Computational Design of Inhibitors Targeting the Catalytic β Subunit of Escherichia coli FOF1-ATP Synthase
Abstract
1. Introduction
2. Results
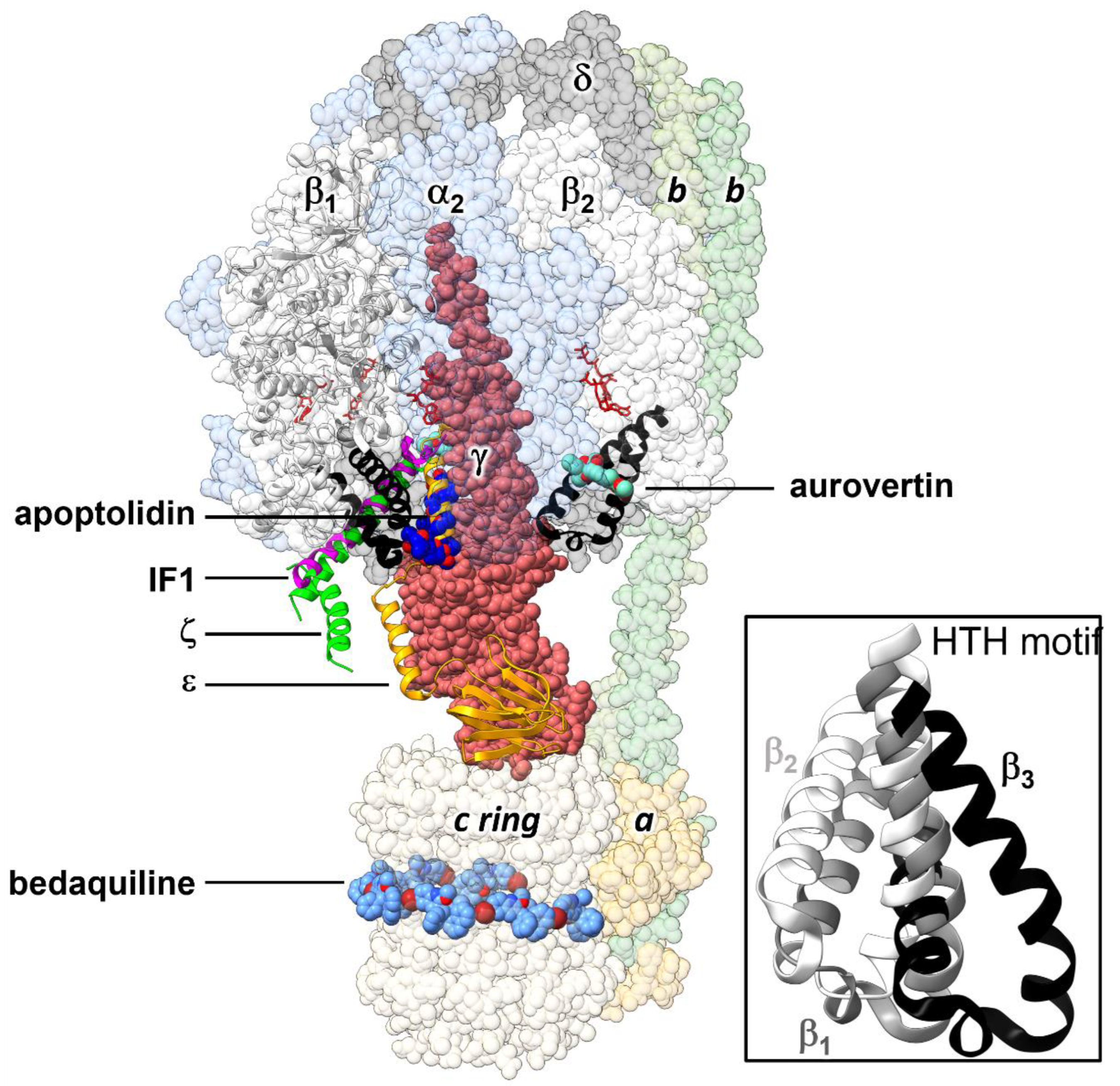
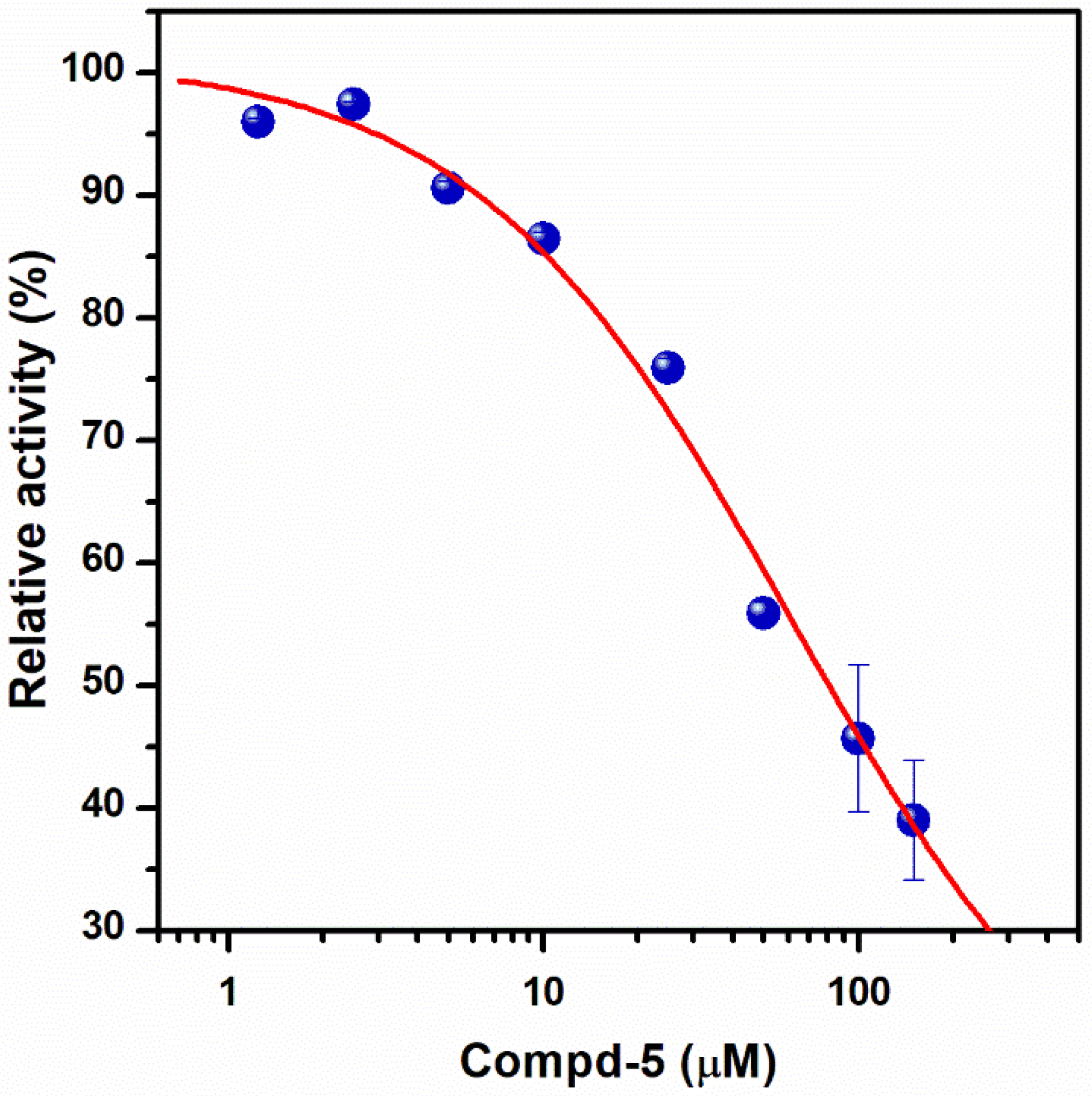
2.1. Structure-Based Design of Small Organic Inhibitors
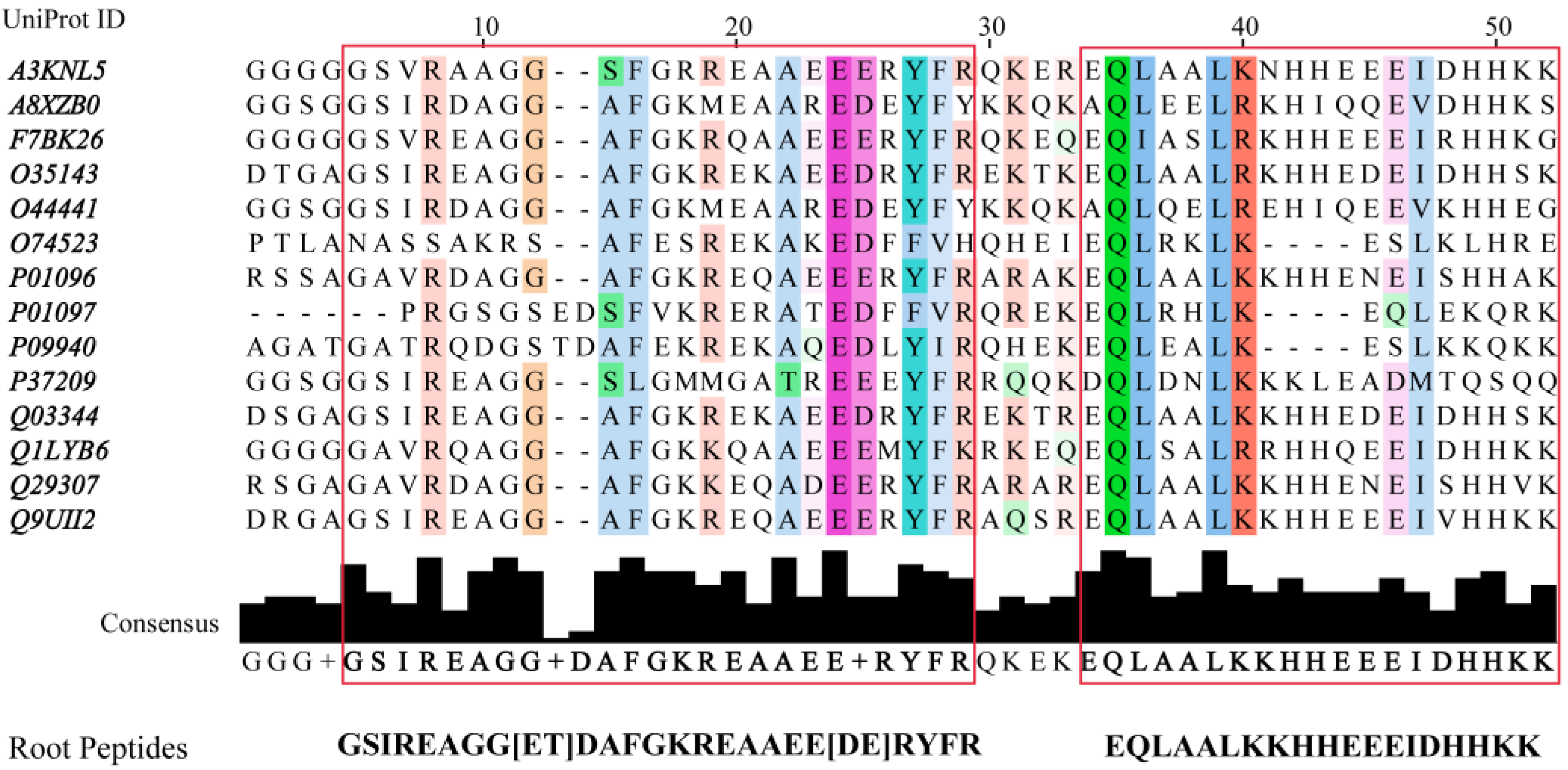
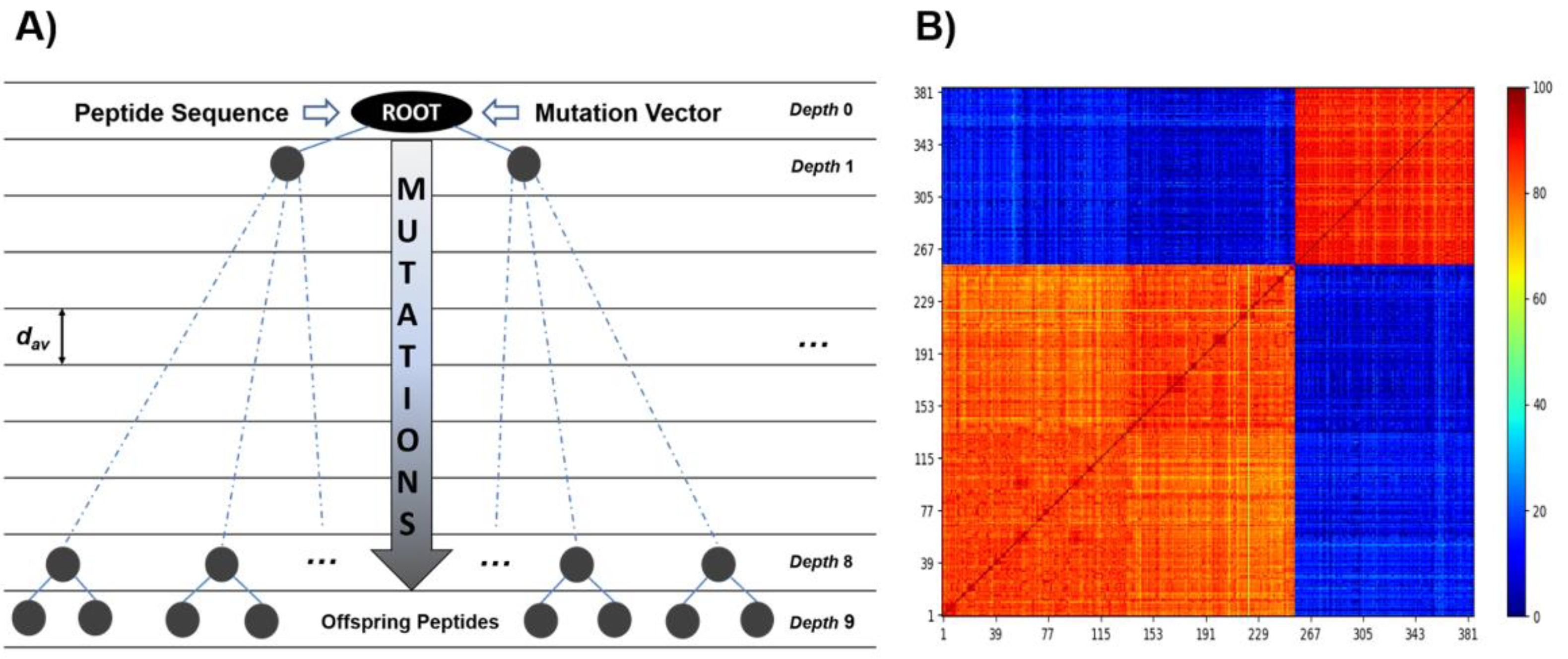
2.2. De Novo Design of Peptide Inhibitors of EcF1 Targeting the βCterm
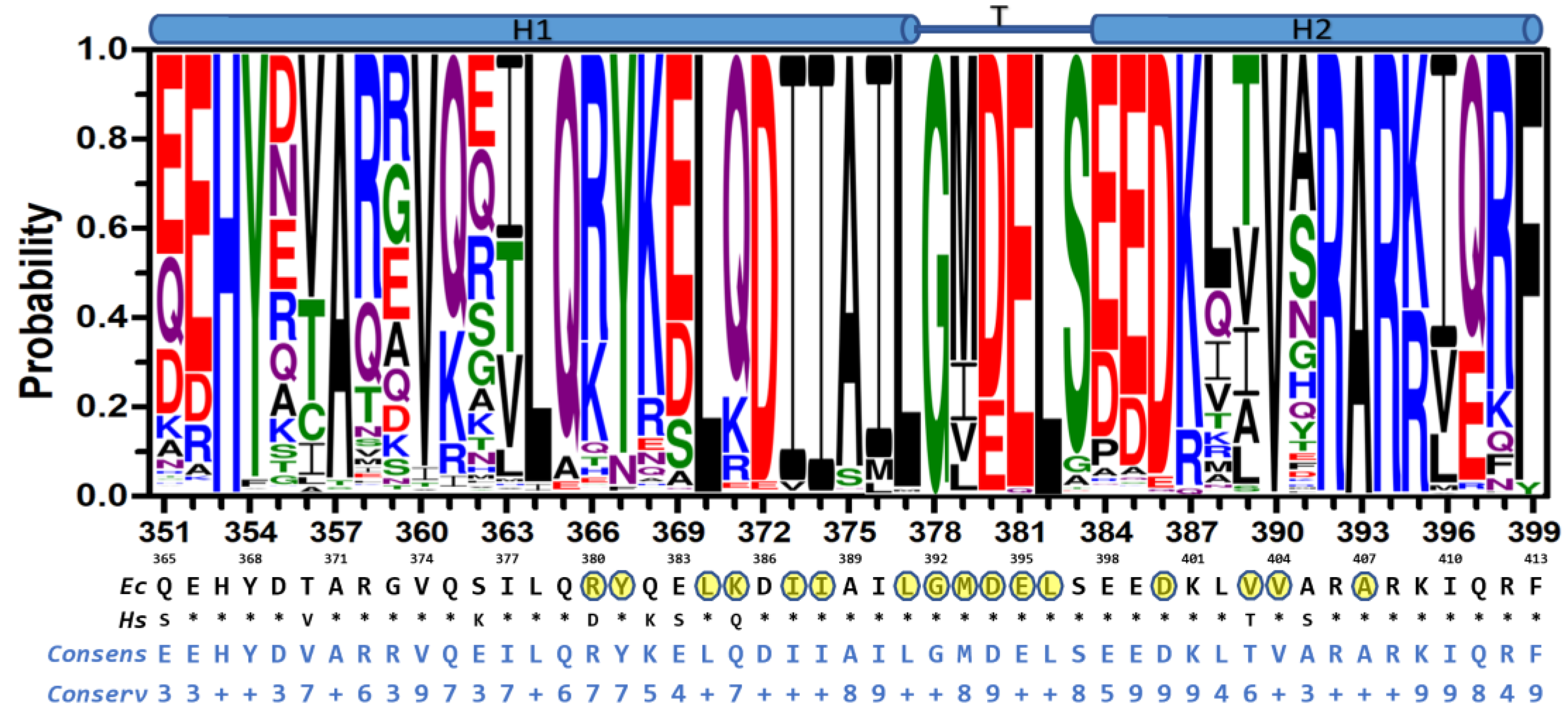
2.3. Sequence Conservation of the HTH Motif in Bacteria
3. Discussion
4. Materials and Methods
4.1. Molecular Dynamics Simulations
4.2. Identification of Solvent Sites, Guided Docking, and Dynamic Undocking
4.3. Engineering Peptide Inhibitors via Evolutionary and Protein–Protein Interaction Algorithms
4.4. Protein Production and Purification
4.5. ATPase Activity Assays
4.6. Sequence Analysis of the HTH Motif
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Casadevall, A. Crisis in infectious diseases: 2 decades later. Clin. Infect. Dis. 2017, 64, 823–828. [Google Scholar] [CrossRef]
- Taylor, J.; Hafner, M.; Yerushalmi, E.; Smith, R.; Bellasio, J.; Vardavas, R.; Bienkowska-gibbs, T.; Rubin, J. Estimating the Economic Costs of Antimicrobial Resistance: Model and Results; RAND Corporation: Cambridge, UK, 2014. [Google Scholar]
- Review on Antimicrobial Resistance (London), & Grande-Bretagne. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations: December 2014. Available online: https://www.naturallivestockfarming.com/wp-content/uploads/2015/09/Antibiotics-UK-dec-2014-Review-paper-on-health-wealth1.pdf (accessed on 6 March 2022).
