Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies
Abstract
1. Introduction
2. Legionella Clinical and Molecular Features
3. Pharmacological Treatment of Legionellosis and Management of LP Resistant Clones
4. Monitoring of Legionella spp. in Hospital Environments and Water Disinfection Strategies
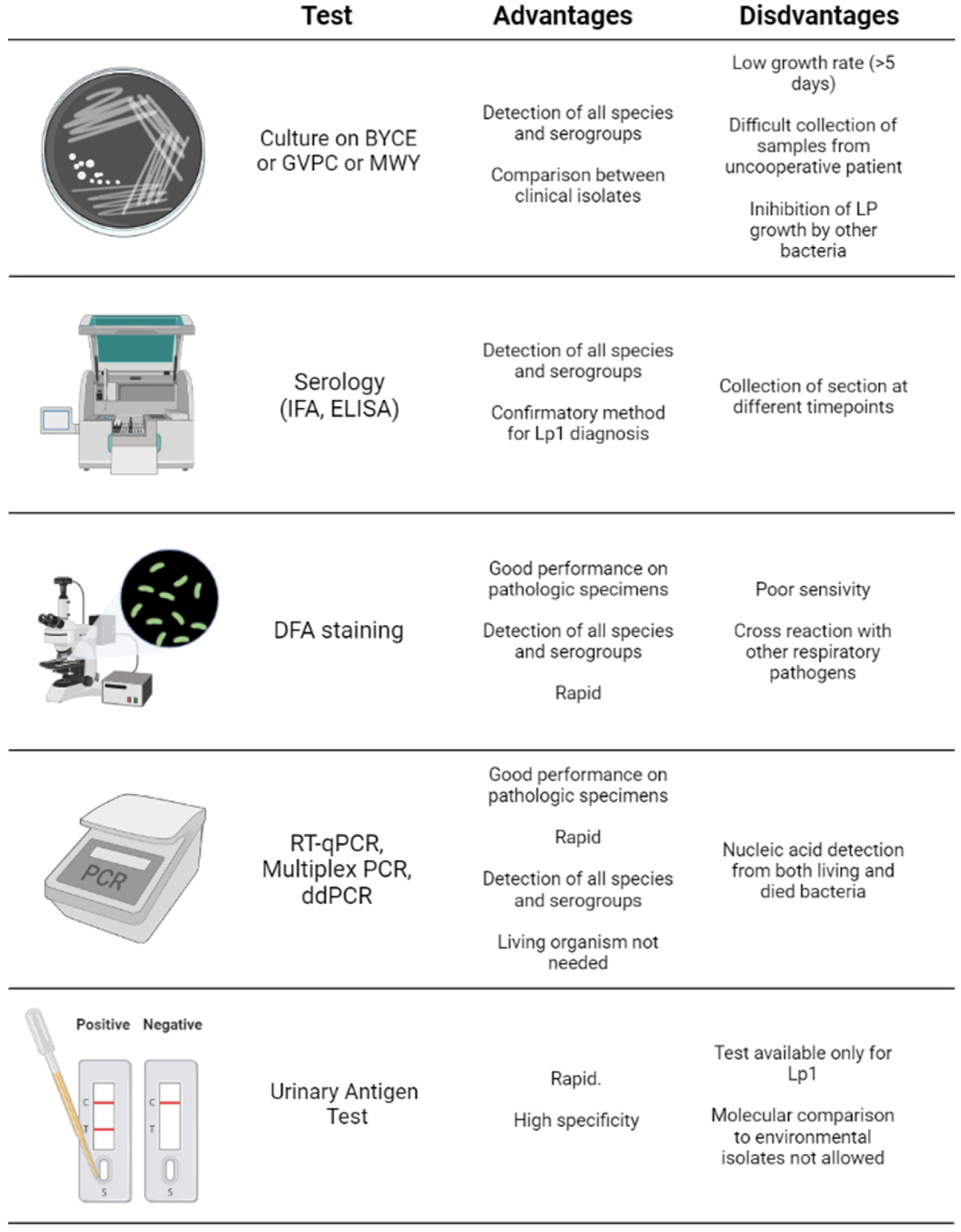
5. Detection of Legionella pneumophila and Diagnosis of Legionellosis
6. Conclusions
- -
- The precise characterization of LP clinical and molecular features is essential to identify potential virulence or drug resistance factors useful for the selection of the most effective antibiotic treatment;
- -
- Disinfection strategies using chemicals, filters or UV lamps are essential in both water tanks, pipes and POU to reduce the risk of LP water contamination;
- -
- The use of molecular high-sensitive diagnostic methods, such as ddPCR, besides the standard culture methods, could be useful to correctly diagnose legionellosis in suspected LP pneumonia.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Garlasco, J.; Zotti, C.M. Comparison of BCYEα+AB agar and MWY agar for detection and enumeration of Legionella spp. in hospital water samples. BMC Microbiol. 2021, 21, 48. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.W.; Tsai, T.R.; Orenstein, W.; Parkin, W.E.; Beecham, H.J.; Sharrar, R.G.; Harris, J.; Mallison, G.F.; Martin, S.M.; McDade, J.E.; et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N. Engl. J. Med. 1977, 297, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.J.; Steigerwalt, A.G.; McDade, J.E. Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 1979, 90, 656–658. [Google Scholar] [CrossRef] [PubMed]
- Potts, A.; Donaghy, M.; Marley, M.; Othieno, R.; Stevenson, J.; Hyland, J.; Pollock, K.G.; Lindsay, D.; Edwards, G.; Hanson, M.F.; et al. Cluster of Legionnaires disease cases caused by Legionella longbeachae serogroup 1. Euro Surveill 2013, 18, 20656. [Google Scholar] [CrossRef]
- Waldron, P.R.; Martin, B.A.; Ho, D.Y. Mistaken identity: Legionella micdadei appearing as acid-fast bacilli on lung biopsy of a hematopoietic stem cell transplant patient. Transpl. Infect. Dis. 2015, 17, 89–93. [Google Scholar] [CrossRef]
- Yu, V.L.; Plouffe, J.F.; Pastoris, M.C.; Stout, J.E.; Schousboe, M.; Widmer, A.; Summersgill, J.; File, T.; Heath, C.M.; Paterson, D.L.; et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J. Infect. Dis. 2002, 186, 127–128. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency|US EPA. 2016. Available online: https://www.epa.gov/ (accessed on 6 January 2022).
