IPA-3: An Inhibitor of Diadenylate Cyclase of Streptococcus suis with Potent Antimicrobial Activity
Abstract
:1. Introduction
2. Results
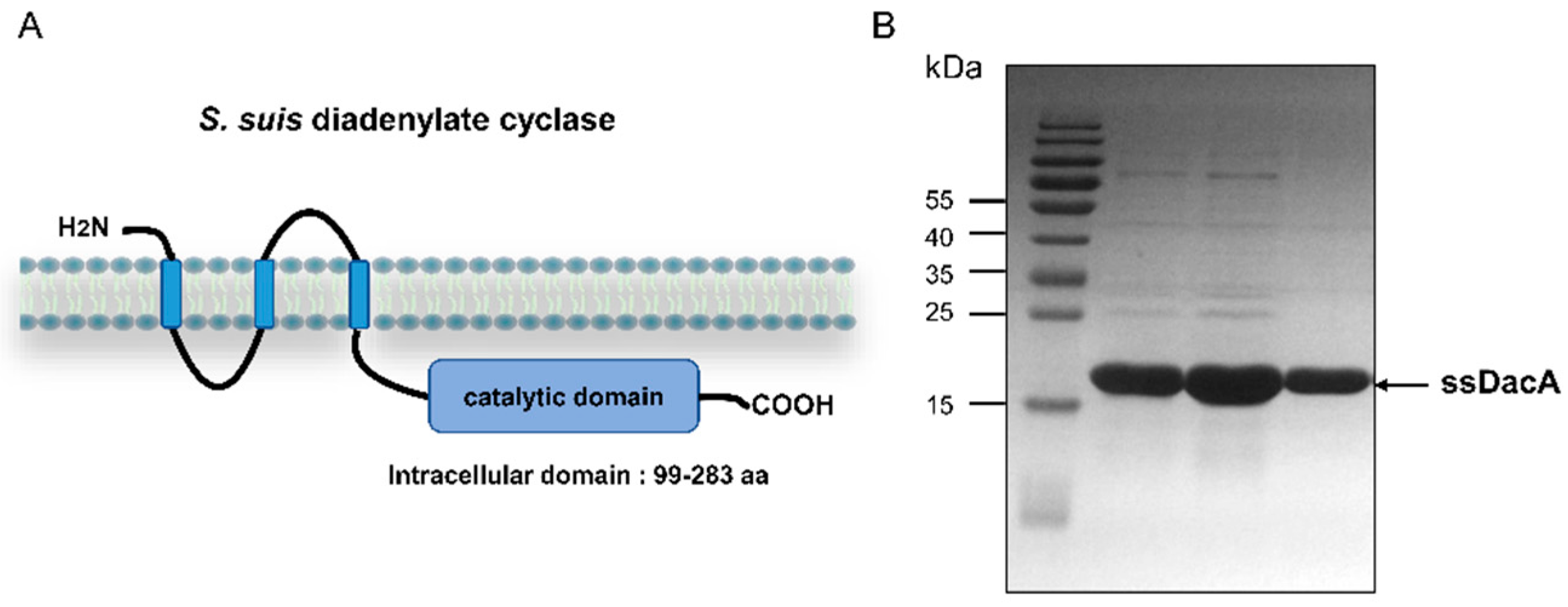
2.1. Purification of ssDacA
2.2. Determination of the Optimal Enzymatic Reaction Conditions for ssDacA
2.3. Screening for ssDacA Inhibitors
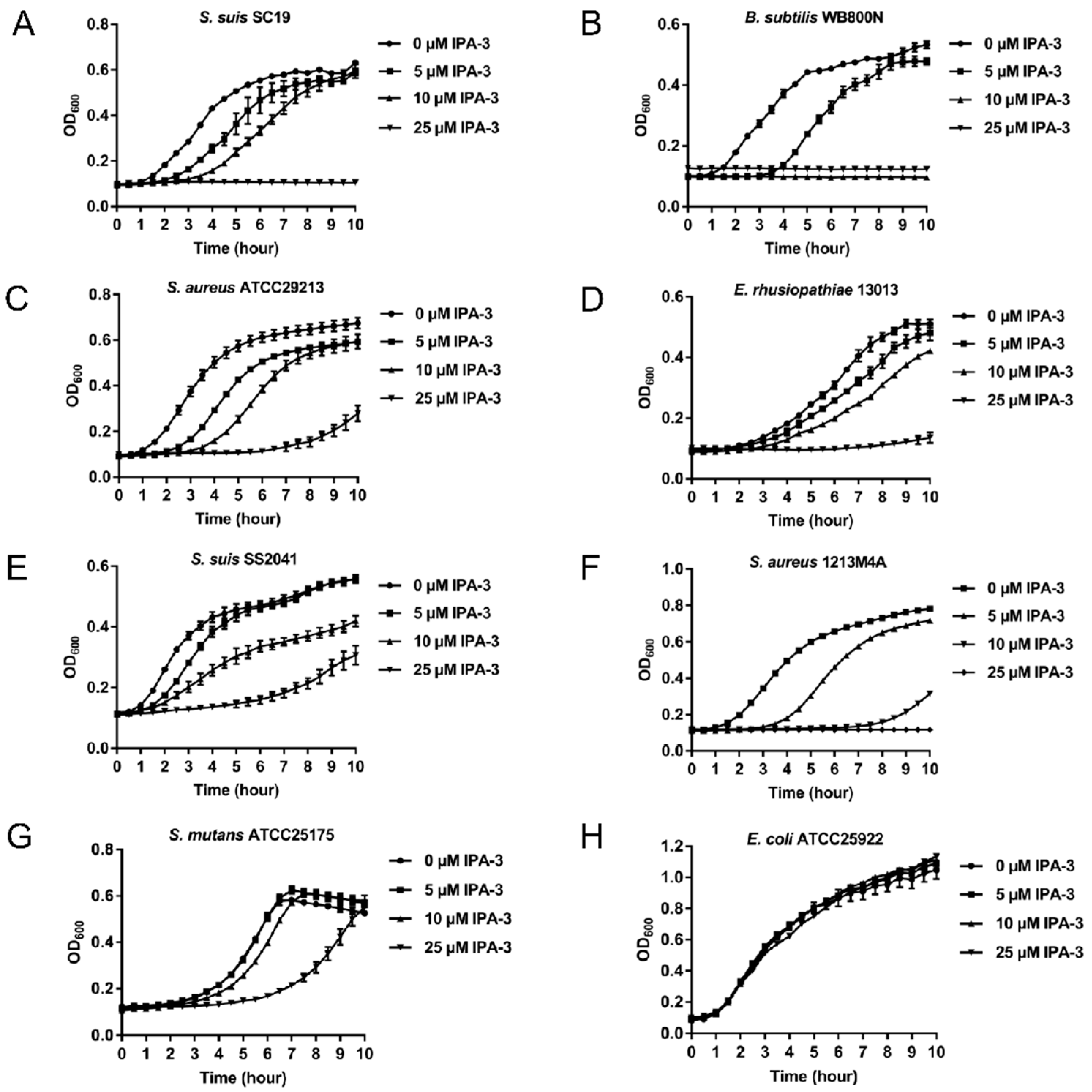
2.4. Antimicrobial Activity of IPA-3
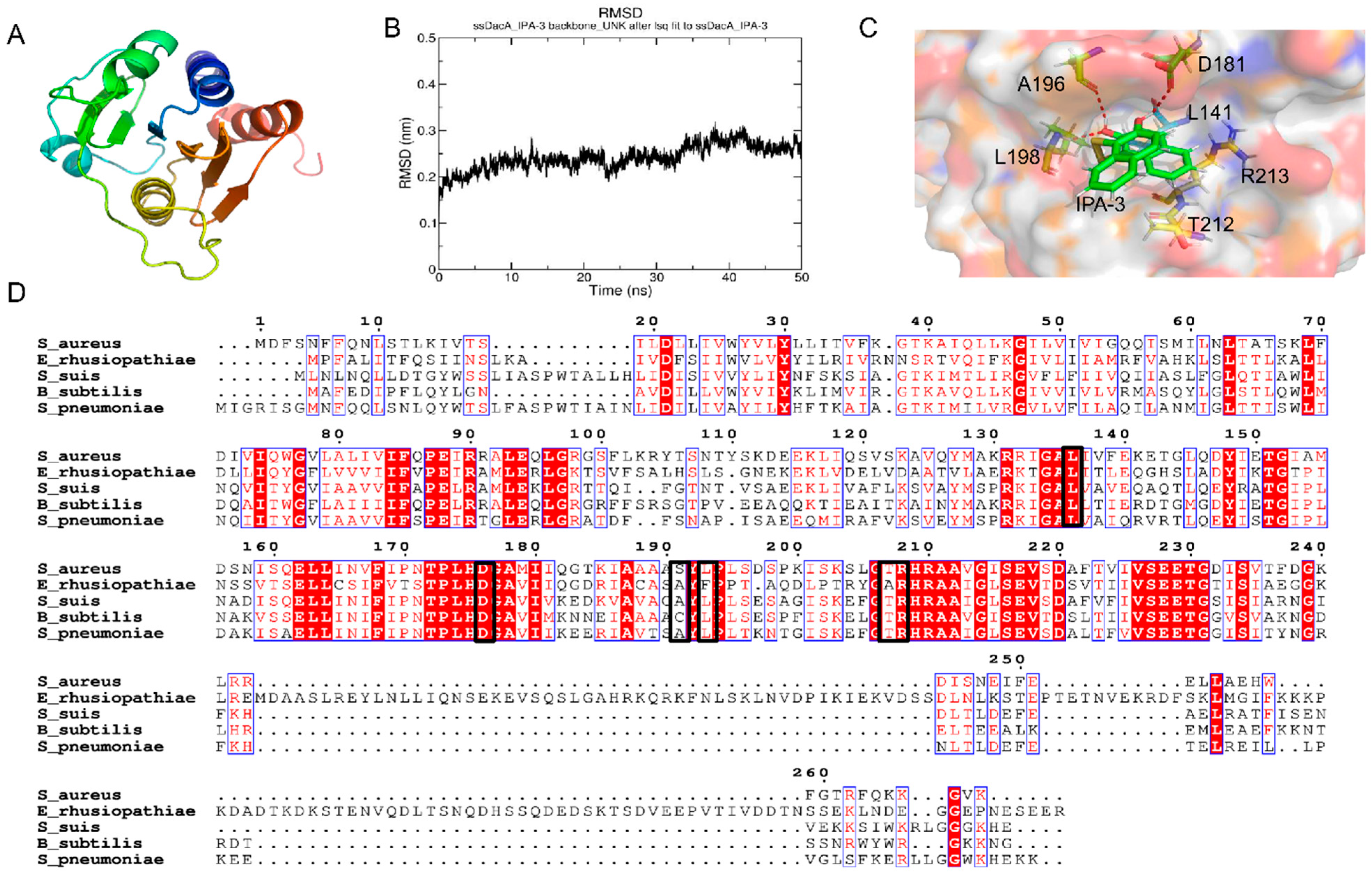
2.5. Potential Binding Mode
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Drug Library
4.2. Protein Expression and Purification
4.3. High-Throughput Screening for ssDacA Inhibitors
4.4. Determination of Half-Maximal Inhibitory Concentration (IC50) of IPA-3 against ssDacA
4.5. High-Performance Liquid Chromatography Analysis
4.6. Bacterial Growth Inhibition Assay
4.7. In Silico Docking
4.8. Molecular Dynamics Simulation
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADME | Absorption, distribution, metabolism, and excretion |
| AMR | Antimicrobial resistance |
| AMP | Adenosine monophosphate |
| BHI | Brain heart infusion |
| c-di-AMP | Cyclic diadenylate monophosphate |
| c-di-GMP | Cyclic diguanylate monophosphate |
| cGAMP | Cyclic GMP-AMP |
| DacA | Diadenylate cyclase |
| GMP | Guanosine monophosphate |
| HPLC | High-performance liquid chromatography |
| IC50 | Half-maximal inhibitory concentration |
| IPA-3 | 2,2′-Dihydroxy-1,1′-dinapthyldisulfide |
| IPTG | Isopropyl-β-D-thiogalactopyranoside |
| LB | Lysogeny broth |
| RLU | Relative light unit |
| RMSD | Root-mean-square deviation |
| SAR | Structure–activity relationship |
| TSA | Tryptic soy agar |
| TSB | Tryptic soy broth |
References
- Brinkac, L.; Voorhies, A.; Gomez, A.; Nelson, K.E. The threat of antimicrobial resistance on the human microbiome. Microb. Ecol. 2017, 74, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Croft, A.C.; D’Antoni, A.V.; Terzulli, S.L. Update on the antibacterial resistance crisis. Med. Sci. Monit. 2007, 13, Ra103–Ra118. [Google Scholar] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.; Grenier, D. Understanding the virulence of Streptococcus suis: A veterinary, medical, and economic challenge. Med. Mal. Infect. 2018, 48, 159–166. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, December 2014. [Google Scholar]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Antimicrobial agents targeting bacterial cell walls and cell membranes. Rev. Sci. Tech. 2012, 31, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Lown, J.W. The mechanism of action of quinone antibiotics. Mol. Cell Biochem. 1983, 55, 17–40. [Google Scholar] [CrossRef]
