Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater
Abstract
1. Introduction
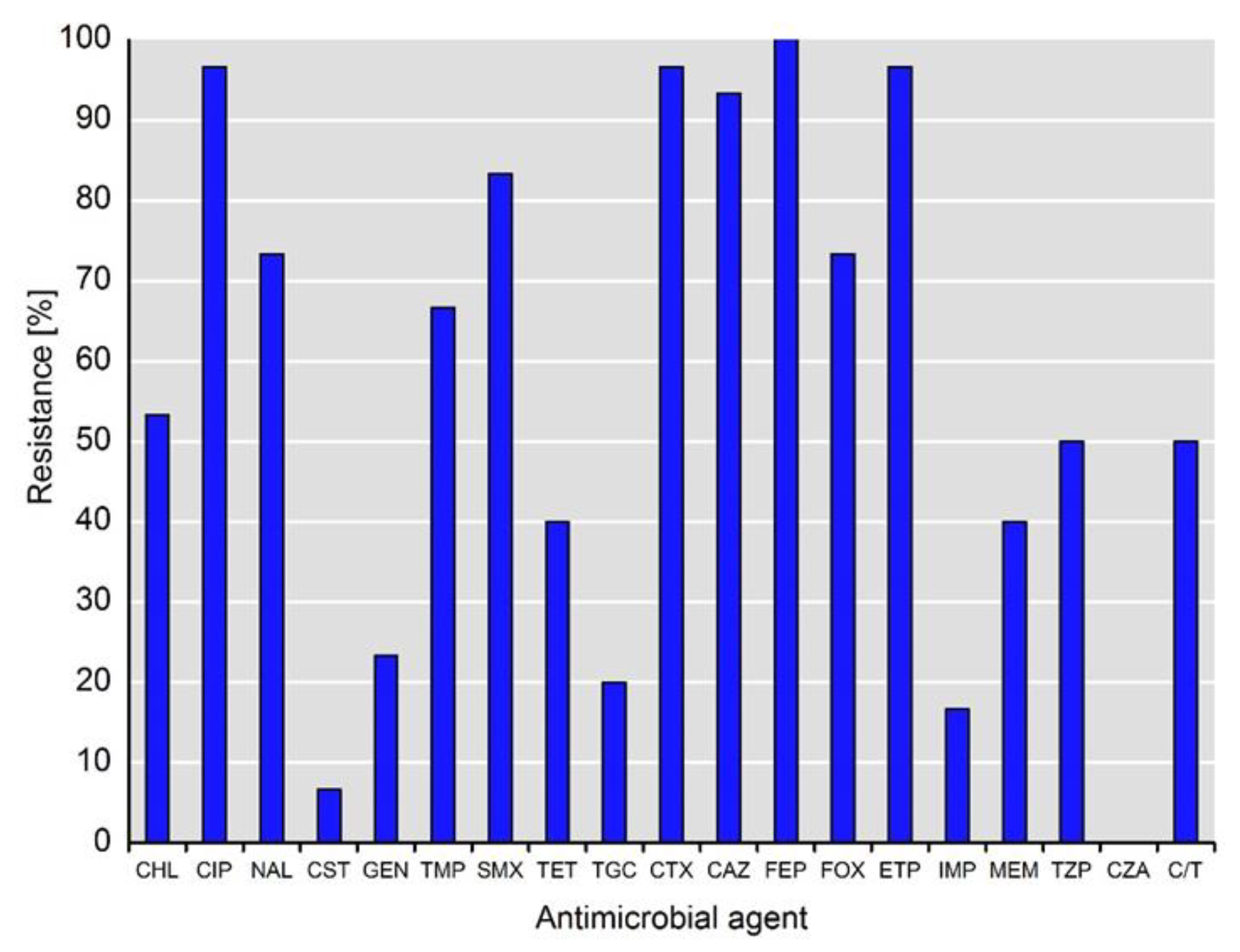
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- Zumla, A. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Lancet Infect. Dis. 2010, 10, 303–304. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Carbapenem-Resistant Enterobacteriaceae, Second Update—26 September 2019. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/carbapenem-resistant-enterobacteriaceae-risk-assessment-rev-2.pdf (accessed on 10 February 2022).
