Phenolic Compound Ethyl 3,4-Dihydroxybenzoate Retards Drug Efflux and Potentiates Antibiotic Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Half-Maximal Inhibitory Concentration (IC50) and Modulation Assay for EDHB against Drug-Resistant E. coli
2.2. Effect of EDHB on Time-Kill Curves
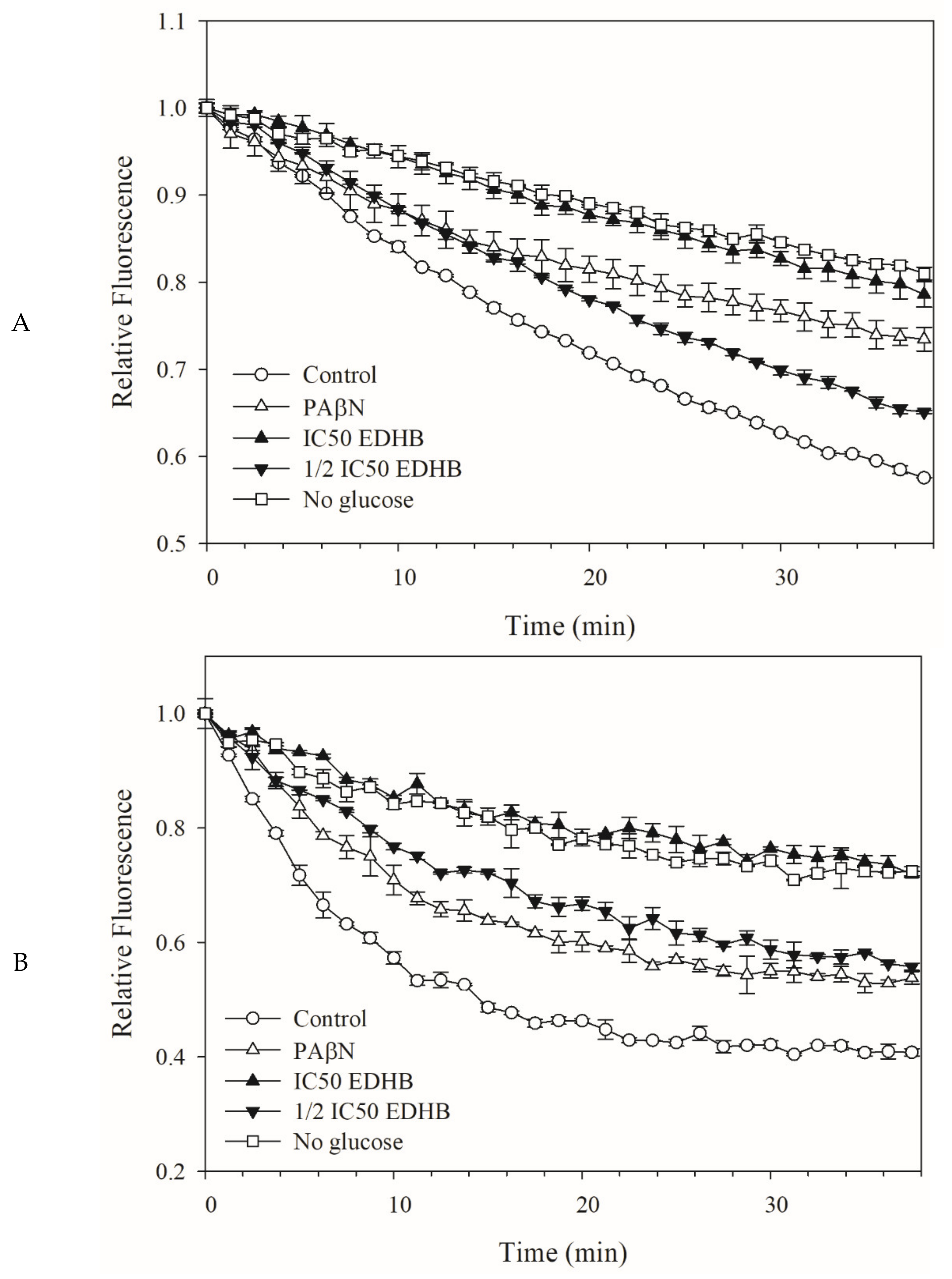
2.3. Fluorescent Dye Accumulation Reduced by EDHB
2.4. Dye Efflux by Kam3-AcrB was Reduced by EDHB
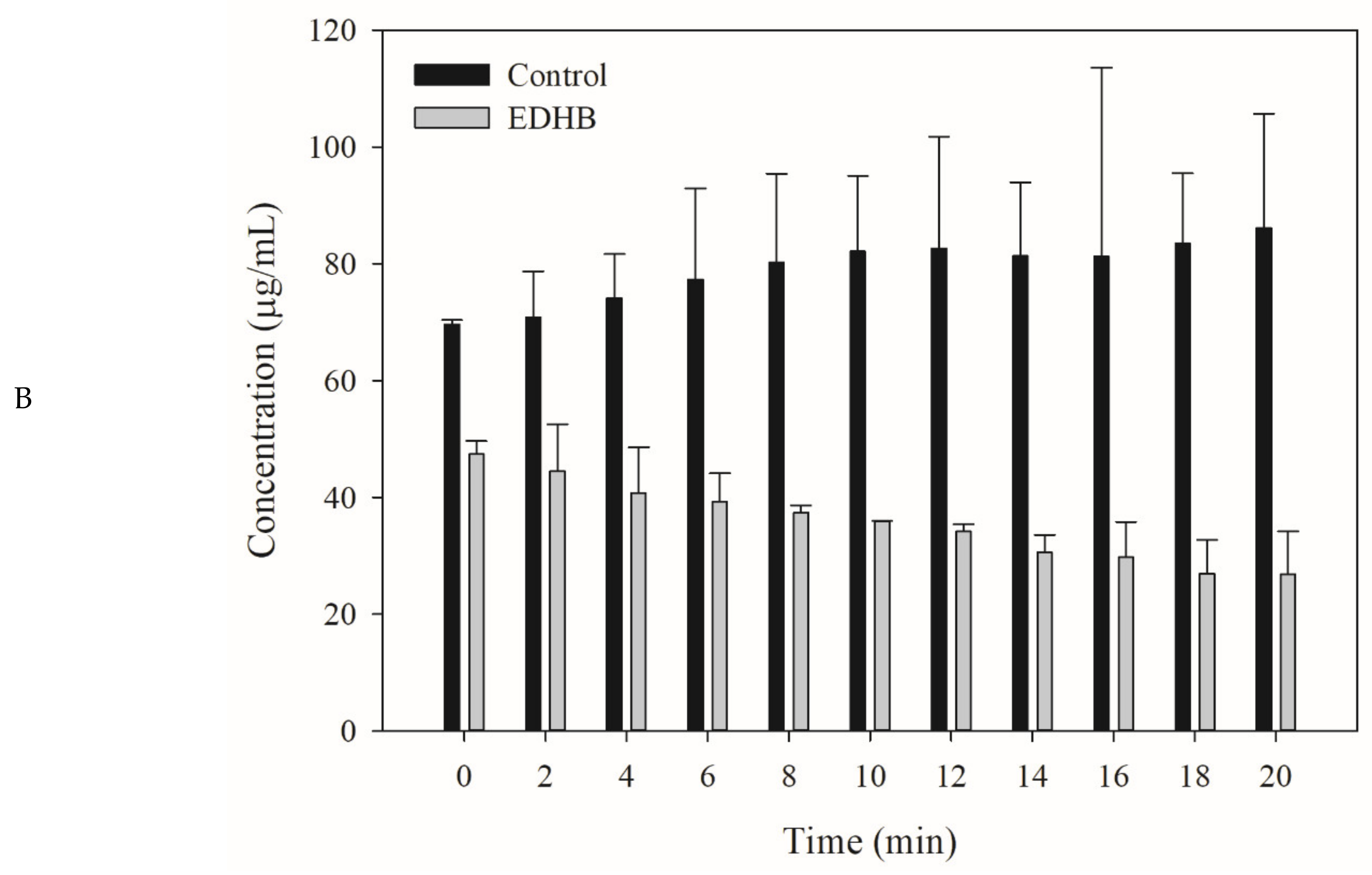
2.5. Drug Efflux Interference by EDHB Monitored Using MALDI-TOF
2.6. Molecular Docking of EDHB
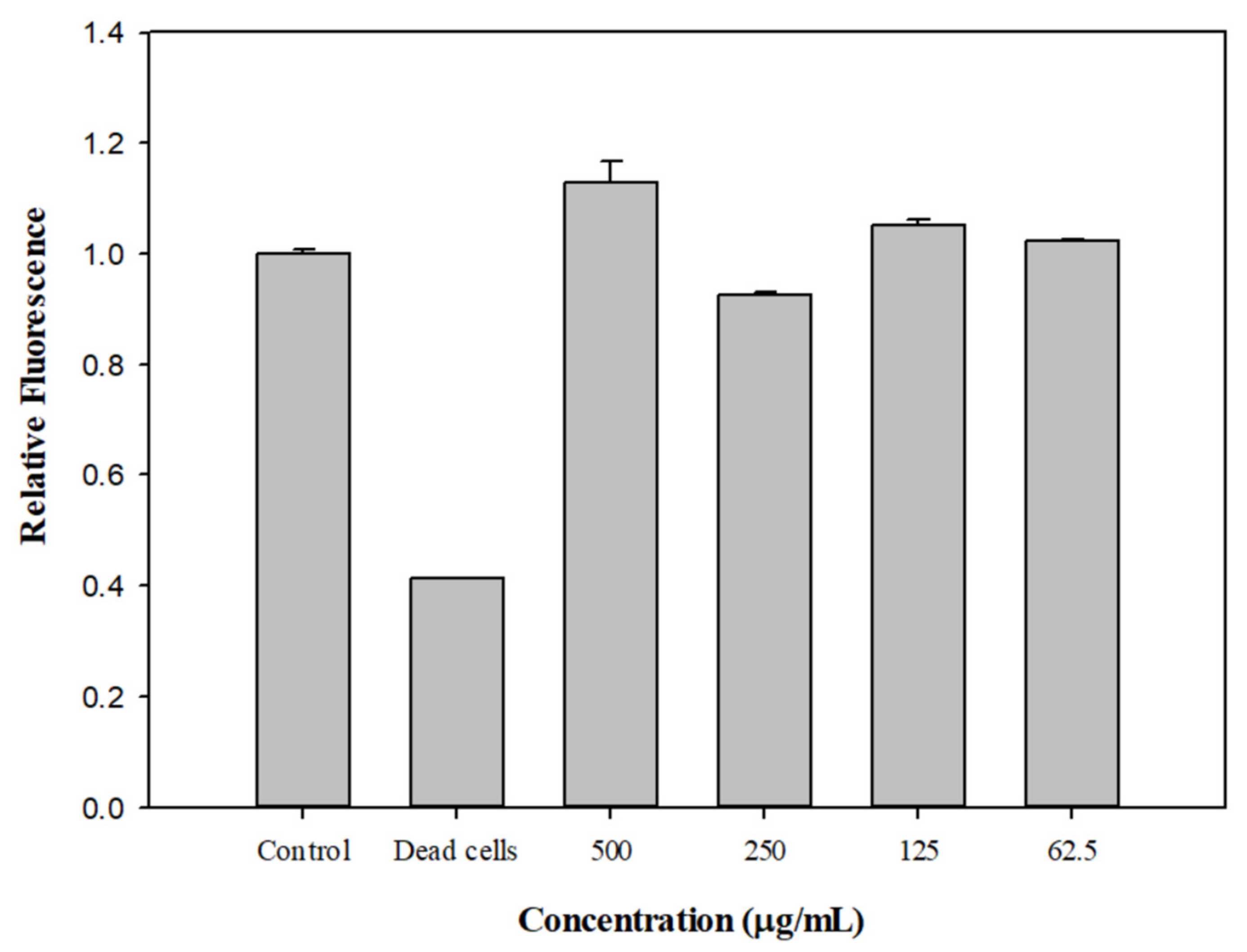
2.7. Effect of EDHB on the Membrane Permeability of Kam3-AcrB
2.8. PAE of EDHB and Antibiotics on E. coli Kam3-AcrB
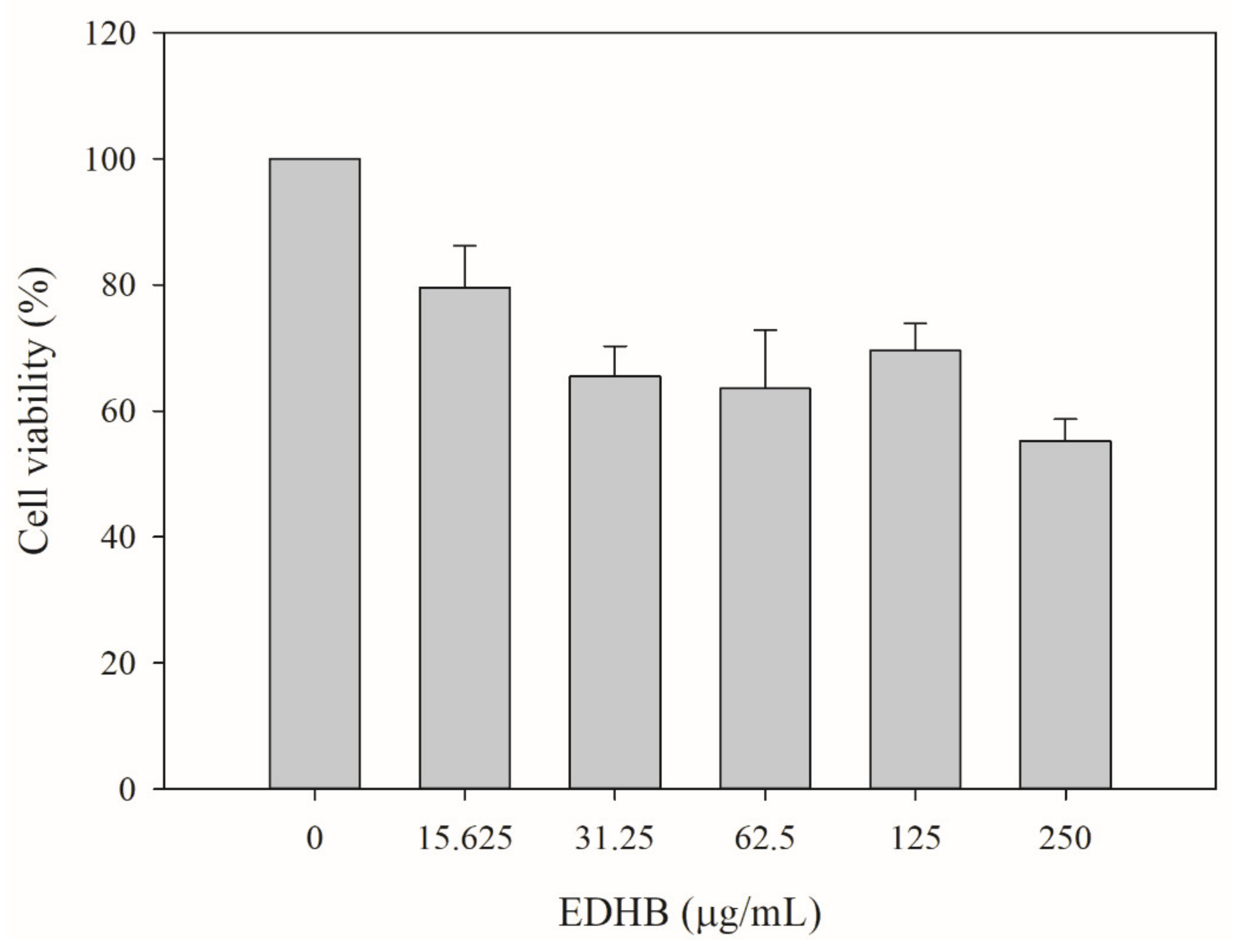
2.9. Cytotoxicity Test of EDHB
3. Materials and Methods
3.1. Bacterial Strains, Constructs, Media, and Chemicals
3.2. IC50 and Modulation Tests
3.3. Time-Kill Assays
3.4. Drug Accumulation Assay
3.5. Drug Efflux Assay
3.6. Monitoring Drug Efflux Using MALDI-TOF Mass Spectrometry
3.7. Molecular Docking
3.8. Membrane Permeability Assay
3.9. Postantibiotic Effect Assay
3.10. Cell Toxicity Assays
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martinez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, S.S.; Sobkowiak, B.; Parreira, R.; Edgeworth, J.D.; Viveiros, M.; Clark, T.G.; Couto, I. Genetic diversity of NorA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 2019, 9, 710. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Kobylka, J.; Kuth, M.S.; Pos, K.M.; Picard, M.; Blair, J.M.A.; Bavro, V.N. Structure, Assembly, and Function of Tripartite Efflux and Type 1 Secretion Systems in Gram-Negative Bacteria. Chem. Rev. 2021, 121, 5479–5596. [Google Scholar] [CrossRef] [PubMed]