- Ryan, K.; Ray, G. Sherris Medical Microbiology, 6th ed.; The McGraw Hill Education Companies: New York, NY, USA, 2015. [Google Scholar]
- Boamah, D.K.; Zhou, G.; Ensminger, A.W.; O’Connor, T.J. From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella. Front. Cell Infect. Microbiol. 2017, 7, 477. [Google Scholar] [CrossRef]
- Abu Khweek, A.; Amer, A.O. Factors Mediating Environmental Biofilm Formation by Legionella pneumophila. Front. Cell Infect. Microbiol. 2018, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, W.J.; Garner, E.; Ji, P.; Zhu, N.; Parks, J.; Schwake, D.O.; Pruden, A.; Edwards, M.A. Distribution System Operational Deficiencies Coincide with Reported Legionnaires’ Disease Clusters in Flint, Michigan. Environ. Sci. Technol. 2017, 51, 11986–11995. [Google Scholar] [CrossRef]
- ECDC, European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/en (accessed on 14 January 2022).
- Rota, M.C.; Bella, A.; Caporali, M.G.; Nicolau, A.; Drasar, V.; Ricci, M.L.; Scaturro, M.; Gumá, M.; Crespi, S. Travel-associated Legionnaires’ disease: Would changing cluster definition lead to the prevention of a larger number of cases? Epidemiol. Infect. 2018, 147, 62. [Google Scholar] [CrossRef]
- Fischer, F.B.; Mäusezahl, D.; Wymann, N.M. Temporal trends in legionellosis national notification data and the effect of COVID-19, Switzerland, 2000–2020. medRxiv 2022. [Google Scholar] [CrossRef]
- Rota, M.C.; Caporali, M.G.; Bella, A.; Scaturro, M.; Giannitelli, S.; Ricci, M.L. Il Sistema di Sorveglianza Della Legionellosi in Italia: I Risultati del 2019. Available online: https://www.epicentro.iss.it/ben/2020/4/sorveglianza-legionellosi-italia-2019 (accessed on 15 January 2022).
- Palazzolo, C.; Maffongelli, G.; D’Abramo, A.; Lepore, L.; Mariano, A.; Vulcano, A.; Bartoli, T.A.; Bevilacqua, N.; Giancola, M.L.; Di Rosa, E.; et al. Legionella pneumonia: Increased risk after COVID-19 lockdown? Italy, May to June 2020. Euro Surveill 2020, 25, 2001372. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Swanson, C.S.; Wang, L.; He, Q. Impact of building closures during the COVID-19 pandemic on Legionella infection risks. Am. J. Infect. Control. 2021, 49, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.M.; Lai, C.C. Increasing legionella in Taiwan during COVID-19 pandemic. Am. J. Infect. Control. 2022, 50, 237–238. [Google Scholar] [CrossRef]
- Agarwal, S.; Abell, V.; File, T.M., Jr. Nosocomial (Health Care-Associated) Legionnaire’s Disease. Infect. Dis. Clin. N. Am. 2017, 31, 155–165. [Google Scholar] [CrossRef]
- Mondino, S.; Schmidt, S.; Rolando, M.; Escoll, P.; Gomez-Valero, L.; Buchrieser, C. Legionnaires’ Disease: State of the Art Knowledge of Pathogenesis Mechanisms of Legionella. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 439–466. [Google Scholar] [CrossRef]
- Kenagy, E.; Priest, P.C.; Cameron, C.M.; Smith, D.; Scott, P.; Cho, V.; Mitchell, P.; Murdoch, D.R. Risk Factors forLegionella longbeachaeLegionnaires’ Disease, New Zealand. Emerg. Infect. Dis. 2017, 23, 1148–1154. [Google Scholar] [CrossRef]
- Brunette, G.W.; Kozarsky, P.E.; Magill, J.A.; Shlim, D.R.; Whatley, A.D. CDC Health Information for International Travel 2010, Chapter 5—Other Infectious Diseases Related to Travel; Elsevier: Amsterdam, The Netherlands, 2009; pp. 290–411. [Google Scholar]
- Poirier, R.; Rodrigue, J.; Villeneuve, J.; Lacasse, Y. Early Radiographic and Tomographic Manifestations of Legionnaires’ Disease. Can. Assoc. Radiol. J. 2017, 68, 328–333. [Google Scholar] [CrossRef]
- Franco-Garcia, A.; Varughese, T.A.; Lee, Y.J.; Papanicolaou, G.; Rosenblum, M.K.; Hollmann, T.J.; Koehne, G.; Boulad, F.; Babady, N.E.; Tang, Y.-W.; et al. Diagnosis of Extrapulmonary Legionellosis in Allogeneic Hematopoietic Cell Transplant Recipients by Direct 16S Ribosomal Ribonucleic Acid Sequencing and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Open Forum Infect. Dis. 2017, 4, 140. [Google Scholar] [CrossRef]