- Culp, E.; Wright, G.D. Bacterial proteases, untapped antimicrobial drug targets. J. Antibiot. 2017, 70, 366–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, A.; Blackledge, M.S. Evaluation of small molecule kinase inhibitors as novel antimicrobial and antibiofilm agents. Chem. Biol. Drug Des. 2021, 98, 1038–1064. [Google Scholar] [CrossRef] [PubMed]
- Steenhuis, M.; van Ulsen, P.; Martin, N.I.; Luirink, J. A ban on BAM: An update on inhibitors of the β-barrel assembly machinery. FEMS Microbiol. Lett. 2021, 368, fnab059. [Google Scholar] [CrossRef] [PubMed]
- Lanyon-Hogg, T. Targeting the bacterial SOS response for new antimicrobial agents: Drug targets, molecular mechanisms and inhibitors. Future Med. Chem. 2021, 13, 143–155. [Google Scholar] [CrossRef]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Zhou, J.; Zheng, Y.; Su, J.; Sintim, H.O. Cyclic dinucleotide (c-di-GMP, c-di-AMP, and cGAMP) signalings have come of age to be inhibited by small molecules. Chem. Commun. 2016, 52, 9327–9342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corrigan, R.M.; Gründling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Römling, U. Great times for small molecules: C-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci. Signal. 2008, 1, pe39. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yang, J.; Zhou, X.; Ding, X.; Eisele, L.E.; Bai, G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS ONE 2012, 7, e35206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, W.; Cai, X.; Ma, H.; Zhu, L.; Zhang, Y.; Chou, S.H.; Galperin, M.Y.; He, J. A decade of research on the second messenger c-di-AMP. FEMS Microbiol. Rev. 2020, 44, 701–724. [Google Scholar] [CrossRef]
- Peng, X.; Li, J.; Xu, X. c-di-AMP regulates bacterial biofilm formation. Sheng Wu Gong Cheng Xue Bao 2017, 33, 1369–1375. [Google Scholar] [CrossRef]