- Talbot, G.H.; Jezek, A.; Murray, B.E.; Jones, R.N.; Ebright, R.H.; Nau, G.J.; Rodvold, K.A.; Newland, J.G.; Boucher, H.W. Infectious Diseases Society of America The Infectious Diseases Society of America’s 10 × ’20 Initiative (10 New Systemic Antibacterial Agents US Food and Drug Administration Approved by 2020): Is 20 × ’20 a Possibility? Clin. Infect. Dis. 2019, 69, 1–11. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial resistance to antimicrobial agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef]
- Hards, K.; Cook, G.M. Targeting bacterial energetics to produce new antimicrobials. Drug Resist. Updat. 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Diacon, A.; Pym, A.; Grobusch, M.; Patientia, R.; Rustomjee, R.; Page-Shipp, L.; Pistorius, C.; Krause, R.; Bogoshi, M.; Churchyard, G.; et al. The diarylquinoline TMC207 for Multidrug-Resistant Tuberculosis. N. Engl. J. Med. 2009, 360, 2397–2405. [Google Scholar] [CrossRef]
- Vestergaard, M.; Nøhr-Meldgaard, K.; Bojer, M.S.; Krogsgård Nielsen, C.; Meyer, R.L.; Slavetinsky, C.; Peschel, A.; Ingmer, H. Inhibition of the ATP Synthase Eliminates the Intrinsic Resistance of Staphylococcus aureus towards Polymyxins. MBio 2017, 8, e01114–e01117. [Google Scholar] [CrossRef]
- Liu, L.; Beck, C.; Nøhr-Meldgaard, K.; Peschel, A.; Kretschmer, D.; Ingmer, H.; Vestergaard, M. Inhibition of the ATP synthase sensitizes Staphylococcus aureus towards human antimicrobial peptides. Sci. Rep. 2020, 10, 11391. [Google Scholar] [CrossRef]
- Liu, A.; Tran, L.; Becket, E.; Lee, K.; Chinn, L.; Park, E.; Tran, K.; Miller, J.H. Antibiotic Sensitivity Profiles Determined with an Escherichia coli Gene Knockout Collection: Generating an Antibiotic Bar Code. Antimicrob. Agents Chemother. 2010, 54, 1393–1403. [Google Scholar] [CrossRef]
- Kinosita, K.; Yasuda, R.; Noji, H.; Adachi, K. A rotary molecular motor that can work at near 100% efficiency. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2000, 355, 473–489. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Structure and Mechanisms of F-Type ATP Synthases. Annu. Rev. Biochem. 2019, 88, 515–549. [Google Scholar] [CrossRef] [PubMed]
- Murphy, B.J.; Klusch, N.; Langer, J.; Mills, D.J.; Yildiz, Ö.; Kühlbrandt, W. Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F1-Fo coupling. Science 2019, 364, eaaw9128. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.M.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA 2012, 109, 11139–11143. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Wilkes, M.; Mills, D.J.; Kühlbrandt, W.; Meier, T. Molecular architecture of the N-type ATP ase rotor ring from Burkholderia pseudomallei. EMBO Rep. 2017, 18, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Sobti, M.; Walshe, J.L.; Wu, D.; Ishmukhametov, R.; Zeng, Y.C.; Robinson, C.V.; Berry, R.M.; Stewart, A.G. Cryo-EM structures provide insight into how E. coli F1Fo ATP synthase accommodates symmetry mismatch. Nat. Commun. 2020, 11, 2615. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, L.; Zong, S.; Guo, R.; Liu, T.; Yi, J.; Wang, P.; Zhuo, W.; Yang, M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 2019, 364, 1068–1075. [Google Scholar] [CrossRef]
- Boyer, P.D. The binding change mechanism for ATP synthase—Some probabilities and possibilities. BBA—Bioenerg. 1993, 1140, 215–250. [Google Scholar] [CrossRef]
- Abrahams, J.P.; Leslie, A.G.W.; Lutter, R.; Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart-mitochondria. Nature 1994, 370, 621–628. [Google Scholar] [CrossRef]
- Usukura, E.; Suzuki, T.; Furuike, S.; Soga, N.; Saita, E.-I.; Hisabori, T.; Kinosita, K.; Yoshida, M.; Yoshida, M. Torque generation and utilization in motor enzyme F0F1-ATP synthase: Half-torque F1 with short-sized pushrod helix and reduced ATP Synthesis by half-torque F0F1. J. Biol. Chem. 2012, 287, 1884–1891. [Google Scholar] [CrossRef]
- Pulido, N.O.; Salcedo, G.; Pérez-Hernández, G.; José-Núñez, C.; Velázquez-Campoy, A.; García-Hernández, E. Energetic effects of magnesium in the recognition of adenosine nucleotides by the F1-ATPase β subunit. Biochemistry 2010, 49, 5258–5268. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, G.; Duncan, T.M. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011, 18, 701–707. [Google Scholar] [CrossRef]
- Morales-Rios, E.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution. Proc. Natl. Acad. Sci. USA 2015, 112, 13231–13236. [Google Scholar] [CrossRef] [PubMed]
- Cabezón, E.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat. Struct. Biol. 2003, 10, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Van Raaij, M.J.; Abrahams, J.P.; Leslie, A.G.W.; Walker, J.E. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc. Natl. Acad. Sci. USA 1996, 93, 6913–6917. [Google Scholar] [CrossRef] [PubMed]
- Reisman, B.J.; Guo, H.; Ramsey, H.E.; Wright, M.T.; Reinfeld, B.I.; Ferrell, P.B.; Sulikowski, G.A.; Rathmell, W.K.; Savona, M.R.; Plate, L.; et al. Apoptolidin family glycomacrolides target leukemia through inhibition of ATP synthase. Nat. Chem. Biol. 2021, 11, 909. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Courbon, G.M.; Bueler, S.A.; Mai, J.; Liu, J.; Rubinstein, J.L. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature 2021, 589, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Pedersen, P.L. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641, Table of Contents. [Google Scholar] [CrossRef]
- Patel, B.A.; D’Amico, T.L.; Blagg, B.S.J. Natural products and other inhibitors of F1FO ATP synthase. Eur. J. Med. Chem. 2020, 207, 112779. [Google Scholar] [CrossRef]
- Gledhill, J.R.; Walker, J.E. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem. J. 2005, 386, 591–598. [Google Scholar] [CrossRef]
- Feniouk, B.A.; Suzuki, T.; Yoshida, M. The role of subunit epsilon in the catalysis and regulation of FOF1-ATP synthase. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 326–338. [Google Scholar] [CrossRef]
- Sielaff, H.; Duncan, T.M.; Börsch, M. The regulatory subunit ε in Escherichia coli FOF1-ATP synthase. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 775–788. [Google Scholar] [CrossRef]
- Zarco-Zavala, M.; Mendoza-Hoffmann, F.; García-Trejo, J.J. Unidirectional regulation of the F1FO-ATP synthase nanomotor by the ζ pawl-ratchet inhibitor protein of Paracoccus denitrificans and related α-proteobacteria. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.S.; Dustin Schaeffer, R.; Kinch, L.; Medvedev, K.E.; Grishin, N.V. Recent advances suggest increased influence of selective pressure in allostery. Curr. Opin. Struct. Biol. 2020, 62, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Shen, Q.; Zhang, J. Allosteric Methods and Their Applications: Facilitating the Discovery of Allosteric Drugs and the Investigation of Allosteric Mechanisms. Acc. Chem. Res. 2019, 52, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Chatzigoulas, A.; Cournia, Z. Rational design of allosteric modulators: Challenges and successes. WIREs Comput. Mol. Sci. 2021, 11, e1529. [Google Scholar] [CrossRef]