- Banderet, F.; Blaich, A.; Soleman, E.; Gaia, V.; Osthoff, M. Septic arthritis due to Legionella cincinnatiensis: Case report and review of the literature. Infection 2017, 45, 551–555. [Google Scholar] [CrossRef]
- Chitasombat, M.N.; Ratchatanawin, N.; Visessiri, Y. Disseminated extrapulmonary Legionella pneumophila infection presenting with panniculitis: Case report and literature review. BMC Infect. Dis. 2018, 18, 467. [Google Scholar] [CrossRef] [PubMed]
- Dooling, K.L.; Toews, K.-A.; Hicks, L.A.; Garrison, L.E.; Bachaus, B.; Zansky, S.; Carpenter, L.R.; Schaffner, B.; Parker, E.; Petit, S.; et al. Active Bacterial Core Surveillance for Legionellosis—United States, 2011–2013. MMWR. Morb. Mortal. Wkly. Rep. 2015, 64, 1190–1193. [Google Scholar] [CrossRef] [PubMed]
- Isenman, H.L.; Chambers, S.T.; Pithie, A.D.; MacDonald, S.L.; Hegarty, J.M.; Fenwick, J.L.; Maze, M.J.; Metcalf, S.C.; Murdoch, D.R. Legionnaires’ disease caused by Legionella longbeachae: Clinical features and outcomes of 107 cases from an endemic area. Respirology 2016, 21, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Sikora, A.; Zahra, F. Nosocomial Infections. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Oliva, G.; Sahr, T.; Buchrieser, C. The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Steiner, B.; Swart, A.L.; Welin, A.; Weber, S.; Personnic, N.; Kaech, A.; Freyre, C.; Ziegler, U.; Klemm, R.; Hilbi, H. ER remodeling by the large GTPase atlastin promotes vacuolar growth of Legionella pneumophila. EMBO Rep. 2017, 18, 1817–1836. [Google Scholar] [CrossRef] [PubMed]
- Steiner, B.; Weber, S.; Hilbi, H. Formation of the Legionella-containing vacuole: Phosphoinositide conversion, GTPase modulation and ER dynamics. Int. J. Med. Microbiol. 2018, 308, 49–57. [Google Scholar] [CrossRef]
- Cianciotto, N.P. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 2001, 291, 331–343. [Google Scholar] [CrossRef]
- Dowling, J.N.; Saha, A.K.; Glew, R.H. Virulence factors of the family Legionellaceae. Microbiol. Rev. 1992, 56, 32–60. [Google Scholar] [CrossRef]
- Heuner, K.; Steinert, M.; Dietrich, C.; Fischer, G.; Kohler, R.F.; Hacker, J. Function and Expression of Legionella pneumophila Surface Factors. In Legionella; John Wiley & Sons, Inc.: New York, NY, USA, 2014; pp. 3–48. [Google Scholar]
- Garduño, R.A.; Garduño, E.; Hoffman, P.S. Surface-Associated Hsp60 Chaperonin of Legionella pneumophila Mediates Invasion in a HeLa Cell Model. Infect. Immun. 1998, 66, 4602–4610. [Google Scholar] [CrossRef]
- Bellinger-Kawahara, C.; Horwitz, M.A. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J. Exp. Med. 1990, 172, 1201–1210. [Google Scholar] [CrossRef]
- Fields, B.S. The molecular ecology of legionellae. Trends Microbiol. 1996, 4, 286–290. [Google Scholar] [CrossRef]
- De Buck, E.; Anné, J.; Lammertyn, E. The role of protein secretion systems in the virulence of the intracellular pathogen Legionella pneumophila. Microbiology 2007, 153, 3948–3953. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jacobi, S.; Heuner, K. Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 2003, 293, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, D.; Zhu, L.; Ren, H.; Li, Y.; Liu, X.; Li, X.; Li, W.; Zhao, N.; Lu, J.; et al. Legionella pneumophila Risk from Cooling Tower Systems in China. Appl. Environ. Microbiol. 2021, 88, e0192121. [Google Scholar] [CrossRef]
- Abu-Zant, A.; Asare, R.; Graham, J.E.; Abu Kwaik, Y. Role for RpoS but Not RelA of Legionella pneumophila in Modulation of Phagosome Biogenesis and Adaptation to the Phagosomal Microenvironment. Infect. Immun. 2006, 74, 3021–3026. [Google Scholar] [CrossRef]
- Bandyopadhyay, P.; Liu, S.; Gabbai, C.B.; Venitelli, Z.; Steinman, H.M. Environmental Mimics and the Lvh Type IVA Secretion System Contribute to Virulence-Related Phenotypes of Legionella pneumophila. Infect. Immun. 2007, 75, 723–735. [Google Scholar] [CrossRef]
- Samrakandi, M.M.; Cirillo, S.L.G.; Ridenour, D.A.; Bermudez, L.E.; Cirillo, J.D. Genetic and Phenotypic Differences between Legionella pneumophila Strains. J. Clin. Microbiol. 2002, 40, 1352–1362. [Google Scholar] [CrossRef]