- Fahmi, T.; Port, G.C.; Cho, K.H. c-di-AMP: An essential molecule in the signaling pathways that regulate the viability and virulence of gram-positive bacteria. Genes 2017, 8, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, T.H.; Liang, Z.X.; Marcellin, E.; Turner, M.S. Replenishing the cyclic-di-AMP pool: Regulation of diadenylate cyclase activity in bacteria. Curr. Genet. 2016, 62, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Zeden, M.S.; Schuster, C.F.; Bowman, L.; Zhong, Q.; Williams, H.D.; Gründling, A. Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J. Biol. Chem. 2018, 293, 3180–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrella, T.M.; Metzger, D.W.; Bai, G. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a Trk Family effector protein in Streptococcus pneumoniae. J. Bacteriol. 2018, 200, e00045-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.; Zhang, A.; Chen, H.; Zhou, R. Recent proceedings on prevalence and pathogenesis of Streptococcus suis. Curr. Issues Mol. Biol. 2019, 32, 473–520. [Google Scholar] [CrossRef] [PubMed]
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Han, L.; Fu, L.; Peng, Y.; Zhang, A. Triggering receptor expressed on myeloid cells-1 signaling: Protective and pathogenic roles on Streptococcal toxic-shock-like syndrome caused by Streptococcus suis. Front. Immunol. 2018, 9, 577. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xu, L.; Lv, W.; Han, L.; Xiang, Y.; Fu, L.; Jin, M.; Zhou, R.; Chen, H.; Zhang, A. An NLRP3 inflammasome-triggered cytokine storm contributes to Streptococcal toxic shock-like syndrome (STSLS). PLoS Pathog. 2019, 15, e1007795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Xie, C.; Chen, H.; Jin, M. Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics 2008, 8, 3506–3515. [Google Scholar] [CrossRef]
- Segura, M. Streptococcus suis vaccines: Candidate antigens and progress. Expert Rev. Vaccines 2015, 14, 1587–1608. [Google Scholar] [CrossRef]
- Devi, M.; Dutta, J.B.; Rajkhowa, S.; Kalita, D.; Saikia, G.K.; Das, B.C.; Hazarika, R.A.; Mahato, G. Prevalence of multiple drug resistant Streptococcus suis in and around Guwahati, India. Vet. World 2017, 10, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.I.; Jeon, A.B.; Jung, B.Y.; Byun, J.W.; Gottschalk, M.; Kim, A.; Kim, J.W.; Kim, H.Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017, 79, 780–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2021, 130, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Entzeroth, M.; Flotow, H.; Condron, P. Overview of high-throughput screening. Curr. Protoc. Pharmacol. 2009, 44, 9.4.1–9.4.27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Wang, D.; Sun, S.; Zhang, H. Issues of Z-factor and an approach to avoid them for quality control in high-throughput screening studies. Bioinformatics 2020. [Google Scholar] [CrossRef]
- Li, W.; Liu, L.; Qiu, D.; Chen, H.; Zhou, R. Identification of Streptococcus suis serotype 2 genes preferentially expressed in the natural host. Int. J. Med. Microbiol. 2010, 300, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Jeong, D.E.; Park, S.H.; Kim, S.J.; Choi, S.K. Complete Genome Sequence of Bacillus subtilis Strain WB800N, an Extracellular Protease-Deficient Derivative of Strain 168. Microbiol. Resour. Announc. 2018, 7, e01380-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Zhu, D.; Zhang, J.; Yang, L.; Wang, X.; Chen, H.; Tan, C. Virulence determinants, antimicrobial susceptibility, and molecular profiles of Erysipelothrix rhusiopathiae strains isolated from China. Emerg. Microbes Infect. 2015, 4, e69. [Google Scholar] [CrossRef]
- Zou, G.; Zhou, J.; Xiao, R.; Zhang, L.; Cheng, Y.; Jin, H.; Li, L.; Zhang, L.; Wu, B.; Qian, P.; et al. Effects of Environmental and Management-Associated Factors on Prevalence and Diversity of Streptococcus suis in Clinically Healthy Pig Herds in China and the United Kingdom. Appl. Environ. Microbiol. 2018, 84, e02590-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, G.; Hartung, S.; Büttner, K.; Hopfner, K.P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 2008, 30, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Krüger, L.; Herzberg, C.; Rath, H.; Pedreira, T.; Ischebeck, T.; Poehlein, A.; Gundlach, J.; Daniel, R.; Völker, U.; Mäder, U.; et al. Essentiality of c-di-AMP in Bacillus subtilis: Bypassing mutations converge in potassium and glutamate homeostasis. PLoS Genet. 2021, 17, e1009092. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Abbott, J.C.; Burhenne, H.; Kaever, V.; Gründling, A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011, 7, e1002217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Commichau, F.M.; Gibhardt, J.; Halbedel, S.; Gundlach, J.; Stülke, J. A delicate connection: C-di-AMP affects cell integrity by controlling osmolyte transport. Trends Microbiol. 