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2019.pdf (accessed on 10 February 2022).
- Baughman, R.P. The use of carbapenems in the treatment of serious infections. J. Intensive Care Med. 2009, 24, 230–241. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities. Available online: https://apps.who.int/iris/bitstream/handle/10665/259462/9789241550178-eng.pdf?sequence=1&isAllowed=y (accessed on 10 February 2022).
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Livorsi, D.J.; Chorazy, M.L.; Schweizer, M.L.; Balkenende, E.C.; Blevins, A.E.; Nair, R.; Samore, M.H.; Nelson, R.E.; Khader, K.; Perencevich, E.N. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control 2018, 7, 55. [Google Scholar] [CrossRef]
- Kehl, K.; Schallenberg, A.; Szekat, C.; Albert, C.; Sib, E.; Exner, M.; Zacharias, N.; Schreiber, C.; Parčina, M.; Bierbaum, G. Dissemination of carbapenem resistant bacteria from hospital wastewater into the environment. Sci. Total Environ. 2022, 806, 151339. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Hellmark, B.; Ehricht, R.; Söderquist, B.; Jass, J. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2241–2251. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Schill, S.; Stoeger, A.; Pekard-Amenitsch, S.; Huhulescu, S.; Inreiter, N.; Hartl, R.; Kerschner, H.; Sorschag, S.; Springer, B.; et al. Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci. Total Environ. 2019, 662, 227–235. [Google Scholar] [CrossRef]
- Bleichenbacher, S.; Stevens, M.J.A.; Zurfluh, K.; Perreten, V.; Endimiani, A.; Stephan, R.; Nüesch-Inderbinen, M. Environmental dissemination of carbapenemase-producing Enterobacteriaceae in rivers in Switzerland. Environ. Pollut. 2020, 265, 115081. [Google Scholar] [CrossRef]
- Teban-Man, A.; Farkas, A.; Baricz, A.; Hegedus, A.; Szekeres, E.; Pârvu, M.; Coman, C. Wastewaters, with or without Hospital Contribution, Harbour MDR, Carbapenemase-Producing, but Not Hypervirulent Klebsiella pneumoniae. Antibiotics 2021, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. Available online: https://www.fao.org/3/i6209e/i6209e.pdf (accessed on 10 February 2022).
- The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, e05598. [CrossRef]
- Hamza, E.; Dorgham, S.M.; Hamza, D.A. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J. Glob. Antimicrob. Resist. 2016, 7, 8–10. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, Y.; Sun, L.; Pang, M.; Zhang, L.; Wang, R. Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu, China. J. Antimicrob. Chemother. 2017, 72, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Diab, M.; Hamze, M.; Bonnet, R.; Saras, E.; Madec, J.-Y.; Haenni, M. OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: Epidemic spread of dominant Klebsiella pneumoniae clones. J. Med. Microbiol. 2017, 66, 1688–1691. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; Reuland, E.A.; Wintermans, B.B.; Al Naiemi, N.; Koek, A.; Abdelwahab, A.M.; Ammar, A.M.; Mohamed, A.A.; Vandenbroucke-Grauls, C.M.J.E. Extended-Spectrum β-Lactamases and/or Carbapenemases-Producing Enterobacteriaceae Isolated from Retail Chicken Meat in Zagazig, Egypt. PLoS ONE 2015, 10, e0136052. [Google Scholar] [CrossRef]
- Wielders, C.C.H.; van Hoek, A.H.A.M.; Hengeveld, P.D.; Veenman, C.; Dierikx, C.M.; Zomer, T.P.; Smit, L.A.M.; van der Hoek, W.; Heederik, D.J.; de Greeff, S.C.; et al. Extended-spectrum β-lactamase- and pAmpC-producing Enterobacteriaceae among the general population in a livestock-dense area. Clin. Microbiol. Infect. 2017, 23, 120.e1–120.e8. [Google Scholar] [CrossRef]