- Huang, B.; Heron, B.A.; Gray, B.R.; Eglezos, S.; Bates, J.R.; Savill, J. A Predominant and Virulent Legionella pneumophila Serogroup 1 Strain Detected in Isolates from Patients and Water in Queensland, Australia, by an Amplified Fragment Length Polymorphism Protocol and Virulence Gene-Based PCR Assays. J. Clin. Microbiol. 2004, 42, 4164–4168. [Google Scholar] [CrossRef][Green Version]
- Huang, B.; Yuan, Z.; Heron, B.A.; Gray, B.R.; Eglezos, S.; Bates, J.R.; Savill, J. Distribution of 19 major virulence genes in Legionella pneumophila serogroup 1 isolates from patients and water in Queensland, Australia. J. Med. Microbiol. 2006, 55, 993–997. [Google Scholar] [CrossRef]
- Kozak, N.A.; Buss, M.; Lucas, C.E.; Frace, M.; Govil, D.; Travis, T.; Olsen-Rasmussen, M.; Benson, R.F.; Fields, B.S. Virulence Factors Encoded by Legionella longbeachae Identified on the Basis of the Genome Sequence Analysis of Clinical Isolate D-4968. J. Bacteriol. 2010, 192, 1030–1044. [Google Scholar] [CrossRef]
- Finsel, I.; Hilbi, H. Formation of a pathogen vacuole according toLegionella pneumophila: How to kill one bird with many stones. Cell. Microbiol. 2015, 17, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Luo, Z.-Q. Legionella and Coxiella effectors: Strength in diversity and activity. Nat. Rev. Genet. 2017, 15, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Nagai, H. The Type IVB secretion system: An enigmatic chimera. Curr. Opin. Microbiol. 2015, 29, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Kubori, T. Type IVB Secretion Systems of Legionella and Other Gram-Negative Bacteria. Front. Microbiol. 2011, 2, 136. [Google Scholar] [CrossRef]
- Copenhaver, A.M.; Casson, C.N.; Nguyen, H.T.; Fung, T.C.; Duda, M.M.; Roy, C.R.; Shin, S. Alveolar Macrophages and Neutrophils Are the Primary Reservoirs for Legionella pneumophila and Mediate Cytosolic Surveillance of Type IV Secretion. Infect. Immun. 2014, 82, 4325–4336. [Google Scholar] [CrossRef]
- Dong, N.; Niu, M.; Hu, L.; Yao, Q.; Zhou, R.; Shao, F. Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat. Microbiol. 2016, 2, 16236. [Google Scholar] [CrossRef]
- Moss, S.M.; Taylor, I.; Ruggero, D.; Gestwicki, J.E.; Shokat, K.M.; Mukherjee, S. A Legionella pneumophila Kinase Phosphorylates the Hsp70 Chaperone Family to Inhibit Eukaryotic Protein Synthesis. Cell Host Microbe 2019, 25, 454–462. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Lucas, M.; Evans, T.R.; Abascal-Palacios, G.; Doms, A.G.; Beauchene, N.A.; Rojas, A.L.; Hierro, A.; Machner, M.P. RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases. PLoS Pathog. 2018, 14, e1006897. [Google Scholar] [CrossRef]
- Rolando, M.; Escoll, P.; Nora, T.; Botti, J.; Boitez, V.; Bedia, C.; Daniels, C.; Abraham, G.; Stogios, P.J.; Skarina, T.; et al. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc. Natl. Acad. Sci. USA 2016, 113, 1901–1906. [Google Scholar] [CrossRef]
- Vogt, S.; Raivio, T.L. Just scratching the surface: An expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 2011, 326, 2–11. [Google Scholar] [CrossRef]
- Altman, E.; Segal, G. The Response Regulator CpxR Directly Regulates Expression of Several Legionella pneumophila icm/dot Components as Well as New Translocated Substrates. J. Bacteriol. 2008, 190, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, G.; Jiménez, N.; Peris-Bondia, F.; Pelaz, C.; Latorre, A.; Moya, A. Virulence factor rtx in Legionella pneumophila, evidence suggesting it is a modular multifunctional protein. BMC Genom. 2008, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.; Abu Kwaik, Y. Expression of Multiple Pili by Legionella pneumophila: Identification and Characterization of a Type IV Pilin Gene and Its Role in Adherence to Mammalian and Protozoan Cells. Infect. Immun. 1998, 66, 1768–1775. [Google Scholar] [CrossRef]
- Sharma, L.; Losier, A.; Tolbert, T.; Cruz, C.S.D.; Marion, C.R. Atypical Pneumonia: Updates on Legionella, Chlamydophila, and Mycoplasma Pneumonia. Clin. Chest Med. 2017, 38, 45–58. [Google Scholar] [CrossRef]