2018, 26, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.E.; Whiteley, A.T.; Burke, T.P.; Sauer, J.D.; Portnoy, D.A.; Woodward, J.J. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. mBio 2013, 4, e00282-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oppenheimer-Shaanan, Y.; Wexselblatt, E.; Katzhendler, J.; Yavin, E.; Ben-Yehuda, S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011, 12, 594–601. [Google Scholar] [CrossRef]
- Zarrella, T.M.; Yang, J.; Metzger, D.W.; Bai, G. Bacterial second messenger cyclic di-AMP modulates the competence state in Streptococcus pneumoniae. J. Bacteriol. 2020, 202, e00691-19. [Google Scholar] [CrossRef]
- Kundra, S.; Lam, L.N.; Kajfasz, J.K.; Casella, L.G.; Andersen, M.J.; Abranches, J.; Flores-Mireles, A.L.; Lemos, J.A. c-di-AMP is essential for the virulence of Enterococcus faecalis. Infect. Immun. 2021, 89, e0036521. [Google Scholar] [CrossRef]
- Commichau, F.M.; Stülke, J. Coping with an essential poison: A genetic suppressor analysis corroborates a key function of c-di-AMP in controlling potassium ion homeostasis in gram-positive bacteria. J. Bacteriol. 2018, 200, e00166-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Sayre, D.A.; Zheng, Y.; Szmacinski, H.; Sintim, H.O. Unexpected complex formation between coralyne and cyclic diadenosine monophosphate providing a simple fluorescent turn-on assay to detect this bacterial second messenger. Anal. Chem. 2014, 86, 2412–2420. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Sintim, H.O. Potent inhibition of cyclic diadenylate monophosphate cyclase by the antiparasitic drug, suramin. Chem. Commun. 2016, 52, 3754–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opoku-Temeng, C.; Sintim, H.O. Inhibition of cyclic diadenylate cyclase, DisA, by polyphenols. Sci. Rep. 2016, 6, 25445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Zhou, J.; Sayre, D.A.; Sintim, H.O. Identification of bromophenol thiohydantoin as an inhibitor of DisA, a c-di-AMP synthase, from a 1000 compound library, using the coralyne assay. Chem. Commun. 