Comparison of a Point-of-Care FilmArray Test to Standard-of-Care Microbiology Test in Diagnosis of Healthcare Associated Infections in a Tertiary Care Pediatric Intensive Care Unit
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Setting and Design
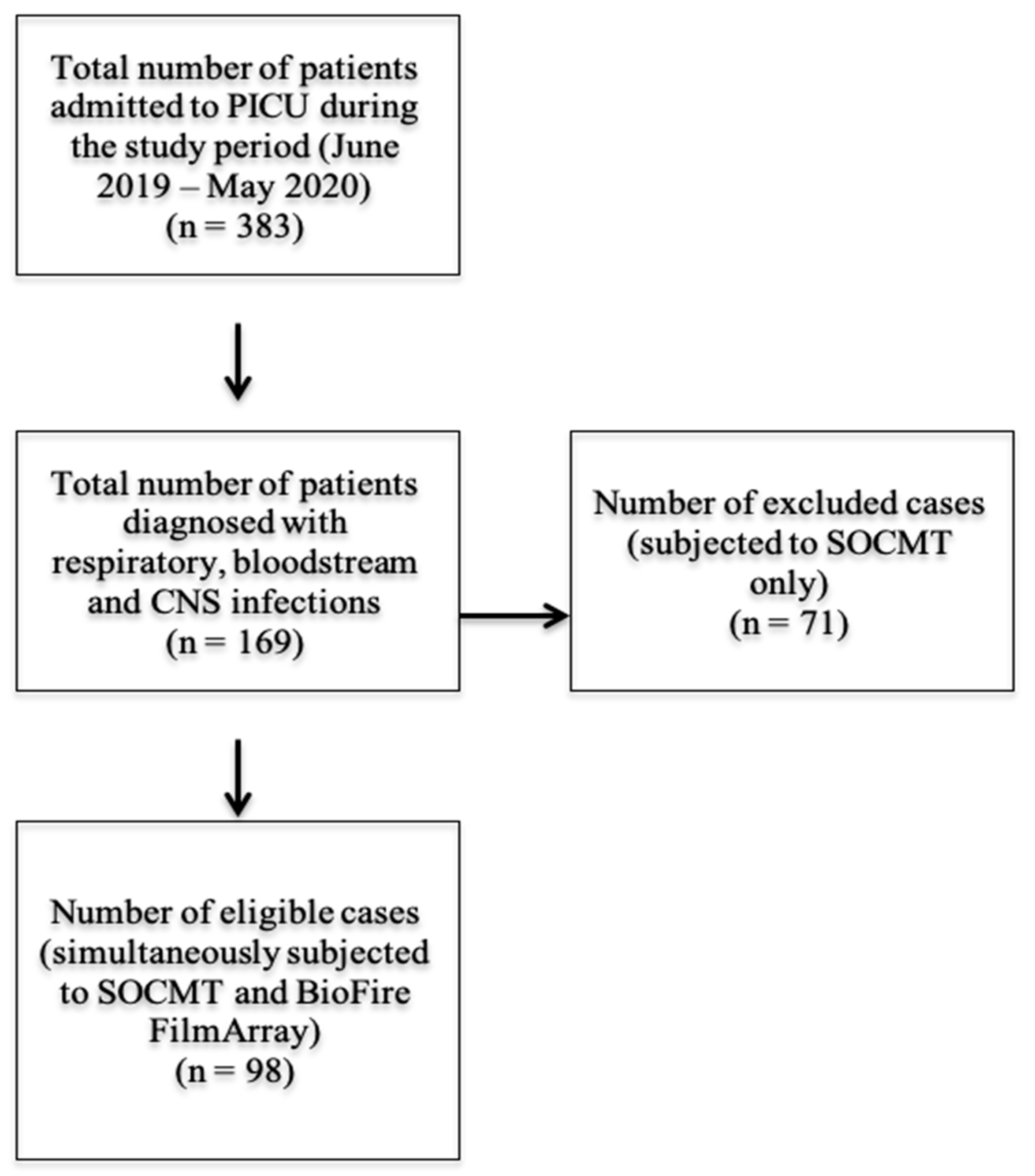
2.2. Patients’ Data
2.3. Standard of Care Microbiology Testing (SOCMT)
2.4. BioFire FilmArray®
2.5. Comparison between the Results of SOCMT and BioFire FilmArray
2.6. Evaluation of the Potential Impact of BioFire Results on Antimicrobial Therapy
2.7. Statistical Analysis
3. Results
3.1. Patients’ and Samples’ Clinical Characteristics
3.2. Diagnostic Performance of SOCMT and BioFire FilmArray
3.3. Screening for Antimicrobial Resistance
3.4. Turn-Around Time (TAT)
3.5. Potential Impacts of BioFire Results on Antimicrobial Therapy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gilligan, P.H.; Gonzalez, M.D.; Jerris, R.C.; Kehl, S.C.; Patel, R.; et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiologya. Clin. Infect. Dis. 2018, 67, e1–e94. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-S.; Tsai, C.-L.; Chang, J.; Hsu, T.-C.; Lin, S.; Lee, C.-C. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: Systematic review and meta-analysis. Clin. Microbiol. Infect. 2018, 24, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Pociute, A.; Kevalas, R.; Malinauskas, M.; Jankauskaite, L. Blood biomarkers differentiating viral versus bacterial pneumonia aetiology: A literature review. Ital. J. Pediatr. 2020, 46, 4. [Google Scholar] [CrossRef] [PubMed]