- Interagency Coordination Group on Antimicrobial Resistance. No Time to Wait: Securing the Funture from Drug-Resistant Infections; Interagency Coordination Group on Antimicrobial Resistance: Geneva, Switzerland, 2019. [Google Scholar]
- Plackett, B. No money for new drugs. Nature 2020, 586, S50–S52. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129–145. [Google Scholar] [CrossRef]
- Pagès, J.M.; Amaral, L. Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 826–833. [Google Scholar] [CrossRef]
- Lu, W.J.; Lin, H.J.; Hsu, P.H.; Lai, M.; Chiu, J.Y.; Lin, H.T.V. Brown and red seaweeds serve as potential efflux pump inhibitors for drug-resistant Escherichia coli. Evid. Based Complement. Altern. Med. 2019, 2019, 1836982. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.A.; Mirza, Z.M.; Kumar, A.; Verma, V.; Qazi, G.N. Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 810–812. [Google Scholar] [CrossRef] [Green Version]
- Markham, P.N.; Westhaus, E.; Klyachko, K.; Johnson, M.E.; Neyfakh, A.A. Multiple novel inhibitors of the NorA multidrug transporter of Staphylococcus aureus. Antimicrob. Agents Chemother. 1999, 43, 2404–2408. [Google Scholar] [CrossRef] [Green Version]
- Piddock, L.J.; Garvey, M.I.; Rahman, M.M.; Gibbons, S. Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Sabatini, S.; Gosetto, F.; Manfroni, G.; Tabarrini, O.; Kaatz, G.W.; Patel, D.; Cecchetti, V. Evolution from a natural flavones nucleus to obtain 2-(4-Propoxyphenyl)quinoline derivatives as potent inhibitors of the S. aureus NorA efflux pump. J. Med. Chem. 2011, 54, 5722–5736. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Kern, W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005, 49, 849–852. [Google Scholar] [CrossRef] [Green Version]