2014, 50, 11234–11237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Ong, C.C.; Gierke, S.; Pitt, C.; Sagolla, M.; Cheng, C.K.; Zhou, W.; Jubb, A.M.; Strickland, L.; Schmidt, M.; Duron, S.G.; et al. Small molecule inhibition of group I p21-activated kinases in breast cancer induces apoptosis and potentiates the activity of microtubule stabilizing agents. Breast Cancer Res. 2015, 17, 59. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Artham, S.; Alwhaibi, A.; Adil, M.S.; Cummings, B.S.; Somanath, P.R. PAK1 inhibitor IPA-3 mitigates metastatic prostate cancer-induced bone remodeling. Biochem. Pharmacol. 2020, 177, 113943. [Google Scholar] [CrossRef]
- Cheng, X.; Zheng, X.; Zhou, X.; Zeng, J.; Ren, Z.; Xu, X.; Cheng, L.; Li, M.; Li, J.; Li, Y. Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans. Environ. Microbiol. 2016, 18, 904–922. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Sun, J.H. Diadenylate cyclase evaluation of ssDacA (SSU98_1483) in Streptococcus suis serotype 2. Genet Mol. Res. 2015, 14, 6917–6924. [Google Scholar] [CrossRef] [PubMed]
- Kuželová, K.; Grebeňová, D.; Holoubek, A.; Röselová, P.; Obr, A. Group I PAK inhibitor IPA-3 induces cell death and affects cell adhesivity to fibronectin in human hematopoietic cells. PLoS ONE 2014, 9, e92560. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Li, T.; Zou, W.; Ni, M.; Hu, Q.; Qiu, X.; Yao, Z.; Fan, J.; Li, L.; Huang, Q.; et al. IPA-3: An Inhibitor of Diadenylate Cyclase of Streptococcus suis with Potent Antimicrobial Activity. Antibiotics 2022, 11, 418. https://doi.org/10.3390/antibiotics11030418
Li H, Li T, Zou W, Ni M, Hu Q, Qiu X, Yao Z, Fan J, Li L, Huang Q, et al. IPA-3: An Inhibitor of Diadenylate Cyclase of Streptococcus suis with Potent Antimicrobial Activity. Antibiotics. 2022; 11(3):418. https://doi.org/10.3390/antibiotics11030418
Chicago/Turabian StyleLi, Haotian, Tingting Li, Wenjin Zou, Minghui Ni, Qiao Hu, Xiuxiu Qiu, Zhiming Yao, Jingyan Fan, Lu Li, Qi Huang, and et al. 2022. "IPA-3: An Inhibitor of Diadenylate Cyclase of Streptococcus suis with Potent Antimicrobial Activity" Antibiotics 11, no. 3: 418. https://doi.org/10.3390/antibiotics11030418
APA StyleLi, H., Li, T., Zou, W., Ni, M., Hu, Q., Qiu, X., Yao, Z., Fan, J., Li, L., Huang, Q., & Zhou, R. (2022). IPA-3: An Inhibitor of Diadenylate Cyclase of Streptococcus suis with Potent Antimicrobial Activity. Antibiotics, 11(3), 418. https://doi.org/10.3390/antibiotics11030418