- Walker, J.E.; Saraste, M.; Runswick, M.J.; Gay, N.J. Distantly related sequences in the α- and—subunits of ATP synthase, myosin, kinases and other ATP-rquiring enzymes and a common nucleotide binding fold. EMBO J. 1982, I, 945–951. [Google Scholar] [CrossRef]
- Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003, 333, 781–815. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.-J. The Different Ways through Which Specificity Works in Orthosteric and Allosteric Drugs. Curr. Pharm. Des. 2012, 18, 1311–1316. [Google Scholar] [CrossRef]
- Hotra, A.; Ragunathan, P.; Shuyi-Ng, P.; Seankongsuk, P.; Harikishore, A.; Sarathy, J.-P.; Saw, W.-G.; Lakshmanan, U.; Sae-Lao, P.; Kalia, N.-P.; et al. Discovery of a novel Mycobacterial F-ATP synthase inhibitor and its potency in combination with diarylquinolines. Angew. Chemie—Int. Ed. 2020, 59, 13295–13304. [Google Scholar] [CrossRef]
- Saw, W.G.; Wu, M.L.; Ragunathan, P.; Biuković, G.; Lau, A.M.; Shin, J.; Harikishore, A.; Cheung, C.Y.; Hards, K.; Sarathy, J.P.; et al. Disrupting coupling within mycobacterial F-ATP synthases subunit ε causes dysregulated energy production and cell wall biosynthesis. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Ma, Y.-X.; Wang, C.-Y.; Li, Y.-Y.; Li, J.; Wan, Q.-Q.; Chen, J.-H.; Tay, F.R.; Niu, L.-N. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef]
- Alvarez-Garcia, D.; Barril, X. Molecular simulations with solvent competition quantify water displaceability and provide accurate interaction maps of protein binding sites. J. Med. Chem. 2014, 57, 8530–8539. [Google Scholar] [CrossRef]
- Ruiz-Carmona, S.; Alvarez-Garcia, D.; Foloppe, N.; Garmendia-Doval, A.B.; Juhos, S.; Schmidtke, P.; Barril, X.; Hubbard, R.E.; Morley, S.D. rDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLoS Comput. Biol. 2014, 10, e1003571. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carmona, S.; Schmidtke, P.; Luque, F.J.; Baker, L.; Matassova, N.; Davis, B.; Roughley, S.; Murray, J.; Hubbard, R.; Barril, X. Dynamic undocking and the quasi-bound state as tools for drug discovery. Nat. Chem. 2017, 9, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Blanco, Y.B.; Ávila-Barrientos, L.P.; Hernández-García, E.; Antunes, A.; Agüero-Chapin, G.; García-Hernández, E. Engineering protein fragments via evolutionary and protein–protein interaction algorithms: De novo design of peptide inhibitors for FOF1-ATP synthase. FEBS Lett. 2021, 595, 183–194. [Google Scholar] [CrossRef]
- Krah, A.; Zarco-Zavala, M.; McMillan, D.G.G. Insights into the regulatory function of the ε subunit from bacterial F-type ATP synthases: A comparison of structural, biochemical and biophysical data. Open Biol. 2018, 8, 170275. [Google Scholar] [CrossRef] [PubMed]
- Shoghi, E.; Fuguet, E.; Bosch, E.; Ràfols, C. Solubility-pH profiles of some acidic, basic and amphoteric drugs. Eur. J. Pharm. Sci. 2013, 48, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Kokubo, H. Exploring the Stability of Ligand Binding Modes to Proteins by Molecular Dynamics Simulations: A Cross-docking Study. J. Chem. Inf. Model. 2017, 57, 2514–2522. [Google Scholar] [CrossRef]
- Stoye, J.; Evers, D.; Meyer, F. Rose: Generating sequence families. Bioinformatics 1998, 14, 157–163. [Google Scholar] [CrossRef]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Harms, M.; Münch, J.; Sanchez-Garcia, E. PPI-Detect: A support vector machine model for sequence-based prediction of protein-protein interactions. J. Comput. Chem. 2019, 40, 1233–1242. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.Z.; Abraham, C.G.; Krishnakumar, A.M.; Weber, J. A functionally important hydrogen-bonding network at the betaDP/alphaDP interface of ATP synthase. J. Biol. Chem. 2008, 283, 24781–24788. [Google Scholar] [CrossRef] [PubMed]
- Mnatsakanyan, N.; Krishnakumar, A.M.; Suzuki, T.; Weber, J. The role of the betaDELSEED-loop of ATP synthase. J. Biol. Chem. 2009, 284, 11336–11345. [Google Scholar] [CrossRef]
- Livingstone, C.D.; Barton, G.J. Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993, 9, 745–756. [Google Scholar] [CrossRef]
- Diacon, A.H.; Donald, P.R.; Pym, A.; Grobusch, M.; Patientia, R.F.; Mahanyele, R.; Bantubani, N.; Narasimooloo, R.; De Marez, T.; van Heeswijk, R.; et al. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: Long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob. Agents Chemother. 2012, 56, 3271–3276. [Google Scholar] [CrossRef]
- Langlois, J.-P.; Millete, G.; Guay, I.; Dubé-Duquette, A.; Chamberland, S.; Jacques, P.-É.; Rodrigue, S.; Bourab, K.; Marsault, É. Malo Bactericidal Activity of the Bacterial ATP Synthase Inhibitor Tomatidine and the Combination of Tomatidine and Aminoglycoside Against Persistent and Virulent Forms of Staphylococcus aureus. Front. Microbiol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Roshanak, S.; Ingmer, H. Targeting the ATP Synthase in Staphylococcus aureus Small Colony Variants, Streptococcus pyogenes and Pathogenic Fungi. Antibiotics 2021, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Milgrom, Y.M.; Duncan, T.M. F-ATP-ase of Escherichia coli membranes: The ubiquitous MgADP-inhibited state and the inhibited state induced by the ε–subunit’s C-terminal domain are mutually exclusive. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148189. [Google Scholar] [CrossRef] [PubMed]
- García-Aguilar, A.; Cuezva, J.M. A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis. Front. Physiol. 2018, 9, 1322. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Astudillo, H.; Zarco-Zavala, M.; García-Trejo, J.J.; González-Halphen, D. Regulation of bacterial ATP synthase activity: A gear-shifting or a pawl–ratchet mechanism? FEBS J. 2021, 288, 3159–3163. [Google Scholar] [CrossRef]
- Syed, H.; Tauseef, M.; Ahmad, Z. A connection between antimicrobial properties of venom peptides and microbial ATP synthase. Int. J. Biol. Macromol. 2018, 119, 23–31. [Google Scholar] [CrossRef]
- Alvarez-Garcia, D.; Barril, X. Relationship between Protein Flexibility and Binding: Lessons for Structure-Based Drug Design. J. Chem. Theory Comput. 2014, 10, 2608–2614. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Okafor, F.; Azim, S.; Laughlin, T.F. ATP synthase: A molecular therapeutic drug target for antimicrobial and antitumor peptides. Curr. Med. Chem. 2013, 20, 1956–1973. [Google Scholar] [CrossRef] [PubMed]