- Carratalà, J.; Garcia-Vidal, C. An update on Legionella. Curr. Opin. Infect. Dis. 2010, 23, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Woodhead, M.; Blasi, F.; Ewig, S.; Garau, J.; Huchon, G.; Ieven, M.; Ortqvist, A.; Schaberg, T.; Torres, A.; van der Heijden, G.; et al. Joint Taskforce of the European Respiratory Society and European Society for Clinical Microbiology and Infectious Diseases. Guidelines for the management of adult lower respiratory tract infections—Full version. Clin. Microbiol. Infect. 2011, 17 (Suppl. 6), E1–E59. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.; Clark, J.; Coote, N.; Fletcher, P.; Harnden, A.; Mckean, M.; Thomson, A.; on behalf of the British Thoracic Society Standards of Care Committee. British Thoracic Society guidelines for the management of community acquired pneumonia in children: Update 2011. Thorax 2011, 66 (Suppl. S2), ii1–ii23. [Google Scholar] [CrossRef]
- Miyashita, N.; Kobayashi, I.; Higa, F.; Aoki, Y.; Kikuchi, T.; Seki, M.; Tateda, K.; Maki, N.; Uchino, K.; Ogasawara, K.; et al. In vitro activity of various antibiotics against clinical strains of Legionella species isolated in Japan. J. Infect. Chemother. 2018, 24, 325–329. [Google Scholar] [CrossRef]
- Bakheit, A.H.; Al-Hadiya, B.M.; Abd-Elgalil, A.A. Profiles Drug Substances Excipients and Related Methodology. Brittain, H.G., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 39, pp. 1–40. [Google Scholar]
- Wimer, S.M.; Schoonover, L.; Garrison, M.W. Levofloxacin: A therapeutic review. Clin. Ther. 1998, 20, 1049–1070. [Google Scholar] [CrossRef]
- Erdogan, H.; Can, F.; Demirbilek, M.; Timurkaynak, F.; Arslan, H. In vitro activity of antimicrobial agents against Legionella isolated from environmental water systems: First results from Turkey. Environ. Monit. Assess. 2010, 171, 689–691. [Google Scholar] [CrossRef]
- Bonaldo, G.; Andriani, L.A.; D’Annibali, O.; Motola, D.; Vaccheri, A. Cardiovascular safety of macrolide and fluoroquinolone antibiotics: An analysis of the WHO database of adverse drug reactions. Pharmacoepidemiol. Drug Saf. 2019, 28, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Postma, D.F.; Spitoni, C.; van Werkhoven, C.H.; van Elden, L.J.R.; Oosterheert, J.J.; Bonten, M.J.M. Cardiac events after macrolides or fluoroquinolones in patients hospitalized for community-acquired pneumonia: Post-hoc analysis of a cluster-randomized trial. BMC Infect. Dis. 2019, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Descours, G.; Ginevra, C.; Jacotin, N.; Forey, F.; Chastang, J.; Kay, E.; Etienne, J.; Lina, G.; Doublet, P.; Jarraud, S. Ribosomal Mutations Conferring Macrolide Resistance in Legionella pneumophila. Antimicrob. Agents Chemother. 2017, 61, e02188-16. [Google Scholar] [CrossRef] [PubMed]
- Massip, C.; Descours, G.; Ginevra, C.; Doublet, P.; Jarraud, S.; Gilbert, C. Macrolide resistance inLegionella pneumophila: The role of LpeAB efflux pump. J. Antimicrob. Chemother. 2017, 72, 1327–1333. [Google Scholar] [CrossRef]
- Jia, X.; Ren, H.; Nie, X.; Li, Y.; Li, J.; Qin, T. Antibiotic Resistance and Azithromycin Resistance Mechanism of Legionella pneumophila Serogroup 1 in China. Antimicrob. Agents Chemother. 2019, 63, e00768-19. [Google Scholar] [CrossRef]
- Vandewalle-Capo, M.; Massip, C.; Descours, G.; Charavit, J.; Chastang, J.; Billy, P.A.; Boisset, S.; Lina, G.; Gilbert, C.; Maurin, M.; et al. Minimum inhibitory concentration (MIC) distribution among wild-type strains of Legionella pneumophila identifies a subpopulation with reduced susceptibility to macrolides owing to efflux pump genes. Int. J. Antimicrob. Agents 2017, 50, 684–689. [Google Scholar] [CrossRef]
- Cocuzza, C.E.; Martinelli, M.; Perdoni, F.; Giubbi, C.; Vinetti, M.E.A.; Calaresu, E.; Frugoni, S.; Scaturro, M.; Ricci, M.L.; Musumeci, R. Antibiotic Susceptibility of Environmental Legionella pneumophila Strains Isolated in Northern Italy. Int. J. Environ. Res. Public Health 2021, 18, 9352. [Google Scholar] [CrossRef]