- Meijs, A.P.; Gijsbers, E.F.; Hengeveld, P.D.; Dierikx, C.M.; De Greeff, S.C.; van Duijkeren, E. ESBL/pAmpC-producing Escherichia coli and Klebsiella pneumoniae carriage among veterinary healthcare workers in the Netherlands. Antimicrob. Resist. Infect. Control 2021, 10, 147. [Google Scholar] [CrossRef]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018, 94, fiy057. [Google Scholar] [CrossRef] [PubMed]
- Kizny Gordon, A.E.; Mathers, A.J.; Cheong, E.Y.L.; Gottlieb, T.; Kotay, S.; Walker, A.S.; Peto, T.E.A.; Crook, D.W.; Stoesser, N. The Hospital Water Environment as a Reservoir for Carbapenem-Resistant Organisms Causing Hospital-Acquired Infections-A Systematic Review of the Literature. Clin. Infect. Dis. 2017, 64, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Surleac, M.; Czobor Barbu, I.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Carbapenemase-producing Gram-negative bacteria in aquatic environments: A review. J. Glob. Antimicrob. Resist. 2021, 25, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Rocha, J.; Ferreira, C.; Mil-Homens, D.; Busquets, A.; Fialho, A.M.; Henriques, I.; Gomila, M.; Manaia, C.M. Third generation cephalosporin-resistant Klebsiella pneumoniae thriving in patients and in wastewater: What do they have in common? BMC Genom. 2022, 23, 72. [Google Scholar] [CrossRef]
- WHO Advisory Group on Integrated Surveillance. Critically Important Antimicrobials for Human Medicine: 6th Revision 2018. Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-Human Use. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 10 February 2022).
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Hubbard, A.T.M.; Mason, J.; Roberts, P.; Parry, C.M.; Corless, C.; van Aartsen, J.; Howard, A.; Bulgasim, I.; Fraser, A.J.; Adams, E.R.; et al. Piperacillin/tazobactam resistance in a clinical isolate of Escherichia coli due to IS26-mediated amplification of blaTEM-1B. Nat. Commun. 2020, 11, 4915. [Google Scholar] [CrossRef]
- Livermore, D.M.; Day, M.; Cleary, P.; Hopkins, K.L.; Toleman, M.A.; Wareham, D.W.; Wiuff, C.; Doumith, M.; Woodford, N. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 2019, 74, 326–333. [Google Scholar] [CrossRef]
- Antunes, N.T.; Fisher, J.F. Acquired Class D β-Lactamases. Antibiotics 2014, 3, 398–434. [Google Scholar] [CrossRef]
- Kotsakis, S.D.; Flach, C.-F.; Razavi, M.; Larsson, D.G.J. Characterization of the First OXA-10 Natural Variant with Increased Carbapenemase Activity. Antimicrob. Agents Chemother. 2019, 63, 4–6. [Google Scholar] [CrossRef]
- Maurya, A.P.; Dhar, D.; Basumatary, M.K.; Paul, D.; Ingti, B.; Choudhury, D.; Talukdar, A.D.; Chakravarty, A.; Mishra, S.; Bhattacharjee, A. Expansion of highly stable bla OXA-10 β-lactamase family within diverse host range among nosocomial isolates of Gram-negative bacilli within a tertiary referral hospital of Northeast India. BMC Res. Notes 2017, 10, 145. [Google Scholar] [CrossRef]
- Arca-Suárez, J.; Lasarte-Monterrubio, C.; Rodiño-Janeiro, B.-K.; Cabot, G.; Vázquez-Ucha, J.C.; Rodríguez-Iglesias, M.; Galán-Sánchez, F.; Beceiro, A.; González-Bello, C.; Oliver, A.; et al. Molecular mechanisms driving the in vivo development of OXA-10-mediated resistance to ceftolozane/tazobactam and ceftazidime/avibactam during treatment of XDR Pseudomonas aeruginosa infections. J. Antimicrob. Chemother. 2021, 76, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Heiden, S.E.; Hübner, N.-O.; Bohnert, J.A.; Heidecke, C.-D.; Kramer, A.; Balau, V.; Gierer, W.; Schaefer, S.; Eckmanns, T.; Gatermann, S.; et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Bathoorn, E.; Rossen, J.W.; Lokate, M.; Friedrich, A.W.; Hammerum, A.M. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill. 2015, 20, 30040. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papagiannitsis, C.C.; Dolejska, M.; Izdebski, R.; Dobiasova, H.; Studentova, V.; Esteves, F.J.; Derde, L.P.G.; Bonten, M.J.M.; Hrabák, J.; Gniadkowski, M. Characterization of pKP-M1144, a Novel ColE1-Like Plasmid Encoding IMP-8, GES-5, and BEL-1 β-Lactamases, from a Klebsiella pneumoniae Sequence Type 252 Isolate. Antimicrob. Agents Chemother. 2015, 59, 5065–5068. [Google Scholar] [CrossRef]
- Kiaei, S.; Moradi, M.; Hosseini-Nave, H.; Ziasistani, M.; Kalantar-Neyestanaki, D. Endemic dissemination of different sequence types of carbapenem-resistant Klebsiella pneumoniae strains harboring blaNDM and 16S rRNA methylase genes in Kerman hospitals, Iran, from 2015 to 2017. Infect. Drug Resist. 2019, 12, 45–54. [Google Scholar] [CrossRef]
- Van Duijkeren, E.; Wielders, C.C.H.; Dierikx, C.M.; van Hoek, A.H.A.M.; Hengeveld, P.; Veenman, C.; Florijn, A.; Lotterman, A.; Smit, L.A.M.; van Dissel, J.T.; et al. Long-term Carriage of Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae in the General Population in The Netherlands. Clin. Infect. Dis. 2018, 66, 1368–1376. [Google Scholar] [CrossRef]
- Yoon, E.-J.; Kim, J.O.; Kim, D.; Lee, H.; Yang, J.W.; Lee, K.J.; Jeong, S.H. Klebsiella pneumoniae Carbapenemase Producers in South Korea between 2013 and 2015. Front. Microbiol. 2018, 9, 56. [Google Scholar] [CrossRef]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef]
- Wei, L.; Feng, Y.; Wen, H.; Ya, H.; Qiao, F.; Zong, Z. NDM-5-producing carbapenem-resistant Klebsiella pneumoniae of sequence type 789 emerged as a threat for neonates: A multicentre, genome-based study. Int. J. Antimicrob. Agents 2021, 59, 106508. [Google Scholar] [CrossRef]
- Kluytmans, J.A.J.W.; Overdevest, I.T.M.A.; Willemsen, I.; Kluytmans-van den Bergh, M.F.Q.; van der Zwaluw, K.; Heck, M.; Rijnsburger, M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M.; Johnston, B.D.; et al. Extended-spectrum β-lactamase-producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 2013, 56, 478–487. [Google Scholar] [CrossRef]
- Kola, A.; Kohler, C.; Pfeifer, Y.; Schwab, F.; Kühn, K.; Schulz, K.; Balau, V.; Breitbach, K.; Bast, A.; Witte, W.; et al. High prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012, 67, 2631–2634. [Google Scholar] [CrossRef] [PubMed]
- Zarfel, G.; Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Kittinger, C.; Grisold, A.J.; Pless, P.; Feierl, G. Multiresistant bacteria isolated from chicken meat in Austria. Int. J. Environ. Res. Public Health 2014, 11, 12582–12593. [Google Scholar] [CrossRef] [PubMed]
- Ghodousi, A.; Bonura, C.; Di Noto, A.M.; Mammina, C. Extended-Spectrum ß-Lactamase, AmpC-Producing, and Fluoroquinolone-Resistant Escherichia coli in Retail Broiler Chicken Meat, Italy. Foodborne Pathog. Dis. 2015, 12, 619–625. [Google Scholar] [CrossRef]
- Egea, P.; López-Cerero, L.; Torres, E.; Del Gómez-Sánchez, M.C.; Serrano, L.; Navarro Sánchez-Ortiz, M.D.; Rodriguez-Baño, J.; Pascual, A. Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int. J. Food Microbiol. 2012, 159, 69–73. [Google Scholar] [CrossRef] [PubMed]
- The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [CrossRef]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef]
- Clegg, S.; Murphy, C.N. Epidemiology and Virulence of Klebsiella pneumoniae. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0, 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 10 February 2022).