- Talibov, V.O.; Fabini, E.; FitzGerald, E.A.; Tedesco, D.; Cederfeldt, D.; Talu, M.J.; Rachman, M.M.; Mihalic, F.; Manoni, E.; Naldi, M.; et al. Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase. ChemBioChem 2021, 22, 1597–1608. [Google Scholar] [CrossRef]
- Arcon, J.P.; Defelipe, L.A.; Lopez, E.D.; Burastero, O.; Modenutti, C.P.; Barril, X.; Marti, M.A.; Turjanski, A.G. Cosolvent-Based Protein Pharmacophore for Ligand Enrichment in Virtual Screening. J. Chem. Inf. Model. 2019, 59, 3572–3583. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Oliveira, A.; Hjort, K.; Bergfors, T.; Gutiérrez-de-Terán, H.; Andersson, D.I.; Sanyal, S.; Åqvist, J. Inhibition of translation termination by small molecules targeting ribosomal release factors. Sci. Rep. 2019, 9, 15424. [Google Scholar] [CrossRef] [PubMed]
- Subiros-Funosas, R.; Ho, V.C.L.; Barth, N.D.; Mendive-Tapia, L.; Pappalardo, M.; Barril, X.; Ma, R.; Zhang, C.-B.; Qian, B.-Z.; Sintes, M.; et al. Fluorogenic Trp(redBODIPY) cyclopeptide targeting keratin 1 for imaging of aggressive carcinomas. Chem. Sci. 2020, 11, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Defelipe, L.; Arcon, J.; Modenutti, C.; Marti, M.; Turjanski, A.; Barril, X. Solvents to Fragments to Drugs: MD Applications in Drug Design. Molecules 2018, 23, 3269. [Google Scholar] [CrossRef] [PubMed]
- Barril, X. Computer-aided drug design: Time to play with novel chemical matter. Expert Opin. Drug Discov. 2017, 12, 977–980. [Google Scholar] [CrossRef] [PubMed]
- Rachman, M.; Bajusz, D.; Hetényi, A.; Scarpino, A.; Merő, B.; Egyed, A.; Buday, L.; Barril, X.; Keserű, G.M. Discovery of a novel kinase hinge binder fragment by dynamic undocking. RSC Med. Chem. 2020, 11, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Zhou, Z.; Han, L.; Karapetyan, K.; Dracheva, S.; Shoemaker, B.A.; et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400. [Google Scholar] [CrossRef]
- Gledhill, J.R.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA 2007, 104, 13632–13637. [Google Scholar] [CrossRef]
- Luo, M.; Zhou, W.; Patel, H.; Srivastava, A.P.; Symersky, J.; Bonar, M.M.; Faraldo-Gómez, J.D.; Liao, M.; Mueller, D.M. Bedaquiline inhibits the yeast and human mitochondrial ATP synthases. Commun. Biol. 2020, 3, 452. [Google Scholar] [CrossRef]
- Degiacomi, G.; Sammartino, J.C.; Sinigiani, V.; Marra, P.; Urbani, A.; Pasca, M.R. In vitro Study of Bedaquiline Resistance in Mycobacterium tuberculosis Multi-Drug Resistant Clinical Isolates. Front. Microbiol. 2020, 11, 2290. [Google Scholar] [CrossRef]
- Lu, S.; Qiu, Y.; Ni, D.; He, X.; Pu, J.; Zhang, J. Emergence of allosteric drug-resistance mutations: New challenges for allosteric drug discovery. Drug Discov. Today 2020, 25, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Vella, P.; Rudraraju, R.S.; Lundbäck, T.; Axelsson, H.; Almqvist, H.; Vallin, M.; Schneider, G.; Schnell, R. A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens. Bioorg. Med. Chem. 2021, 30, 115898. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.M.; Sourimant, J.; Toots, M.; Yoon, J.-J.; Ikegame, S.; Govindarajan, M.; Watkinson, R.E.; Thibault, P.; Makhsous, N.; Lin, M.J.; et al. Orally efficacious broad-spectrum allosteric inhibitor of paramyxovirus polymerase. Nat. Microbiol. 2020, 5, 1232–1246. [Google Scholar] [CrossRef]
- Ketchum, C.J.; Al-Shawi, M.K.; Nakamoto, R.K. Intergenic suppression of the gammaM23K uncoupling mutation in F0F1 ATP synthase by betaGlu-381 substitutions: The role of the beta380DELSEED386 segment in energy coupling. Biochem. J. 1998, 330, 707–712. [Google Scholar] [CrossRef]
- Hara, K.Y.; Noji, H.; Bald, D.; Yasuda, R.; Kinosita, K.; Yoshida, M. The role of the DELSEED motif of the beta subunit in rotation of F1-ATPase. J. Biol. Chem. 2000, 275, 14260–14263. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.Y.; Kato-Yamada, Y.; Kikuchi, Y.; Hisabori, T.; Yoshida, M. The role of the betaDELSEED motif of F1-ATPase: Propagation of the inhibitory effect of the epsilon subunit. J. Biol. Chem. 2001, 276, 23969–23973. [Google Scholar] [CrossRef]
- Scanlon, J.A.B.; Al-Shawi, M.K.; Nakamoto, R.K. A rotor-stator cross-link in the F1-ATPase blocks the rate-limiting step of rotational catalysis. J. Biol. Chem. 2008, 283, 26228–26240. [Google Scholar] [CrossRef] [PubMed]
- Mnatsakanyan, N.; Kemboi, S.K.; Salas, J.; Weber, J. The beta subunit loop that couples catalysis and rotation in ATP synthase has a critical length. J. Biol. Chem. 2011, 286, 29788–29796. [Google Scholar] [CrossRef] [PubMed]
- Tanigawara, M.; Tabata, K.V.; Ito, Y.; Ito, J.; Watanabe, R.; Ueno, H.; Ikeguchi, M.; Noji, H. Role of the DELSEED loop in torque transmission of F1-ATPase. Biophys. J. 2012, 103, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Koyasu, K.; You, H.; Tanigawara, M.; Noji, H. Torque transmission mechanism via DELSEED loop of F1-ATPase. Biophys. J. 2015, 108, 1144–1152. [Google Scholar] [CrossRef] [PubMed]
- La, T.; Clark-Walker, G.D.; Wang, X.; Wilkens, S.; Chen, X.J. Mutations on the N-terminal edge of the DELSEED loop in either the α or β subunit of the mitochondrial F1-ATPase enhance ATP hydrolysis in the absence of the central γ rotor. Eukaryot. Cell 2013, 12, 1451–1461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salcedo, G.; Cano-Sánchez, P.; De Gómez-Puyou, M.T.M.T.; Velázquez-Campoy, A.; García-Hernández, E. Isolated noncatalytic and catalytic subunits of F1-ATPase exhibit similar, albeit not identical, energetic strategies for recognizing adenosine nucleotides. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 44–50. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rao, S.P.S.; Alonso, S.; Rand, L.; Dick, T.; Pethe, K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2008, 105, 11945–11950. [Google Scholar] [CrossRef] [PubMed]
- Nuskova, H.; Mikesova, J.; Efimova, I.; Pecinova, A.; Pecina, P.; Drahota, Z.; Houstek, J.; Mracek, T. Biochemical thresholds for pathological presentation of ATP synthase deficiencies. Biochem. Biophys. Res. Commun. 2020, 521, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Nesci, S.; Trombetti, F.; Algieri, C.; Pagliarani, A. A Therapeutic Role for the F1FO-ATP Synthase. SLAS DISCOVERY: Adv. Sci. Drug Discov. 2019, 24, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, M.; Ózsvári, B.; Sotgia, F.; Lisanti, M.P. High ATP Production Fuels Cancer Drug Resistance and Metastasis: Implications for Mitochondrial ATP Depletion Therapy. Front. Oncol. 2021, 11, 3875. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Berryman, J.T.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; et al. AMBER 2014; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Molecular Operating Environment (MOE), 2014.09; Chemical Computing Group ULC: Montreal, QC, Canada, 2015.