- Shadoud, L.; Almahmoud, I.; Jarraud, S.; Etienne, J.; Larrat, S.; Schwebel, C.; Timsit, J.-F.; Schneider, D.; Maurin, M. Hidden Selection of Bacterial Resistance to Fluoroquinolones In Vivo: The Case of Legionella pneumophila and Humans. EBioMedicine 2015, 2, 1179–1185. [Google Scholar] [CrossRef]
- AlMahmoud, I.; Kay, E.; Schneider, M.; Maurin, M. Mutational paths towards increased fluoroquinolone resistance in Legionella pneumophila. J. Antimicrob. Chemother. 2009, 64, 284–293. [Google Scholar] [CrossRef]
- Hennebique, A.; Bidart, M.; Jarraud, S.; Beraud, L.; Schwebel, C.; Maurin, M.; Boisset, S. Digital PCR for Detection and Quantification of Fluoroquinolone Resistance in Legionella pneumophila. Antimicrob. Agents Chemother. 2017, 61, e00628-17. [Google Scholar] [CrossRef]
- Pashaei-Asl, R.; Khodadadi, K.; Pashaei-Asl, F.; Haqshenas, G.; Ahmadian, N.; Pashaiasl, M.; Baghdadabadi, R.H. Legionella Pneumophila and Dendrimers-Mediated Antisense Therapy. Adv. Pharm. Bull. 2017, 7, 179–187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mobarez, A.M.; Ahmadrajabi, R.; Khoramabadi, N.; Salmanian, A.H. Recombinant flagellin-PAL fusion protein of Legionella pneumophila induced cell-mediated and protective immunity against bacteremia in BALB/c mice. World J. Microbiol. Biotechnol. 2017, 33, 175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Z.; Dong, Y.; Chen, Y. Recombinant PAL/PilE/FlaA DNA vaccine provides protective immunity against Legionella pneumophila in BALB/c mice. BMC Biotechnol. 2020, 20, 28. [Google Scholar] [CrossRef]
- Benedict, K.M.; Reses, H.; Vigar, M.; Roth, D.M.; Roberts, V.A.; Mattioli, M.; Cooley, L.A.; Hilborn, E.D.; Wade, T.J.; Fullerton, K.E.; et al. Surveillance for Waterborne Disease Outbreaks Associated with Drinking Water—United States, 2013–2014. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 1216–1221. [Google Scholar] [CrossRef] [PubMed]
- Granseth, G.; Bhattarai, R.; Sylvester, T.; Prasai, S.; Livar, E. Notes from the Field: Two Cases of Legionnaires’ Disease in Newborns After Water Births—Arizona, 2016. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 590–591. [Google Scholar] [CrossRef]
- Weiss, D.; Boyd, C.; Rakeman, J.L.; Greene, S.K.; Fitzhenry, R.; McProud, T.; Musser, K.; Huang, L.; Kornblum, J.; Nazarian, E.J.; et al. A Large Community Outbreak of Legionnaires’ Disease Associated With a Cooling Tower in New York City, 2015. Public Health Rep. 2017, 132, 241–250. [Google Scholar] [CrossRef]
- Gaikwad, U.N.; Jinna, S. Environmental surveillance of Legionella pneumophila in distal water supplies of a hospital for early identification & prevention of hospital-acquired legionellosis. Indian J. Med Res. 2018, 147, 611–614. [Google Scholar] [CrossRef]
- Sikora, A.; Wójtowicz-Bobin, M.; Kozioł-Montewka, M.; Magryś, A.; Gładysz, I. Prevalence of Legionella pneumophila in water distribution systems in hospitals and public buildings of the Lublin region of eastern Poland. Ann. Agric. Environ. Med. 2015, 22, 195–201. [Google Scholar] [CrossRef]
- Correia, A.M.; Ferreira, J.S.; Borges, V.; Nunes, A.; Gomes, B.; Capucho, R.; Gonçalves, J.; Antunes, D.M.; Almeida, S.; Mendes, A.; et al. Probable Person-to-Person Transmission of Legionnaires’ Disease. N. Engl. J. Med. 2016, 374, 497–498. [Google Scholar] [CrossRef]
- Rota, M.C.; Caporali, M.G.; Bella, A.; Scaturro, M.; Giannitelli, S.; Ricci, M.L. I risultati del sistema di sorveglianza della legionellosi in Italia nel 2020 durante la pandemia di COVID-19. Boll. Epidemiol. Naz. 2021, 2, 9–16. [Google Scholar] [CrossRef]
- Lagana, P.; Soraci, L.; Gambuzza, M.E.; Mancuso, G.; Delia, S.A. Innate Immune Surveillance in the Central Nervous System Following Legionella pneumophila Infection. CNS Neurol. Disord. Drug Targets 2017, 16, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- WHO—World Health Organization—Legionella and the Prevention of Legionellosis. Available online: https://www.who.int/ (accessed on 7 January 2022).