- Christena, L.R.; Subramaniam, S.; Vidhyalakshmi, M.; Mahadevan, V.; Sivasubramanian, A.; Nagarajan, S. Dual role of pinostrobin-a flavonoid nutraceutical as an efflux pump inhibitor and antibiofilm agent to mitigate food borne pathogens. RSC Adv. 2015, 5, 61881–61887. [Google Scholar] [CrossRef]
- Stermitz, F.R.; Scriven, L.N.; Tegos, G.; Lewis, K. Two flavonols from Artemisa annua which potentiate the activity of berberine and norfloxacin against a resistant strain of Staphylococcus aureus. Planta Med. 2002, 68, 1140–1141. [Google Scholar] [CrossRef]
- Oliveira-Tintino, C.D.D.; Tintino, S.R.; Limaverde, P.W.; Figueredo, F.G.; Campina, F.F.; da Cunha, F.A.B.; da Costa, R.H.S.; Pereira, P.S.; Lima, L.F.; de Matos, Y.M.L.S.; et al. Inhibition of the essential oil from Chenopodium ambrosioides L. and alpha-terpinene on the NorA efflux-pump of Staphylococcus aureus. Food Chem. 2018, 262, 72–77. [Google Scholar] [CrossRef]
- Prasch, S.; Bucar, F. Plant derived inhibitors of bacterial efflux pumps: An update. Phytochem. Rev. 2015, 14, 961–974. [Google Scholar] [CrossRef]
- Stavri, M.; Piddock, L.J.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef] [Green Version]
- Baderschneider, B.; Winterhalter, P. Isolation and characterization of novel benzoates, cinnamates, flavonoids, and lignans from Riesling wine and screening for antioxidant activity. J. Agric. Food Chem. 2001, 49, 2788–2798. [Google Scholar] [CrossRef]
- Merkl, R.; Hradkova, I.; Filip, V.; Smidrkal, J. Antimicrobial and Antioxidant Properties of Phenolic Acids Alkyl Esters. Czech J. Food Sci. 2010, 28, 275–279. [Google Scholar] [CrossRef] [Green Version]
- Otreebska-Machaj, E.; Chevalier, J.; Handzlik, J.; Szymanska, E.; Schabikowski, J.; Boyer, G.; Bolla, J.M.; Kiec-Kononowicz, K.; Pages, J.M.; Alibert, S. Efflux Pump Blockers in Gram-Negative Bacteria: The New Generation of Hydantoin Based-Modulators to Improve Antibiotic Activity. Front. Microbiol. 2016, 7, 622. [Google Scholar] [CrossRef]
- Poole, K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005, 56, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikaido, H. Multidrug efflux pumps of gram negative bacteria. J. Bacteriol. 1996, 178, 5853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savoia, D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, N.P.; Mahajan, P.; Mehra, R.; Nargotra, A.; Sharma, J.P.; Koul, S.; Khan, I.A. Capsaicin, a novel inhibitor of the NorA efflux pump, reduces the intracellular invasion of Staphylococcus aureus. J. Antimicrob. Chemother. 2012, 67, 2401–2408. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, N.; Nishino, K.; Yamaguchi, A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 2001, 183, 5639–5644. [Google Scholar] [CrossRef] [Green Version]
- Paixao, L.; Rodrigues, L.; Couto, I.; Martins, M.; Fernandes, P.; de Carvalho, C.C.; Monteiro, G.A.; Sansonetty, F.; Amaral, L.; Viveiros, M. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 2009, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Richmond, G.E.; Chua, K.L.; Piddock, L.J.V. Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzamide). J. Antimicrob. Chemother. 2013, 68, 1594–1600. [Google Scholar] [CrossRef]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.-S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 722–733. [Google Scholar] [CrossRef] [Green Version]
- Aparna, V.; Dineshkumar, K.; Mohanalakshmi, N.; Velmurugan, D.; Hopper, W. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS ONE 2014, 9, e101840. [Google Scholar] [CrossRef] [Green Version]
- Blair, J.M.; Piddock, L.J. How to measure export via bacterial multidrug resistance efflux pumps. MBio 2016, 7, e00840-16. [Google Scholar] [CrossRef] [Green Version]
- Li, X.L.; Hu, Y.J.; Wang, H.; Yu, B.Q.; Yue, H.L. Molecular Spectroscopy Evidence of Berberine Binding to DNA: Comparative Binding and Thermodynamic Profile of Intercalation. Biomacromolecules 2012, 13, 873–880. [Google Scholar] [CrossRef]
- Dumont, E.; Vergalli, J.; Conraux, L.; Taillier, C.; Vassort, A.; Pajovic, J.; Refregiers, M.; Mourez, M.; Pages, J.M. Antibiotics and efflux: Combined spectrofluorimetry and mass spectrometry to evaluate the involvement of concentration and efflux activity in antibiotic intracellular accumulation. J. Antimicrob. Chemother. 2019, 74, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.J.; Lin, H.J.; Hsu, P.H.; Lin, H.T.V. Determination of drug efflux pump efficiency in drug-resistant bacteria using MALDI-TOF MS. Antibiotics 2020, 9, 639. [Google Scholar] [CrossRef]
- Zwama, M.; Yamasaki, S.; Nakashima, R.; Sakurai, K.; Nishino, K.; Yamaguchi, A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Zwama, M.; Yamaguchi, A. Molecular mechanisms of AcrB-mediated multidrug export. Res. Microbiol. 2018, 169, 372–383. [Google Scholar] [CrossRef]
- Muller, R.T.; Travers, T.; Cha, H.J.; Phillips, J.L.; Gnanakaran, S.; Pos, K.M. Switch Loop Flexibility Affects Substrate Transport of the AcrB Efflux Pump. J. Mol. Biol. 2017, 429, 3863–3874. [Google Scholar] [CrossRef]
- Eicher, T.; Cha, H.J.; Seeger, M.A.; Brandstatter, L.; El-Delik, J.; Bohnert, J.A.; Kern, W.V.; Verrey, F.; Grutter, M.G.; Diederichs, K.; et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA 2012, 109, 5687–5692. [Google Scholar] [CrossRef] [Green Version]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Nikaido, H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, 20637–20642. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Halim, H.; Al Dajani, A.; Abdelhalim, A.; Abdelmalek, S. The search of potential inhibitors of the AcrAB-TolC system of multidrug-resistant Escherichia coli: An in silico approach. Appl. Microbiol. Biotechnol. 2019, 103, 6309–6318. [Google Scholar] [CrossRef]
- Anoushiravani, M.; Falsafi, T.; Niknam, V. Proton motive force-dependent efflux of tetracycline in clinical isolates of Helicobacter pylori. J. Med. Microbiol. 2009, 58, 1309–1313. [Google Scholar] [CrossRef]
- Fenosa, A.; Fuste, E.; Ruiz, L.; Veiga-Crespo, P.; Vinuesa, T.; Guallar, V.; Villa, T.G.; Vinas, M. Role of TolC in Klebsiella oxytoca resistance to antibiotics. J. Antimicrob. Chemother. 2009, 63, 668–674. [Google Scholar] [CrossRef] [Green Version]
- Osei Sekyere, J.; Amoako, D.G. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front. Microbiol. 2017, 8, 228. [Google Scholar] [CrossRef] [Green Version]
- Srimani, J.K.; Huang, S.; Lopatkin, A.J.; You, L. Drug detoxification dynamics explain the postantibiotic effect. Mol. Syst. Biol. 2017, 13, 948. [Google Scholar] [CrossRef]
- Grimsey, E.M.; Piddock, L.J.V. Do phenothiazines possess antimicrobial and efflux inhibitory properties? FEMS Microbiol. Rev. 2019, 43, 577–590. [Google Scholar] [CrossRef]
- Machado, D.; Fernandes, L.; Costa, S.S.; Cannalire, R.; Manfroni, G.; Tabarrini, O.; Couto, I.; Sabatini, S.; Viveiros, M. Mode of action of the 2-phenylquinoline efflux inhibitor PQQ4R against Escherichia coli. PeerJ 2017, 5, e3168. [Google Scholar] [CrossRef] [Green Version]
- Jafri, A.; Siddiqui, S.; Rais, J.; Ahmad, M.S.; Kumar, S.; Jafar, T.; Afzal, M.; Arshad, M. Induction of apoptosis by piperine in human cervical adenocarcinoma via ROS mediated mitochondrial pathway and caspase-3 activation. Excli J. 2019, 18, 154–164. [Google Scholar] [CrossRef]
- Lu, W.J.; Hsu, P.H.; Chang, C.J.; Su, C.K.; Huang, Y.J.; Lin, H.J.; Lai, M.; Ooi, G.X.; Dai, J.Y.; Lin, H.V. Identified Seaweed Compound Diphenylmethane Serves as an Efflux Pump Inhibitor in Drug-Resistant Escherichia coli. Antibiotics 2021, 10, 1378. [Google Scholar] [CrossRef]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782. [Google Scholar] [CrossRef] [Green Version]
- Soothill, J.S.; Ward, R.; Girling, A.J. The IC50: An exactly defined measure of antibiotic sensitivity. J. Antimicrob. Chemother. 1992, 29, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, G.R.; Tiwari, N.; Singh, A.; Kumar, A.; Roy, S.; Negi, A.S.; Pal, A.; Chanda, D.; Sharma, A.; Darokar, M.P. Gallic acid-based indanone derivative interacts synergistically with tetracycline by inhibiting efflux pump in multidrug resistant E. coli. Appl. Microbiol. Biotechnol. 2016, 100, 2311–2325. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.J.; Lin, H.J.; Janganan, T.K.; Li, C.Y.; Chin, W.C.; Bavro, V.N.; Lin, H.V. ATP-binding cassette transporter VcaM from Vibrio cholerae is dependent on the outer membrane factor family for its function. Int. J. Mol. Sci. 2018, 19, 1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.R.; Ettefagh, K.A.; Todd, D.; Cole, P.S.; Egan, J.M.; Foil, D.H.; Graf, T.N.; Schindler, B.D.; Kaatz, G.W.; Cech, N.B. A mass spectrometry-based assay for improved quantitative measurements of efflux pump inhibition. PLoS ONE 2015, 10, e0124814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnert, J.A.; Karamian, B.; Nikaido, H. Optimized Nile Red Efflux Assay of AcrAB-TolC Multidrug Efflux System Shows Competition between Substrates. Antimicrob. Agents Chemother. 2010, 54, 3770–3775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Tambat, R.; Jangra, M.; Mahey, N.; Chandal, N.; Kaur, M.; Chaudhary, S.; Verma, D.K.; Thakur, K.G.; Raje, M.; Jachak, S.; et al. Microbe-derived indole metabolite demonstrates potent multidrug efflux pump inhibition in Staphylococcus aureus. Front. Microbiol. 2019, 10, 2153. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015, 111, A.3B1–A.3B2. [Google Scholar] [CrossRef]




| Antibiotics | EDHB Concentration (µg/mL) | IC50 (µg/mL) | Modulation Factor | |||
|---|---|---|---|---|---|---|
| Alone | With EDHB | With PAβN | EDHB | PAβN | ||
| Clarithromycin | 125 | 175 | 43.75 | 21.87 | 4 | 8 |
| Erythromycin | 31.25 | 125 | 31.25 | 31.25 | 4 | 4 |
| Ciprofloxacin | 3.9 | 0.06 | 0.03 | - | 2 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.-J.; Huang, Y.-J.; Lin, H.-J.; Chang, C.-J.; Hsu, P.-H.; Ooi, G.-X.; Huang, M.-Y.; Lin, H.-T.V. Phenolic Compound Ethyl 3,4-Dihydroxybenzoate Retards Drug Efflux and Potentiates Antibiotic Activity. Antibiotics 2022, 11, 497. https://doi.org/10.3390/antibiotics11040497
Lu W-J, Huang Y-J, Lin H-J, Chang C-J, Hsu P-H, Ooi G-X, Huang M-Y, Lin H-TV. Phenolic Compound Ethyl 3,4-Dihydroxybenzoate Retards Drug Efflux and Potentiates Antibiotic Activity. Antibiotics. 2022; 11(4):497. https://doi.org/10.3390/antibiotics11040497
Chicago/Turabian StyleLu, Wen-Jung, Yan-Jyun Huang, Hsuan-Ju Lin, Chun-Ju Chang, Pang-Hung Hsu, Gui-Xia Ooi, Mei-Ying Huang, and Hong-Ting Victor Lin. 2022. "Phenolic Compound Ethyl 3,4-Dihydroxybenzoate Retards Drug Efflux and Potentiates Antibiotic Activity" Antibiotics 11, no. 4: 497. https://doi.org/10.3390/antibiotics11040497
APA StyleLu, W.-J., Huang, Y.-J., Lin, H.-J., Chang, C.-J., Hsu, P.-H., Ooi, G.-X., Huang, M.-Y., & Lin, H.-T. V. (2022). Phenolic Compound Ethyl 3,4-Dihydroxybenzoate Retards Drug Efflux and Potentiates Antibiotic Activity. Antibiotics, 11(4), 497. https://doi.org/10.3390/antibiotics11040497