- Savin, M.; Bierbaum, G.; Schmithausen, R.M.; Heinemann, C.; Kreyenschmidt, J.; Schmoger, S.; Akbaba, I.; Käsbohrer, A.; Hammerl, J.A. Slaughterhouse wastewater as a reservoir for extended-spectrum β-lactamase (ESBL)-producing, and colistin-resistant Klebsiella spp. and their impact in a “One Health” perspective. Sci. Total Environ. 2021, 804, 150000. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
| Isolate | Species | Origin | Resistance Phenotype a | Combinations of β-Lactam–β-Lactamase Inhibitor | Antimicrobial Resistance Genes to Β-Lactams | MLST |
|---|---|---|---|---|---|---|
| 05/11-30 | K. oxytoca | Effluent mWWTP | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, ETP | TZP, C/T | blaOXY-2-8-like c | - d |
| 05/11-32 | K. oxytoca | Effluent mWWTP | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP | TZP, C/T | blaOXY-2-8-like | - |
| 05/10-58 | K. oxytoca | Influent mWWTP | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP | TZP, C/T | blaOXY-2-8-like | - |
| 05/10-60 | K. oxytoca | Influent mWWTP | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP | TZP, C/T | blaOXY-2-8-like | - |
| 03/12-04Bki | K. oxytoca | On-site preflooder downstream | CIP, NAL, GEN, SMX, CTX, CAZ, FEP, FOX, ETP | TZP, C/T | blaCTX-M-9, blaOXA-4, blaOXY-2-8-like | - |
| 05/13-23 | K. oxytoca | On-site preflooder upstream | CIP, TMP, SMX, TET, CTX, CAZ, FEP, FOX, ETP | - b | blaCTX-M-15, blaOXY-2-5 c | - |
| 05/13-25 | K. oxytoca | On-site preflooder upstream | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP | TZP, C/T | blaOXY-2-8-like | - |
| 03/11-12 | K. pneumoniae | Effluent mWWTP | CIP, NAL, TET, CTX, CAZ, FEP, FOX, ETP, MEM | - | blaOKP-B-3-like c | - |
| 03/11-28 | K. pneumoniae | Effluent mWWTP | CIP, NAL, TMP, SMX, CTX, CAZ, FEP, ETP | - | blaCTX-M-15, blaOXA-1, blaSHV-28, blaTEM-1B | ST307 |
| 03/11-38 | K. pneumoniae | Effluent mWWTP | CHL, CIP, NAL, CTX, CAZ, FEP, FOX, ETP, IMI, MEM | - | blaGES-5-like, blaSHV-2-like | - |
| 05/11-29 | K. pneumoniae | Effluent mWWTP | CHL, CIP, NAL, CST, CTX, CAZ, FEP, FOX, ETP, IMI | TZP | blaCTX-M-15, blaOXA-1, blaSHV-1 c, blaSHV-148-like | ST16 |
| 05/11-43 | K. pneumoniae | Effluent mWWTP | CHL, CIP, NAL, GEN, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP | TZP | blaCTX-M-15, blaOXY-2-2-like c, blaTEM-1B | - |
| 04/08-35 | K. pneumoniae | Poultry Eviscerators | CIP, CTX, CAZ, FEP, FOX, ETP | - | blaSHV-25 | ST789 |
| 03/06-23 | K. pneumoniae | Pig Holding Pens | CHL, CIP, TMP, SMX, TET, CTX, CAZ, FEP, ETP | TZP, C/T | blaCTX-M-1, blaSHV-27-like, blaTEM-1B | ST873 |
| 03/01-52 | K. pneumoniae | Influent in-house chemical-physical WWTP | CIP, TMP, SMX, CTX, FEP, FOX, ETP, MEM | - | blaSHV-33 | ST1948 |