- Buchan, B.W.; Windham, S.; Balada-Llasat, J.-M.; Leber, A.; Harrington, A.; Relich, R.; Murphy, C.; Bard, J.D.; Naccache, S.; Ronen, S.; et al. Practical Comparison of the BioFire FilmArray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J. Clin. Microbiol. 2020, 58, e00135-20. [Google Scholar] [CrossRef] [PubMed]
- Mayer, L.M.; Kahlert, C.; Rassouli, F.; Vernazza, P.; Albrich, W.C. Impact of viral multiplex real-time PCR on management of respiratory tract infection: A retrospective cohort study. Pneumonia 2017, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, P.; Bryson, A.L.; Binnicker, M.J.; Pritt, B.S.; Patel, R. Syndromic Panel-Based Testing in Clinical Microbiology. Clin. Microbiol. Rev. 2017, 31, e00024-17. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.N.; Fowler, R.; Balada-Llasat, J.M.; Carroll, A.; Stone, H.; Akerele, O.; Buchan, B.; Windham, S.; Hopp, A.; Ronen, S.; et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J. Clin. Microbiol. 2020, 58, e00128-20. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention/National Healthcare Safety Network. Identifying Healthcare-Associated Infections (HAI) for NHSN Surveillance. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/2psc_identifyinghais_nhsncurrent.pdf (accessed on 3 January 2020).
- Centers for Disease Control and Prevention/National Healthcare Safety Network. CDC/NHSN Surveillance Definitions for Specific Types of Infections. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/17pscnosinfdef_current.pdf (accessed on 3 January 2020).