- Gamage, S.D.; Ambrose, M.; Kralovic, S.M.; Roselle, G.A. Water Safety and Legionella in Health Care: Priorities, Policy, and Practice. Infect. Dis. Clin. N. Am. 2016, 30, 689–712. [Google Scholar] [CrossRef] [PubMed]
- Orlikowski, J.; Ryl, J.; Jazdzewska, A.; Krakowiak, S. Effect of Thermal Shock During Legionella Bacteria Removal on the Corrosion Properties of Zinc-Coated Steel Pipes. J. Mater. Eng. Perform. 2015, 25, 2711–2719. [Google Scholar] [CrossRef]
- Cates, E.L.; Torkzadeh, H. Can incorporation of UVC LEDs into showerheads prevent opportunistic respiratory pathogens? Microbial behavior and device design considerations. Water Res. 2019, 168, 115163. [Google Scholar] [CrossRef] [PubMed]
- Springston, J.P.; Yocavitch, L. Existence and control of Legionella bacteria in building water systems: A review. J. Occup. Environ. Hyg. 2017, 14, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, T.; Xie, X. Rationally designed tubular coaxial-electrode copper ionization cells (CECICs) harnessing non-uniform electric field for efficient water disinfection. Environ. Int. 2019, 128, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Cloutman-Green, E.; Barbosa, V.L.; Jimenez, D.; Wong, D.; Dunn, H.; Needham, B.; Ciric, L.; Hartley, J.C. Controlling Legionella pneumophila in water systems at reduced hot water temperatures with copper and silver ionization. Am. J. Infect. Control 2019, 47, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Casini, B.; Aquino, F.; Totaro, M.; Miccoli, M.; Galli, I.; Manfredini, L.; Giustarini, C.; Costa, A.L.; Tuvo, B.; Valentini, P.; et al. Application of Hydrogen Peroxide as an Innovative Method of Treatment for Legionella Control in a Hospital Water Network. Pathogens 2017, 6, 15. [Google Scholar] [CrossRef]
- Sciuto, E.; Laganà, P.; Filice, S.; Scalese, S.; Libertino, S.; Corso, D.; Faro, G.; Coniglio, M. Environmental Management of Legionella in Domestic Water Systems: Consolidated and Innovative Approaches for Disinfection Methods and Risk Assessment. Microorganisms 2021, 9, 577. [Google Scholar] [CrossRef]
- Paranjape, K.; Bédard, É.; Whyte, L.G.; Ronholm, J.; Prévost, M.; Faucher, S.P. Presence of Legionella spp. in cooling towers: The role of microbial diversity, Pseudomonas, and continuous chlorine application. Water Res. 2020, 169, 115252. [Google Scholar] [CrossRef]
- Mouchtouri, V.A.; Goutziana, G.; Kremastinou, J.; Hadjichristodoulou, C. Legionella species colonization in cooling towers: Risk factors and assessment of control measures. Am. J. Infect. Control 2010, 38, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Digiano, F.A.; Zhang, W. Pipe Section Reactor to Evaluate Chlorine-Wall Reaction. J. Am. Water Work. Assoc. 2005, 97, 74–85. [Google Scholar] [CrossRef]
- Kirmeyer, G.J.; LeChevallier, M.; Barbeau, H.; Martel, K.; Thompson, G.; Radder, L.; Klement, W.; Flores, A. Optimizing Chloramine Treatment, 2nd ed.; Prepared for the Water Research Foundation: Denver, CO, USA, 2004. [Google Scholar]
- Vincenti, S.; de Waure, C.; Raponi, M.; Teleman, A.A.; Boninti, F.; Bruno, S.; Boccia, S.; Damiani, G.; Laurenti, P. Environmental surveillance of Legionella spp. colonization in the water system of a large academic hospital: Analysis of the four–year results on the effectiveness of the chlorine dioxide disinfection method. Sci. Total Environ. 2019, 657, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Muraca, P.W.; Goetz, A.; Yu, V.L. Disinfection of Water Distribution Systems for Legionella: A Review of Application Procedures and Methodologies. Infect. Control Hosp. Epidemiol. 1990, 11, 79–88. [Google Scholar] [CrossRef]
- Buse, H.Y.; Hall, J.S.; Hunter, G.L.; Goodrich, J.A. Differences in UV-C LED Inactivation of Legionella pneumophila Serogroups in Drinking Water. Microorganisms 2022, 10, 352. [Google Scholar] [CrossRef]
- Bertolino, G.; Marras, L.; Sanna, C.; Carrucciu, G.; Schintu, M.; Coroneo, V. Ten-Year Retrospective Analysis of Legionella Diffusion in Hospital Water Systems and Its Serogroup Seasonal Variation. Adv. Exp. Med. Biol. 2020, 1282, 93–103. [Google Scholar] [CrossRef]
- Percival, S.L.; Williams, D.W. Microbiology of Waterborne Diseases, 2nd ed.; Percival, S.L., Yates, M.V., Williams, W.D., Chalmers, R.M., Gray, N.F., Eds.; Academic Press: Cambridge, MA, USA, 2014; Chapter 8; pp. 155–175. [Google Scholar] [CrossRef]
- Ditommaso, S.; Giacomuzzi, M.; Memoli, G.; Garlasco, J.; Zotti, C.M. The use of BCYE medium for the detection of Legionella in environmental water samples: An appropriate update to ISO 11731:2017 standard? Diagn. Microbiol. Infect. Dis. 2022, 102, 115593. [Google Scholar] [CrossRef]
- ISO 11731:2017; Water Quality-Enumeration of Legionella. International Organization for Standardization: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/61782.html (accessed on 20 January 2022).