| 03/10-26 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP, MEM | TZP, C/T | blaCTX-M-15, blaOXA-1, blaSHV-1 | ST2459 |
| 03/10-27 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, TMP, SMX, CTX, CAZ, FEP, FOX, ETP, MEM | TZP, C/T | blaCTX-M-15, blaOXA-1, blaSHV-1 | ST2459 |
| 03/10-46 | K. pneumoniae | Influent mWWTP | CIP, TMP, SMX, CTX, CAZ, FEP, IMI | - | blaCTX-M-15, blaSHV-1-like | ST219 |
| 05/10-20 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, GEN, SMX, TET, CTX, CAZ, FEP, FOX, ETP, IMI, MEM | TZP, C/T | blaOXA-10, blaSHV-31 | ST252 |
| 05/10-21 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, GEN, SMX, TET, CTX, CAZ, FEP, FOX, ETP, IMI, MEM | TZP, C/T | blaOXA-10, blaSHV-31 | ST252 |
| 05/10-59 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, GEN, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP | C/T | blaCTX-M-15, blaSHV-11, blaTEM-1A | ST268 |
| 05/10-69A | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, SMX, CTX, CAZ, FEP, FOX, ETP, MEM | TZP, C/T | blaOXA-10, blaSHV-69-like, blaTEM-1B | ST503 |
| 05/10-69B | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, SMX, FEP, ETP, MEM | TZP, C/T | blaOXA-10, blaSHV-69-like, blaTEM-1B | ST503 |
| 05/10-71 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, CST, SMX, CTX, CAZ, FEP, ETP, MEM | TZP, C/T | blaOXA-10, blaSHV-69-like | ST503 |
| 05/10-83 | K. pneumoniae | Influent mWWTP | CHL, CIP, NAL, GEN, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP | TZP | blaCTX-M-15, blaOXA-1, blaSHV-38-like, blaTEM-1B | ST441 |
| 03/13-21 | K. pneumoniae | On-site preflooder upstream | CHL, CIP, NAL, TMP, CTX, CAZ, FEP, ETP | - | blaCTX-M-15, blaSHV-1 | ST2459 |
| 05/13-31 | K. pneumoniae | On-site preflooder upstream | CHL, CIP, NAL, GEN, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP | - | blaCTX-M-15, blaSHV-11, blaTEM-1A | ST268 |
| 03/05-22 | K. pneumoniae | Pig Transporters | CHL, TMP, SMX, TET, CTX, CAZ, FEP, ETP | - | blaCTX-M-1, blaSHV-27-like, blaTEM-1B | ST873 |
| 01/07-40 | K. pneumoniae | Poultry Stunning Facilities | CIP, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP, MEM | TZP | blaSHV-28-like, blaTEM-1B | ST458 |
| 01/07-41 | K. pneumoniae | Poultry Stunning Facilities | CIP, TMP, SMX, TET, TGC, CTX, CAZ, FEP, FOX, ETP, MEM | - | blaSHV-28-like, blaTEM-1B | ST458 |
| Antimicrobial Class | Genes | Percentage [%] |
|---|---|---|
| β-lactams | blaCTX-M-15 | 36.7 |
| blaTEM-1B | 30.0 | |
| blaOXY-2-8-like a | 20.0 | |
| blaOXA-1 | 16.7 | |
| blaSHV-1 a | 16.7 | |
| blaOXA-10 | 16.7 | |
| blaSHV-69-like | 10.0 | |
| blaTEM-1B, blaSHV-27-like, blaSHV-27-like, blaSHV-11, blaTEM-1A | each 6.7 | |
| blaOKP-B-3-likea, blaSHV-28, blaGES-5-like, blaSHV-2-like, blaSHV-148-like, blaOXY-2-2-likea, blaSHV-25, blaSHV-33, blaSHV-38-like, blaCTX-M-9, blaOXA-4, blaOXY-2-5a, blaSHV-28-like | each 3.3 | |
| Aminoglycosides | strB | 40.0 |
| strA | 36.7 | |
| aadA5 | 23.3 | |
| aac(3)-I-like | 16.7 | |
| aadA1 | 16.7 | |
| aadA2 | 16.7 | |
| strA-like | 13.3 | |
| strB-like | 13.3 | |
| aadB | 10.0 | |
| aac(3)-IId-like, aacA4, aadA24-like, aph(3′)-Ia | each 6.7 | |
| aac(3)-IIa-like, aacA4-like, aadA22, aph(3′)-XV | each 3.3 | |
| Phenicols | catB3-like | 20.0 |
| floR-like | 6.7 | |
| catB2 | 3.3 | |
| Fluoroquinolones and aminoglycosides | aac(6’)Ib-cr | 20.0 |
| aac(6’)Ib-cr-like | 10.0 | |
| Diaminopyrimidines (Trimethoprim) | dfrA14-like | 26.7 |
| dfrA17 | 23.3 | |
| dfrA1 | 13.3 | |
| dfrA12 | 6.7 | |
| Sulfonamides | sul1 | 56.7 |
| sul2 | 33.3 | |
| sul2-like | 16.7 | |
| Phosphonic Acid (Fosfomycin) | fosA-like a | 56.7 |
| fosAa | 13.3 | |
| Quinolones | oqxA-like a | 70.0 |
| oqxB-like a | 70.0 | |
| qnrB66-like | 13.3 | |
| qnrS1 | 13.3 | |
| qnrA1-like | 10.0 | |
| qnrB1 | 6.7 | |
| Tetracyclines | tet(A) | 13.3 |
| tet(A)-like | 6.7 | |
| tet(B) | 6.7 | |
| Macrolides | mph(A) | 30.0 |
| erm(B)-like | 6.7 | |
| Lincosamides | lnu(F) | 3.3 |
| Virulence Factor | Genes | Percentage a |
|---|---|---|
| Enterobactin | ent | 96.7 |
| Yersiniabactin | ybt, irp1, irp2, fyuA | 40.0 |
| Salmochelin | iroN, iroBCD | 40.0 |
| Aerobactin | iucABCD, iutA | 33.3 |
| Colibactin | clbA-R | 0 |
| Regulators of mucoid phenotype | rmpA, rmpA2, rmpB | 0 |
| K1 capsule synthesis | magA | 0 |
| Chromosomal capsule production | cps | 0 |
| Fimbriae type 1 | fim | 90.0 |
| Fimbriae type 3 | mrk | 63.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savin, M.; Bierbaum, G.; Mutters, N.T.; Schmithausen, R.M.; Kreyenschmidt, J.; García-Meniño, I.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics 2022, 11, 435. https://doi.org/10.3390/antibiotics11040435
Savin M, Bierbaum G, Mutters NT, Schmithausen RM, Kreyenschmidt J, García-Meniño I, Schmoger S, Käsbohrer A, Hammerl JA. Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics. 2022; 11(4):435. https://doi.org/10.3390/antibiotics11040435
Chicago/Turabian StyleSavin, Mykhailo, Gabriele Bierbaum, Nico T. Mutters, Ricarda Maria Schmithausen, Judith Kreyenschmidt, Isidro García-Meniño, Silvia Schmoger, Annemarie Käsbohrer, and Jens Andre Hammerl. 2022. "Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater" Antibiotics 11, no. 4: 435. https://doi.org/10.3390/antibiotics11040435
APA StyleSavin, M., Bierbaum, G., Mutters, N. T., Schmithausen, R. M., Kreyenschmidt, J., García-Meniño, I., Schmoger, S., Käsbohrer, A., & Hammerl, J. A. (2022). Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics, 11(4), 435. https://doi.org/10.3390/antibiotics11040435