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Hahn-Herrera, O.; Salcedo, G.; Barril, X.; García-Hernández, E. Inherent conformational flexibility of F1-ATPase α-subunit. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1392–1402. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2020, 30, 70–82. [Google Scholar] [CrossRef]
- Romero-Molina, S.; Ruiz-Blanco, Y.B.; Green, J.R.; Sanchez-Garcia, E. ProtDCal-Suite: A web server for the numerical codification and functional analysis of proteins. Protein Sci. 2019, 28, 1734–1743. [Google Scholar] [CrossRef]
- Senior, A.E.; Lee, R.S.F.; Al-shawi, M.K.; Weber, J. Catalytic properties of Escherichia coli F1-ATPase depleted of endogenous nucleotides. Arch. Biochem. Biophys. 1992, 297, 340–344. [Google Scholar] [CrossRef]
- Van Veldhoven, P.P.; Mannaerts, G.P. Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 1987, 161, 45–48. [Google Scholar] [CrossRef]
- Kornberg, A.; Pricer, W.E. Enzymatic Phosphorylation of Adenosine and 2,6-Diaminopurine Riboside. J. Biol. Chem. 1951, 193, 481–495. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Bowler, M.W.; Montgomery, M.G.; Leslie, A.G.W.; Walker, J.E. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 2007, 282, 14238–14242. [Google Scholar] [CrossRef] [PubMed]


| Structure a | ΔGrDock b (kcal/mol) | WQB c (kcal/mol) | ΔGPB d (kcal/mol) | Residual ATPase Activity (%) e | logS f | |
|---|---|---|---|---|---|---|
| Compd-5 | 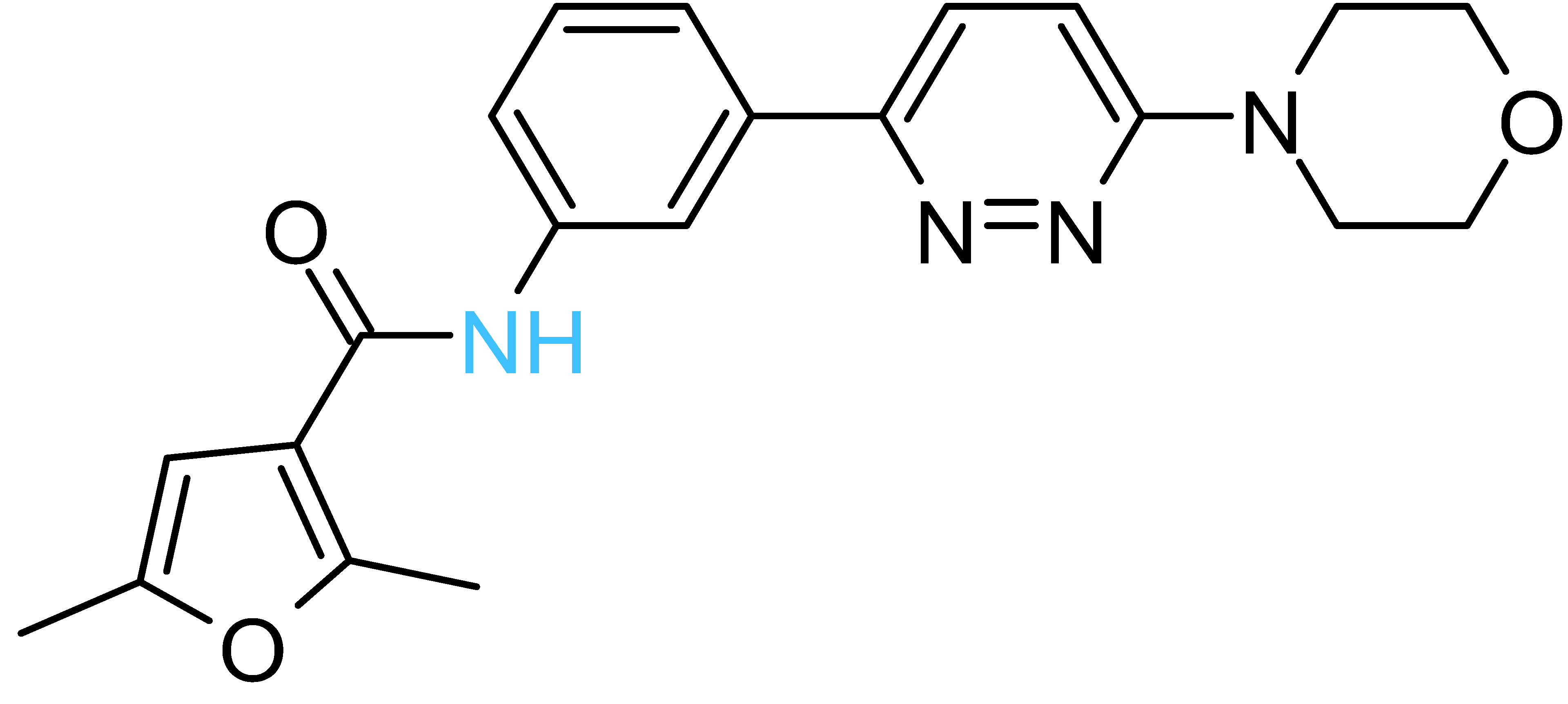 | −6.0 | 8.7 | −29 | 43 ± 6 (50 ± 5%) | −4.7 |
| Compd-7 | 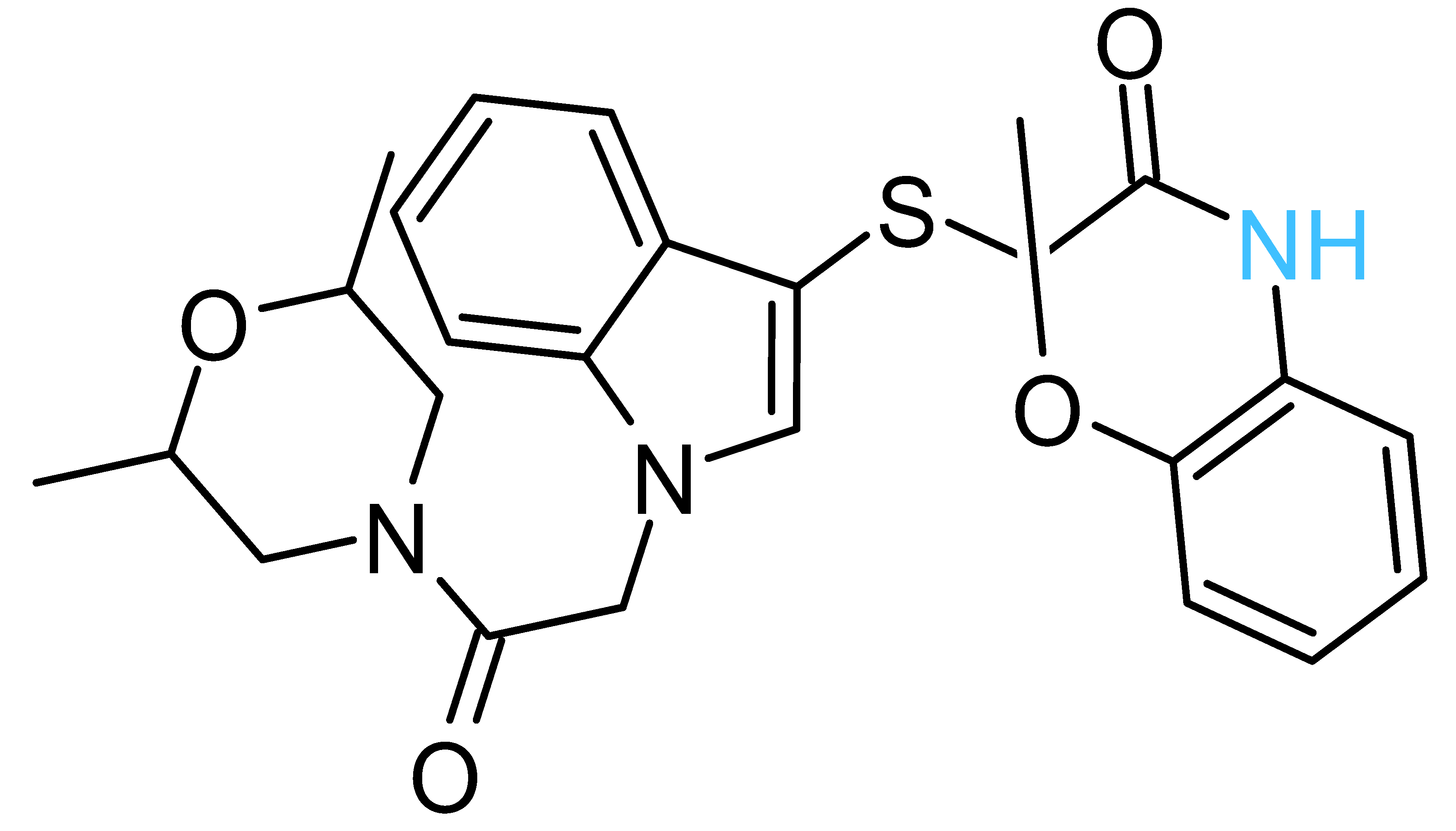 | −5.5 | 6.0 | −30 | 64 ± 12 (70 ± 10%) | −5.1 |
| Compd-14 | 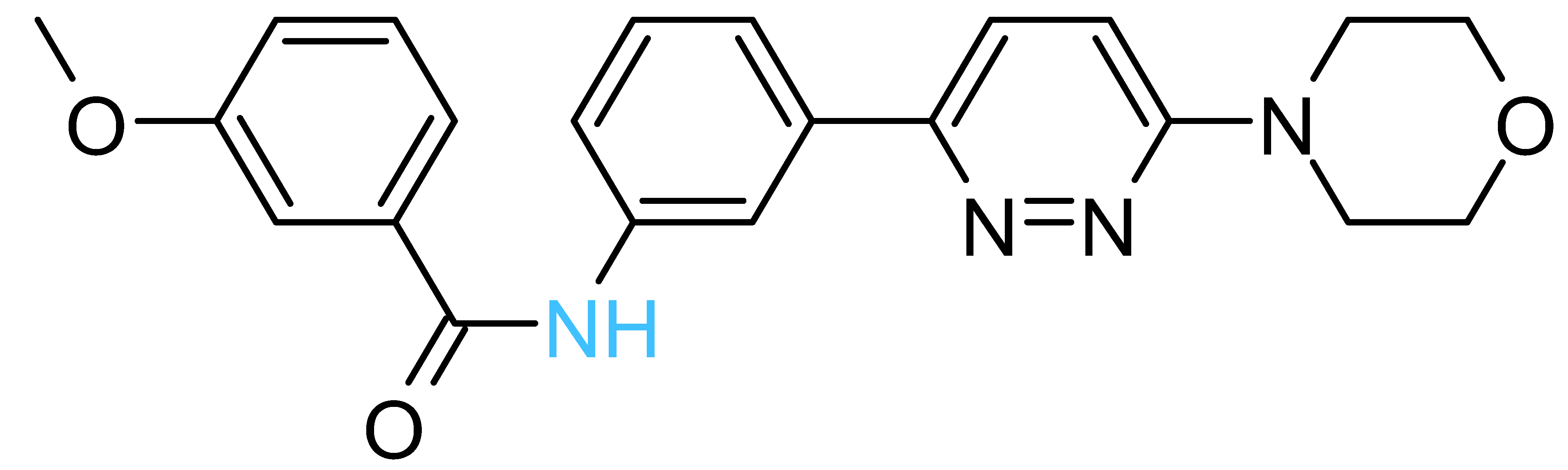 | −4.5 | 6.7 | −25 | 75 ± 8 (73 ± 2%) | −5.3 |
| Compd-15 |  | −4.3 | 6.0 | −24 | 67 ± 5 (ND) | −4.9 |
| Compd-19 |  | −4.0 | 6.0 | −29 | 77 ± 7 (70 ± 10%) | −2.8 |
| Peptide | Sequence | Score a (EcF1) | Score a (HsF1) | Charge (pI) b | GRAVY b |
|---|---|---|---|---|---|
| Pept1-IF1 | GSIREAGGTHAFGKRESAEEERYFR | 0.512 | 0.402 | 0 (6.78) | −1.292 |
| Pept2-IF1 | GSIREAGGTDGFGKREAAEEEKYGR | 0.561 | 0.420 | −1 (5.11) | −1.392 |
| Pept3-IF1 | GSVREAGGTGAFGKRESAEEERYFR | 0.580 | 0.479 | 0 (6.34) | −1.192 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila-Barrientos, L.P.; Cofas-Vargas, L.F.; Agüero-Chapin, G.; Hernández-García, E.; Ruiz-Carmona, S.; Valdez-Cruz, N.A.; Trujillo-Roldán, M.; Weber, J.; Ruiz-Blanco, Y.B.; Barril, X.; et al. Computational Design of Inhibitors Targeting the Catalytic β Subunit of Escherichia coli FOF1-ATP Synthase. Antibiotics 2022, 11, 557. https://doi.org/10.3390/antibiotics11050557
Avila-Barrientos LP, Cofas-Vargas LF, Agüero-Chapin G, Hernández-García E, Ruiz-Carmona S, Valdez-Cruz NA, Trujillo-Roldán M, Weber J, Ruiz-Blanco YB, Barril X, et al. Computational Design of Inhibitors Targeting the Catalytic β Subunit of Escherichia coli FOF1-ATP Synthase. Antibiotics. 2022; 11(5):557. https://doi.org/10.3390/antibiotics11050557
Chicago/Turabian StyleAvila-Barrientos, Luis Pablo, Luis Fernando Cofas-Vargas, Guillermin Agüero-Chapin, Enrique Hernández-García, Sergio Ruiz-Carmona, Norma A. Valdez-Cruz, Mauricio Trujillo-Roldán, Joachim Weber, Yasser B. Ruiz-Blanco, Xavier Barril, and et al. 2022. "Computational Design of Inhibitors Targeting the Catalytic β Subunit of Escherichia coli FOF1-ATP Synthase" Antibiotics 11, no. 5: 557. https://doi.org/10.3390/antibiotics11050557
APA StyleAvila-Barrientos, L. P., Cofas-Vargas, L. F., Agüero-Chapin, G., Hernández-García, E., Ruiz-Carmona, S., Valdez-Cruz, N. A., Trujillo-Roldán, M., Weber, J., Ruiz-Blanco, Y. B., Barril, X., & García-Hernández, E. (2022). Computational Design of Inhibitors Targeting the Catalytic β Subunit of Escherichia coli FOF1-ATP Synthase. Antibiotics, 11(5), 557. https://doi.org/10.3390/antibiotics11050557