- Tille, P. Bailey and Scott’s Diagnostic Microbiology, 14th ed.; Mosby Elsevier: St Louis, MO, USA, 2017. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Kang, C.M.; Chen, X.J.; Chih, C.C.; Hsu, C.C.; Chen, P.H.; Fen Lee, T.; Teng, L.-J.; Hsueh, P.-R. Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel. J. Microbiol. Immunol. Infect. 2020, 53, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Tansarli, G.S.; Chapin, K.C. Diagnostic test accuracy of the BioFire® FilmArray® meningitis/encephalitis panel: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Ozongwu, C.; Personne, Y.; Platt, G.; Jeanes, C.; Aydin, S.; Kozato, N.; Gant, V.; O’Grady, J.; Enne, V.I. The Unyvero P55 ‘sample-in, answer-out’ pneumonia assay: A performance evaluation. Biomol. Detect. Quantif. 2017, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Beal, S.G.; Cherabuddi, K.; Couturier, B.; Lingenfelter, B.; Rindlisbacher, C.; Jones, J.; Houck, H.J.; Lessard, K.J.; Tremblay, E.E. Performance of a Semiquantitative Multiplex Bacterial and Viral PCR Panel Compared with Standard Microbiological Laboratory Results: 396 Patients Studied With the BioFire Pneumonia Panel. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2020; p. 8. [Google Scholar] [CrossRef]
- Blaschke, A.J.; Holmberg, K.M.; Daly, J.A.; Leber, A.L.; Dien Bard, J.; Korgenski, E.K.; Bourzac, K.M.; Kanack, K.J. Retrospective Evaluation of Infants Aged 1 to 60 Days with Residual Cerebrospinal Fluid (CSF) Tested Using the FilmArray Meningitis/Encephalitis (ME) Panel. J. Clin. Microbiol. 2018, 56, e00277-18. [Google Scholar] [CrossRef] [PubMed]
- Webber, D.M.; Wallace, M.A.; Burnham, C.A.; Anderson, N.W. Evaluation of the BioFire FilmArray Pneumonia Panel for Detection of Viral and Bacterial Pathogens in Lower Respiratory Tract Specimens in the Setting of a Tertiary Care Academic Medical Center. J. Clin. Microbiol. 2020, 58, e00343-20. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.A.; West, J.E.; Balada-Llasat, J.M.; Pancholi, P.; Stevenson, K.B.; Goff, D.A. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin. Infect. Dis. 2010, 51, 1074–1080. [Google Scholar] [CrossRef]
- Walker, T.; Dumadag, S.; Lee, C.J.; Lee, S.H.; Bender, J.M.; Cupo Abbott, J.; She, R.C. Clinical Impact of Laboratory Implementation of Verigene BC-GN Microarray-Based Assay for Detection of Gram-Negative Bacteria in Positive Blood Cultures. J. Clin. Microbiol. 2016, 54, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
| SOCMT (n = 111) | No. | % |
|---|---|---|
| miniBAL culture (n = 72) | 72 | 100 |
| No growth | 51 | 70.8 |
| Klebsiella spp. * | 7 | 9.7 |
| Pseudomonas spp. * | 6 | 8.3 |
| Acinetobacter spp. * | 5 | 7.0 |
| Klebsiella spp., pseudomonas spp. * | 2 | 2.8 |
| Staphylococcus aureus (MRSA) ^ | 1 | 1.4 |
| Total # of positive cases | 21 | 29.2 |
| Blood culture (n = 16) | 16 | 100 |
| No growth | 6 | 37.5 |
| Klebsiella spp. * | 7 | 43.8 |
| E. coli * | 1 | 6.2 |
| Acinetobacter spp. * | 1 | 6.2 |
| Candida albicans # | 1 | 6.2 |
| Total # of positive cases | 10 | 62.5 |
| CSF culture (n = 23) | 23 | 100 |
| No growth | 23 | 100 |
| BioFire FimArray Results | No. | % |
|---|---|---|
| FilmArray® Blood Culture Identification (BCID) Panel (n = 16) | ||
| Negative | 5 | 31.3 |
| Klebsiella pneumoniae * | 7 | 43.8 |
| E. coli * | 1 | 6.3 |
| Acinetobacter baumannii * | 1 | 6.3 |
| Neisseria meningitidis * | 1 | 6.3 |
| Candida albicans # | 1 | 6.3 |
| Total number of positive cases | 11 | 68.7 |
| FilmArray® Pneumonia panel plus (n = 72) | ||
| Negative (n = 6) and viral etiology (n = 21) | 27 | 37.5 |
| Klebsiella pneumoniae * | 15 | 20.8 |
| Pseudomonas aeuroginosa * | 7 | 9.7 |
| Haemophilus influenzae * | 6 | 8.3 |
| Acinetobacter calcoaceticus-baumannii complex * | 4 | 5.6 |
| Streptococcus pneumoniae ^ | 4 | 5.6 |
| Klebsiella pneumoniae/Pseudomonas aeruginosa * | 3 | 4.2 |
| Staphylococcus aureus ^ | 1 | 1.4 |
| Hemophilus influenzae */Staphylococcus aureus ^ | 1 | 1.4 |
| Stahylococcus aureus ^/Streptoccocus.pneumoniae ^/Hemophilus influenzae * | 1 | 1.4 |
| Pseudomonas aeruginosa/Hemophilus influenzae * | 1 | 1.4 |
| Klebsiella pneumoniae */Streptococcus pneumoniae ^ | 1 | 1.4 |
| Acinetobacter calcoaceticus-baumannii complex/Pseudomonas aeruginosa * | 1 | 1.4 |
| Total number of positive cases | 45 | 62.5 |
| Total number of identified bacterial organisms | 54 | |
| FilmArray® Meningitis/Encephalitis (ME) Panel (n = 23) | ||
| Negative | 18 | 78.3 |
| Streptococcus pneumoniae ^ | 4 | 17.4 |
| Hemophilus influenzae * | 1 | 4.3 |
| Total number of positive cases | 5 | 21.7 |
| Organism | Number of Pathogens Identified | p Value | |||
|---|---|---|---|---|---|
| Blood Culture (n = 10) | FilmArray® Blood Culture Identification (BCID) Panel (n = 11) | ||||
| No. | % | No. | % | ||
| Klebsiella pneumoniae | 7 | 70.0 | 7 | 63.6 | 1.000 |
| E. coli | 1 | 10.0 | 1 | 9.1 | 1.000 |
| Acinetobacter calcoaceticus-baumannii complex | 1 | 10.0 | 1 | 9.1 | 1.000 |
| Candida albicans | 1 | 10.0 | 1 | 9.1 | 1.000 |
| Neisseria meningitidis | 0 | 0.0 | 1 | 9.1 | 1.000 |
| miniBAL culture (n = 23) | FilmArray® Pneumonia Panel plus (n = 54) | ||||
| No. | % | No. | % | ||
| Klebsiella pneumoniae | 9 | 39.1 | 19 | 35.2 | 0.742 |
| Pseudomonas aeruginosa | 8 | 34.8 | 12 | 22.2 | 0.250 |
| Acinetobacter calcoaceticus-baumannii complex | 5 | 21.7 | 5 | 9.3 | 0.154 |
| Staphylococcus aureus (MRSA) | 1 | 4.3 | 3 | 5.6 | 1.000 |
| Haemophilus influenzae | 0 | 0.0 | 9 | 16.7 | 0.051 |
| Streptococcus pneumoniae | 0 | 0.0 | 6 | 11.1 | 0.170 |
| CSF culture (n = 0) | FilmArray® Meningitis/Encephalitis (ME) Panel (n = 5) | ||||
| No. | % | No. | % | ||
| Streptococcus pneumoniae | 0 | 0.0 | 4 | 80.0 | - |
| Hemophilus influenzae | 0 | 0.0 | 1 | 20.0 | - |
| Organism | BioFire FilmArray Pneumonia Panel plus Copy Number (Copies/mL) | |||
|---|---|---|---|---|
| 104 | 105 | 106 | ≥107 | |
| Klebsiella pneumoniae (n = 19) | ||||
| Culture positive (n = 9) | - | 1 | 6 | 2 |
| Culture negative (n = 10) | 2 | 1 | 4 | 3 |
| Pseudomonas aeruginosa (n = 12) | ||||
| Culture positive (n = 8) | 1 | 2 | 1 | 4 |
| Culture negative (n = 4) | - | 2 | 1 | 1 |
| Acinetobacter spp. (n = 5) | ||||
| Culture positive (n = 5) | 2 | 1 | - | 2 |
| Culture negative (n = 0) | - | - | - | - |
| Staphylococcus aureus (MRSA) (n = 3) | ||||
| Culture positive (n = 1) | - | - | 1 | - |
| Culture negative (n = 2) | - | 2 | - | - |
| Haemophilus influenzae (n = 9) | ||||
| Culture positive (n = 0) | - | - | - | - |
| Culture negative (n = 9) | 1 | - | 3 | 5 |
| Streptococcus pneumoniae (n = 6) | ||||
| Culture positive (n = 0) | - | - | - | - |
| Culture negative (n = 6) | 2 | - | 4 | - |
| Viruses | Number of miniBAL Specimens | % |
|---|---|---|
| Human Rhinovirus/Enterovirus | 21 | 29.2 |
| Respiratory Syncytial Virus | 19 | 26.4 |
| Influenza A | 8 | 11.1 |
| Adenovirus | 7 | 9.7 |
| Parainfluenza Virus | 5 | 6.9 |
| Coronavirus | 2 | 2.7 |
| Human Metapneumovirus | 1 | 1.4 |
| Influenza B | 1 | 1.4 |
| MERS-CoV | 0 | 0 |
| None detected | 8 | 11.1 |
| Total | 72 | 100 |
| Bacterial/ Fungal Targets | Number of Samples | PPA (95% CI) | NPA (95% CI) | OPA (95% CI) | |||
|---|---|---|---|---|---|---|---|
| SOCMT Positive/BioFire Positive | SOCMT Positive/BioFire Negative | SOCMT Negative/BioFire Positive | SOCMT Negative/BioFire Negative | ||||
| Klebsiella pneumoniae | 16 | 0 | 10 | 72 | 100 (80.6–100.0) | 87.8 (79.0–93.2) | 89.8 (82.2–94.4) |
| E. coli | 1 | 0 | 0 | 97 | 100 (20.7–100.0) | 100 (96.2–100.0) | 100 (96.2–100.0) |
| Pseudomonas aeruginosa | 8 | 0 | 4 | 86 | 100 (67.6–100.0) | 95.6 (89.1–98.3) | 95.9 (90.0–98.4) |
| Acinetobacter spp. | 6 | 0 | 0 | 92 | 100 (61.0–100.0) | 100 (96.0–100.0) | 100 (96.2–100.0) |
| Haemophilus influenzae | 0 | 0 | 10 | 88 | - | 89.8 (82.2–94.4) | 89.8 (82.2–94.4) |
| Neisseria meningitidis | 0 | 0 | 1 | 97 | - | 99.0 (94.4–99.8) | 99.0 (94.4–99.8) |
| Streptococcus pneumoniae | 0 | 0 | 10 | 88 | - | 89.8 (82.2–94.4) | 89.8 (82.2–94.4) |
| Staphylococcus aureus | 1 | 0 | 2 | 95 | 100 (20.7–100.0) | 97.9 (92.8–99.4) | 98.0 (92.9–99.4) |
| Candida albicans | 1 | 0 | 0 | 97 | 100 (20.7–100.0) | 100 (96.2–100.0) | 100 (96.2–100.0) |
| Total | 33 | 0 | 37 | 812 | 100 (89.6–100.0) | 95.6 (94.1–96.8) | 95.8 (94.3–96.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Nawawy, A.A.; Antonios, M.A.; Tawfik, M.E.; Meheissen, M.A. Comparison of a Point-of-Care FilmArray Test to Standard-of-Care Microbiology Test in Diagnosis of Healthcare Associated Infections in a Tertiary Care Pediatric Intensive Care Unit. Antibiotics 2022, 11, 453. https://doi.org/10.3390/antibiotics11040453
El-Nawawy AA, Antonios MA, Tawfik ME, Meheissen MA. Comparison of a Point-of-Care FilmArray Test to Standard-of-Care Microbiology Test in Diagnosis of Healthcare Associated Infections in a Tertiary Care Pediatric Intensive Care Unit. Antibiotics. 2022; 11(4):453. https://doi.org/10.3390/antibiotics11040453
Chicago/Turabian StyleEl-Nawawy, Ahmed A., Manal A. Antonios, Medhat E. Tawfik, and Marwa A. Meheissen. 2022. "Comparison of a Point-of-Care FilmArray Test to Standard-of-Care Microbiology Test in Diagnosis of Healthcare Associated Infections in a Tertiary Care Pediatric Intensive Care Unit" Antibiotics 11, no. 4: 453. https://doi.org/10.3390/antibiotics11040453
APA StyleEl-Nawawy, A. A., Antonios, M. A., Tawfik, M. E., & Meheissen, M. A. (2022). Comparison of a Point-of-Care FilmArray Test to Standard-of-Care Microbiology Test in Diagnosis of Healthcare Associated Infections in a Tertiary Care Pediatric Intensive Care Unit. Antibiotics, 11(4), 453. https://doi.org/10.3390/antibiotics11040453