- Kimura, S.; Tateda, K.; Ishii, Y.; Horikawa, M.; Miyairi, S.; Gotoh, N.; Ishiguro, M.; Yamaguchi, K. Pseudomonas aeruginosa Las quorum sensing autoinducer suppresses growth and biofilm production in Legionella species. Microbiology 2009, 155, 1934–1939. [Google Scholar] [CrossRef]
- Chaudhry, R.; Sreenath, K.; Agrawal, S.K.; Valavane, A. Legionella and Legionnaires’ Disease: Time to Explore in India. Indian J. Med. Microbiol. 2018, 36, 324–333. [Google Scholar] [CrossRef]
- Mercante, J.W.; Winchell, J.M. Current and Emerging Legionella Diagnostics for Laboratory and Outbreak Investigations. Clin. Microbiol. Rev. 2015, 28, 95–133. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef] [PubMed]
- Musso, N.; Falzone, L.; Stracquadanio, S.; Bongiorno, D.; Salerno, M.; Esposito, M.; Sessa, F.; Libra, M.; Stefani, S.; Pomara, C. Post-Mortem Detection of SARS-CoV-2 RNA in Long-Buried Lung Samples. Diagnostics 2021, 11, 1158. [Google Scholar] [CrossRef] [PubMed]
- Stout, J.E.; Yu, V.L. Legionellosis. N. Engl. J. Med. 1997, 337, 682–687. [Google Scholar] [CrossRef]
- Lindsay, D.S.J.; Abraham, W.H.; Findlay, W.; Christie, P.; Johnston, F.; Edwards, G.F.S. Laboratory diagnosis of legionnaires’ disease due to Legionella pneumophila serogroup 1: Comparison of phenotypic and genotypic methods. J. Med. Microbiol. 2004, 53, 457. [Google Scholar] [CrossRef]
- Rantakokko-Jalava, K.; Jalava, J. Development of Conventional and Real-Time PCR Assays for Detection of Legionella DNA in Respiratory Specimens. J. Clin. Microbiol. 2001, 39, 2904–2910. [Google Scholar] [CrossRef]
- Bonetta, S.; Ferretti, E.; Balocco, F.; Carraro, E. Evaluation ofLegionella pneumophilacontamination in Italian hotel water systems by quantitative real-time PCR and culture methods. J. Appl. Microbiol. 2010, 108, 1576–1583. [Google Scholar] [CrossRef]
- Touron-Bodilis, A.; Pougnard, C.; Frenkiel-Lebossé, H.; Hallier-Soulier, S. Usefulness of real-time PCR as a complementary tool to the monitoring of Legionella spp. and Legionella pneumophila by culture in industrial cooling systems. J. Appl. Microbiol. 2011, 111, 499–510. [Google Scholar] [CrossRef]
- Collins, S.; Stevenson, D.; Walker, J.; Bennett, A. Evaluation of Legionella real-time PCR against traditional culture for routine and public health testing of water samples. J. Appl. Microbiol. 2017, 122, 1692–1703. [Google Scholar] [CrossRef]
- Benitez, A.J.; Winchell, J.M. Clinical application of a multiplex real-time PCR assay for simultaneous detection of Legionella species, Legionella pneumophila, and Legionella pneumophila serogroup 1. J. Clin. Microbiol. 2014, 52, 709. [Google Scholar] [CrossRef]
- Benitez, A.J.; Winchell, J.M. Rapid detection and typing of pathogenic nonpneumophila Legionella spp. isolates using a multiplex real-time PCR assay. Diagn. Microbiol. Infect. Dis. 2016, 84, 298–303. [Google Scholar] [CrossRef] [PubMed]
- El Basha, N.R.; Shaaban, H.H.; El Atroush, H.A.; Sherif, M.M.; El Kholy, A.A. The use of multiplex PCR for the detection of atypical pathogens in Egyptian children with CAP: A high rate of Bordetella pertussis in early infancy. J. Egypt. Public Health Assoc. 2019, 94, 5. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Gattuso, G.; Lombardo, C.; Lupo, G.; Grillo, C.M.; Spandidos, D.A.; Libra, M.; Salmeri, M. Droplet digital PCR for the detection and monitoring of Legionella pneumophila. Int. J. Mol. Med. 2020, 46, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Logan-Jackson, A.; Rose, J.B. Cooccurrence of Five Pathogenic Legionella spp. and Two Free-Living Amoebae Species in a Complete Drinking Water System and Cooling Towers. Pathogens 2021, 10, 1407. [Google Scholar] [CrossRef]
- Logan-Jackson, A.R.; Rose, J.B. Water Age Effects on the Occurrence and Concentration of Legionella Species in the Distribution System, Premise Plumbing, and the Cooling Towers. Microorganisms 2021, 10, 81. [Google Scholar] [CrossRef]
- Marinelli, L.; Cottarelli, A.; Solimini, A.G.; Del Cimmuto, A.; De Giusti, M. Evaluation of timing of re-appearance of VBNC Legionella for risk assessment in hospital water distribution systems. Ann. Ig. 2017, 29, 431–439. [Google Scholar] [CrossRef]
- Eble, D.; Gehrig, V.; Schubert-Ullrich, P.; Köppel, R.; Füchslin, H. Comparison of the culture method with multiplex PCR for the confirmation of Legionella spp. and Legionella pneumophila. J. Appl. Microbiol. 2021, 131, 2600–2609. [Google Scholar] [CrossRef]
- Peci, A.; Winter, A.-L.; Gubbay, J.B. Evaluation and Comparison of Multiple Test Methods, Including Real-time PCR, for Legionella Detection in Clinical Specimens. Front. Public Health 2016, 4, 175. [Google Scholar] [CrossRef]
- Ueda, A.; Oki, M.; Yanagi, H.; Ozawa, H.; Takagi, A. Clinical Characteristics of Legionella Pneumonia Diagnosed with Legionella Urinary Antigen Test. Tokai J. Exp. Clin. Med. 2016, 41, 8–13. [Google Scholar]
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and Legionnaires’ Disease: 25 Years of Investigation. Clin. Microbiol. Rev. 2002, 15, 506–526. [Google Scholar] [CrossRef]
- