS ONE 2015, 4, e0123206. [Google Scholar] [CrossRef]
- Kim, H.-R.; Eom, Y.-B. Antifungal and Anti-Biofilm Effects of 6-Shogaol against Candida auris. J. Appl. Microbiol. 2020, 130, 1142–1153. [Google Scholar] [CrossRef]
- Bayan, L.; Koulivand, P.H.; Gorji, A. Garlic: A Review of Potential Therapeutic Effects. Avicenna J. Phytomedicine 2014, 4, 1. [Google Scholar]
- Wu, X.; Santos, R.R.; Fink-Gremmels, J.; Johanna Fink-Gremmels, C. Analyzing the Antibacterial Effects of Food Ingredients: Model Experiments with Allicin and Garlic Extracts on Biofilm Formation and Viability of Staphylococcus epidermidis. Food Sci. Nutr. 2015, 3, 158–168. [Google Scholar] [CrossRef]
- Choo, S.; Chin, V.K.; Wong, H.; Madhavan, P.; Tay, S.T.; Voon, P.; Yong, C.; Chong, P.P. Antimicrobial Properties of Allicin Used Alone or in Combination with Other Medications. Folia Microbiol. 2020, 65, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Leontiev, R.; Hohaus, N.; Jacob, C.; Gruhlke, M.C.H.; Slusarenko, A.J. A Comparison of the Antibacterial and Antifungal Activities of Thiosulfinate Analogues of Allicin. Sci. Rep. 2018, 8, 6763. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, K.S.; Han, I.; Kim, M.-H.; Jung, M.H. Quantitative and Qualitative Analysis of the Antifungal Activity of Allicin Alone and in Combination with Antifungal Drugs. PLoS ONE 2012, 7, 38242. [Google Scholar] [CrossRef]
- Bachrach, G.; Jamil, A.; Naor, R.; Tal, G.; Ludmer, Z.; Steinberg, D. Garlic Allicin as a Potential Agent for Controlling Oral Pathogens. J. Med. Food 2011, 14, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.; Wilson, E.D.; et al. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-Review. J. Engg. Tech. 2017, 6, 1–42. [Google Scholar]
- Bor, B.; Cen, L.; Agnello, M.; Shi, W.; He, X. Morphological and Physiological Changes Induced by Contact-Dependent Interaction between Candida albicans and Fusobacterium nucleatum. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, S.; De Bok, F.A.M.; Mols, E.; De Vos, W.M.; Van Hylckama Vlieg, J.E.T. A Simple and Fast Method for Determining Colony Forming Units. Lett. Appl. Microbiol. 2008, 47, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Shah, S.; Tammela, P. Defining Conditions for Biofilm Inhibition and Eradication Assays for Gram-Positive Clinical Reference Strains. BMC Microbiol. 2018, 18, 173. [Google Scholar] [CrossRef]
- Herten, M.; Bisdas, T.; Knaack, D.; Becker, K.; Osada, N.; Torsello, G.B.; Idelevich, E.A. Rapid in Vitro Quantification of S. Aureus Biofilms on Vascular Graft Surfaces. Front. Microbiol. 2017, 8, 2333. [Google Scholar] [CrossRef]
- Park, S.J.; Han, K.-H.; Park, J.Y.; Choi, S.J.; Lee, K.-H. Influence of Bacterial Presence on Biofilm Formation of Candida Albicans. YMJ 2014, 55, 449–458. [Google Scholar] [CrossRef]
- Bandara, H.M.; Cheung, B.P.K.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Secretory Products of Escherichia Coli Biofilm Modulate Candida Biofilm Formation and Hyphal Development. J. Investig. Clin. Dent. 2013, 4, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Sammons, R.L.; Addison, O.; Dehghani, H.; Walmsley, A.D. A Quantitative Method to Measure Biofilm Removal Efficiency from Complex Biomaterial Surfaces Using SEM and Image Analysis. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.; Yu, A.; Fidel, P.L.; Noverr, M.C. Transcription Factors Efg1 and Bcr1 Regulate Biofilm Formation and Virulence during Candida albicans-Associated Denture Stomatitis. PLoS ONE 2016, 11, e0159692. [Google Scholar] [CrossRef]
- Kang, J.; Li, Q.; Liu, L.; Jin, W.; Wang, J.; Sun, Y. The Specific Effect of Gallic Acid on Escherichia coli Biofilm Formation by Regulating PgaABCD Genes Expression. Appl. Microbiol. Biotechnol. 2018, 102, 1837–1846. [Google Scholar] [CrossRef]
- Catto, C.; Secundo, F.; James, G.; Villa, F.; Cappitelli, F. α-Chymotrypsin Immobilized on a Low-Density Polyethylene Surface Successfully Weakens Escherichia coli Biofilm Formation. Intl. J. Mol. Sci. 2018, 19, 4003. [Google Scholar] [CrossRef]
- Watters, C.; Fleming, D.; Bishop, D.; Rumbaugh, K.P. Host Responses to Biofilm. Prog. Mol. Biol. Transl. Sci. 2016, 142, 193–239. [Google Scholar] [CrossRef]
- Batoni, G.; Martinez-Pomares, L.; Esin, S. Editorial: Immune Response to Biofilms. Front. Immunol. 2021, 12, 696356. [Google Scholar] [CrossRef]
- Miquel, S.; Lagrafeuille, R.; Souweine, B.; Forestier, C. Anti-Biofilm Activity as a Health Issue. Front. Microbiol. 2016, 7, 592. [Google Scholar] [CrossRef]
- Vidal-Acuna, M.R.; Ruiz-Perez de Pipaon, M.; Torres-Sanchez, M.J.; Aznar, J. Identification of Clinical Isolates of Aspergillus, Including Cryptic Species, by Matrix Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). Med. Mycol. 2018, 56, 838–846. [Google Scholar] [CrossRef]
- Dahiya, P.; Purkayastha, S. Phytochemical Screening and Antimicrobial Activity of Some Medicinal Plants against Multi-Drug Resistant Bacteria from Clinical Isolates. Ind. J. Pharm. Sci. 2012, 74, 443. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Ligozzi, M.; Bernini, C.; Bonora, M.G.; De Fatima, M.; Zuliani, J.; Fontana, R. Evaluation of the VITEK 2 System for Identification and Antimicrobial Susceptibility Testing of Medically Relevant Gram-Positive Cocci. J. Clin. Microbiol. 2002, 40, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Mukherjee, P.K.; Leidich, S.D.; Faddoul, F.F.; Hoyer, L.L.; Douglas, L.J.; Ghannoum, M.A. Antifungal Resistance of Candidal Biofilms Formed on Denture Acrylic In Vitro. J. Dent. Res. 2001, 80, 903–908. [Google Scholar] [CrossRef]
- Nagashima, M.; Yamagishi, Y.; Mikamo, H. Antifungal Susceptibilities of Candida Species Isolated from the Patients with Vaginal Candidiasis. J. Infect. Chemother. 2016, 22, 124–126. [Google Scholar] [CrossRef]
- Trafny, E.A.; Lewandowski, R.; Zawistowska-Marciniak, I.; Stepińska, M. Use of MTT Assay for Determination of the Biofilm Formation Capacity of Microorganisms in Metalworking Fluids. World J. Microbiol. Biotechnol. 2013, 29, 1635–1643. [Google Scholar] [CrossRef]
- Montelongo-Jauregui, D.; Srinivasan, A.; Ramasubramanian, A.; Lopez-Ribot, J. An In Vitro Model for Candida Albicans–Streptococcus Gordonii Biofilms on Titanium Surfaces. J. Fungi 2018, 4, 66. [Google Scholar] [CrossRef]

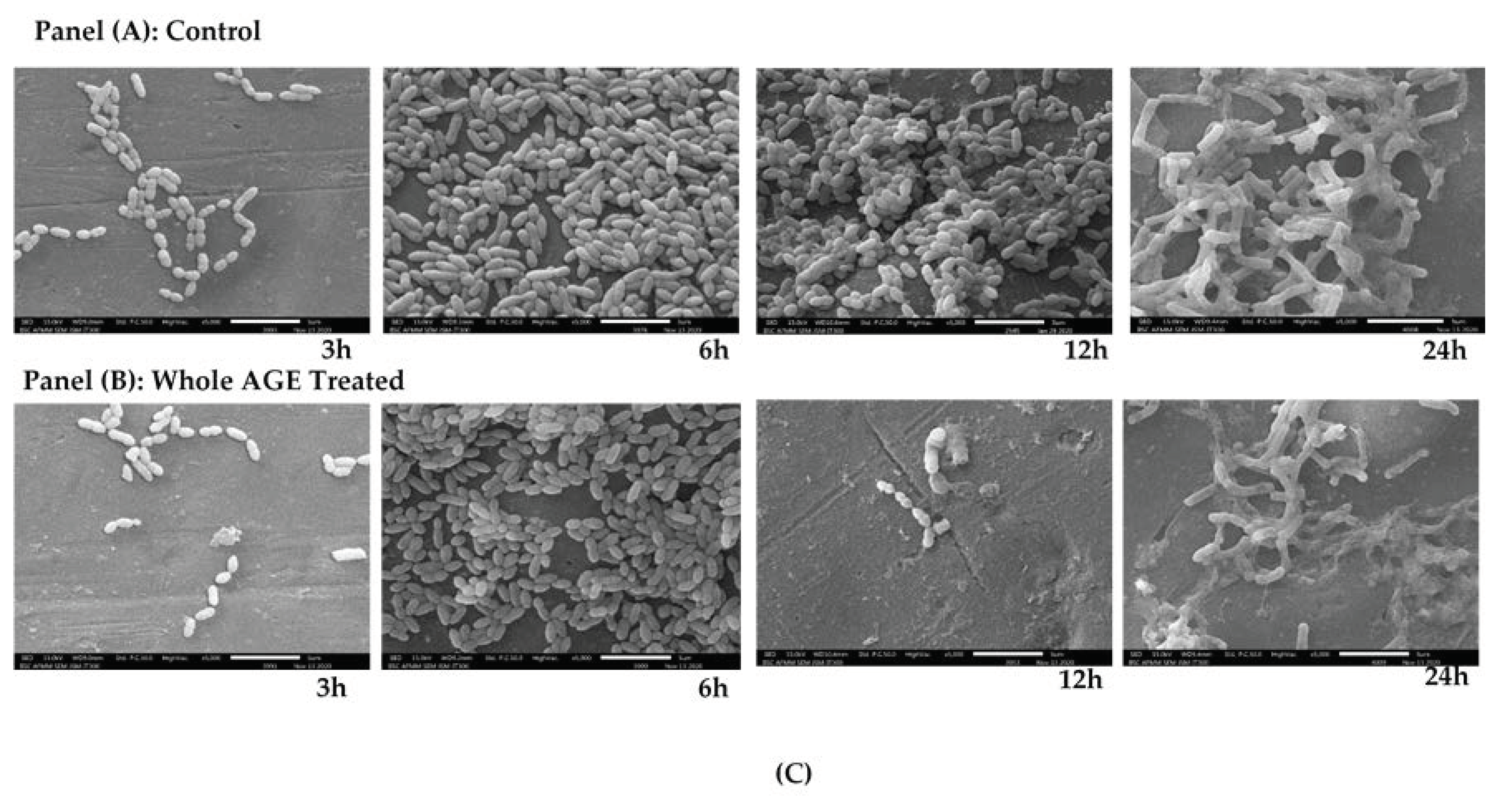
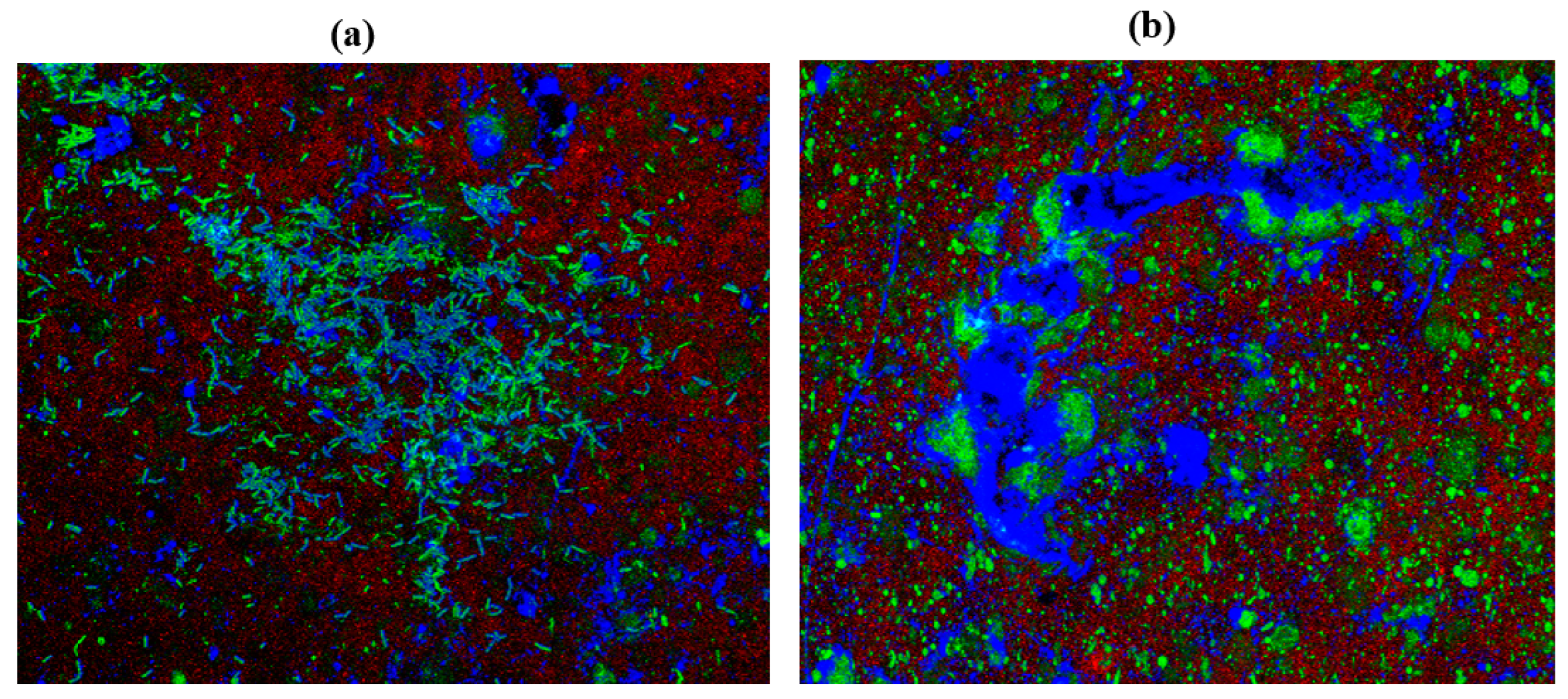
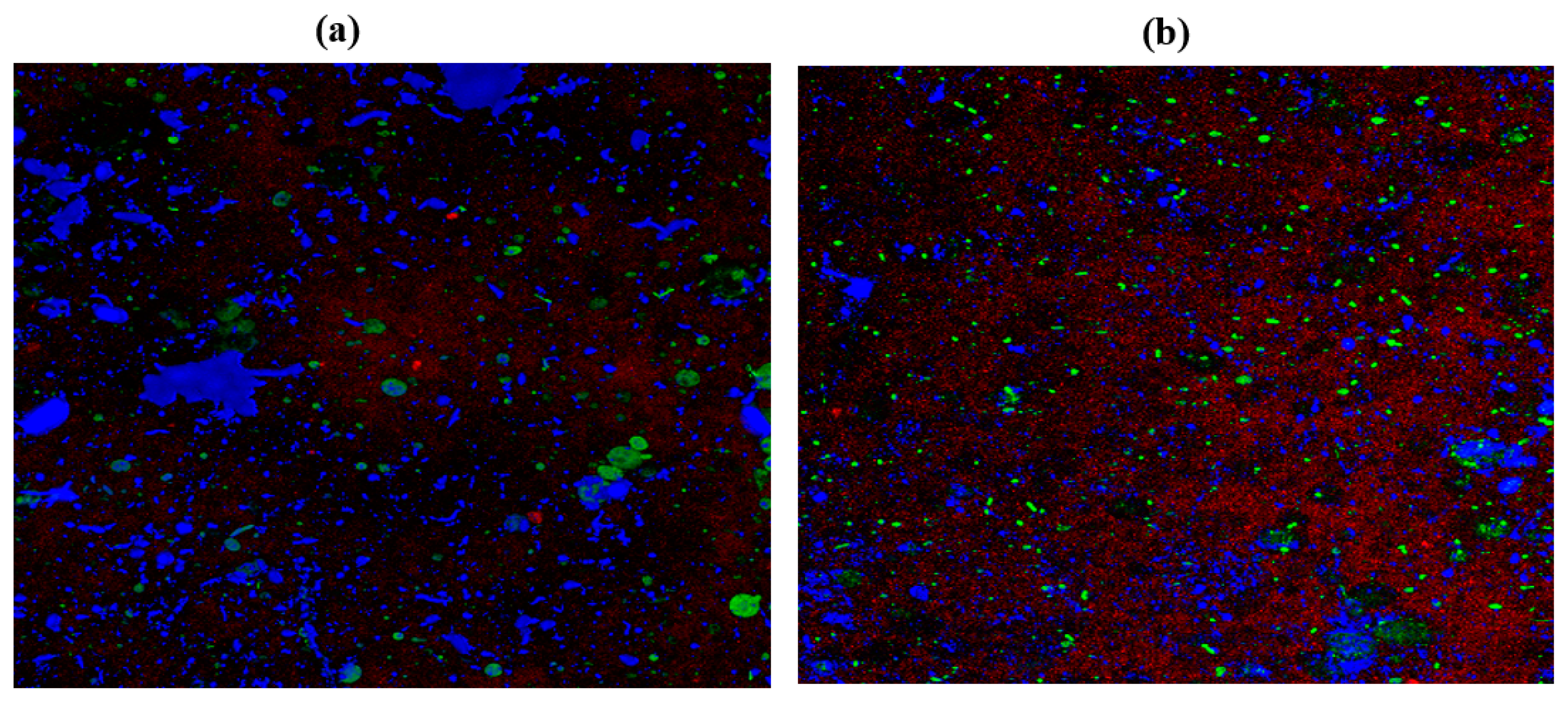
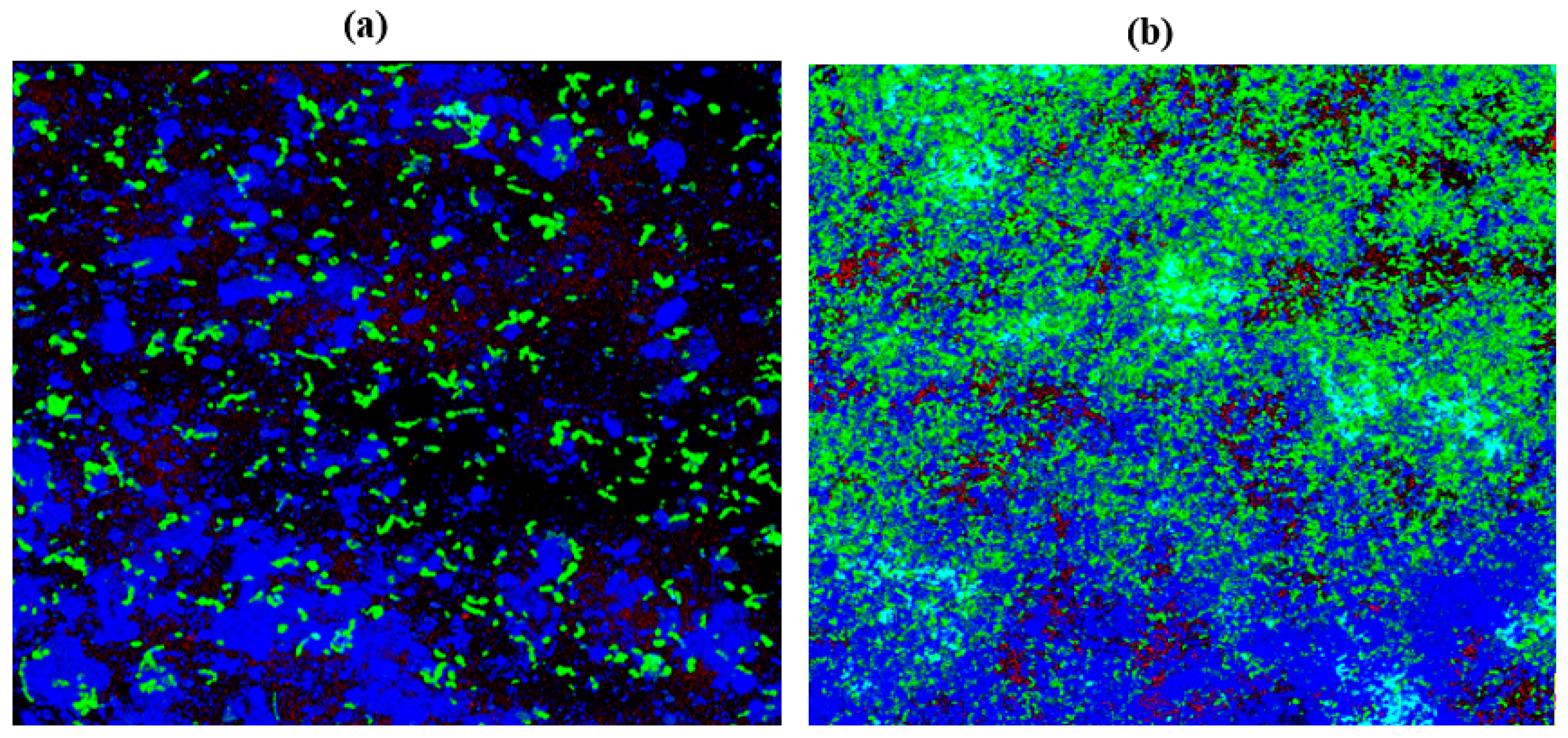
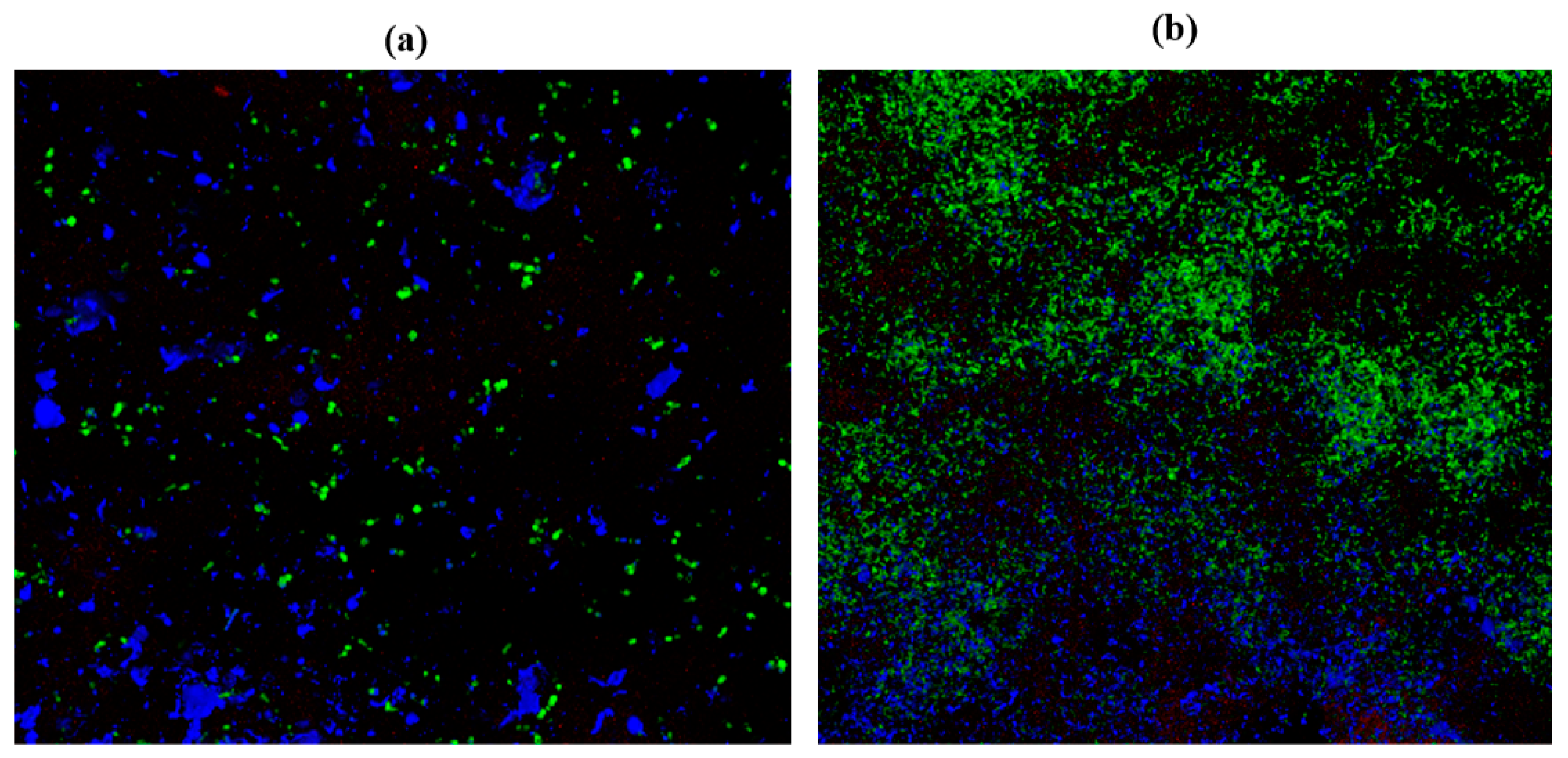
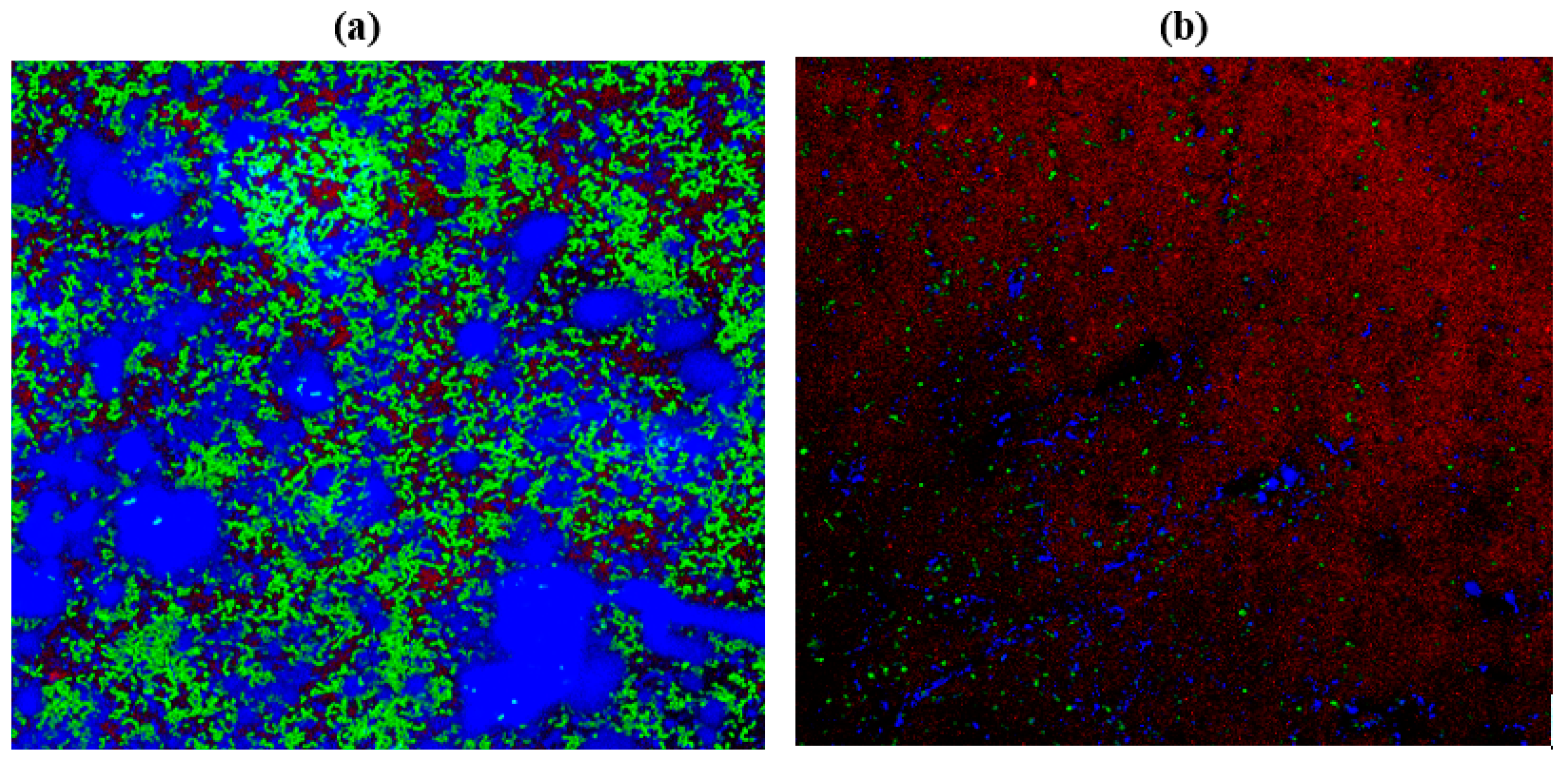

| Cultures | ZOI for Garlic (Radii Measurements in mm) | ZOI for Gooseberry (Radii Measurements in mm) | ZOI for Clove (Radii Measurements in mm) |
|---|---|---|---|
| C. albicans M207 | 13 | - | 07 |
| E. coli ATCC 39936 | 12 | - | 5.5 |
| C. albicans M207+ E. coli ATCC 39936 | 11 | - | 06 |
| Sl. No | Cultures | No of Colonies (CFUmL−1)-Control Samples | No of Colonies (CFUmL−1) AGE Treated |
|---|---|---|---|
| 1 | C. albicans M207 | 354 × 106 | 0 |
| 2 | E. coli ATCC 39936 | 417 × 106 | 7 × 106 |
| 3 | C. albicans M207+ E. coli 39936 | 122 × 106 | 41 × 106 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashrit, P.; Sadanandan, B.; Shetty, K.; Vaniyamparambath, V. Polymicrobial Biofilm Dynamics of Multidrug-Resistant Candida albicans and Ampicillin-Resistant Escherichia coli and Antimicrobial Inhibition by Aqueous Garlic Extract. Antibiotics 2022, 11, 573. https://doi.org/10.3390/antibiotics11050573
Ashrit P, Sadanandan B, Shetty K, Vaniyamparambath V. Polymicrobial Biofilm Dynamics of Multidrug-Resistant Candida albicans and Ampicillin-Resistant Escherichia coli and Antimicrobial Inhibition by Aqueous Garlic Extract. Antibiotics. 2022; 11(5):573. https://doi.org/10.3390/antibiotics11050573
Chicago/Turabian StyleAshrit, Priya, Bindu Sadanandan, Kalidas Shetty, and Vijayalakshmi Vaniyamparambath. 2022. "Polymicrobial Biofilm Dynamics of Multidrug-Resistant Candida albicans and Ampicillin-Resistant Escherichia coli and Antimicrobial Inhibition by Aqueous Garlic Extract" Antibiotics 11, no. 5: 573. https://doi.org/10.3390/antibiotics11050573
APA StyleAshrit, P., Sadanandan, B., Shetty, K., & Vaniyamparambath, V. (2022). Polymicrobial Biofilm Dynamics of Multidrug-Resistant Candida albicans and Ampicillin-Resistant Escherichia coli and Antimicrobial Inhibition by Aqueous Garlic Extract. Antibiotics, 11(5), 573. https://doi.org/10.3390/antibiotics11050573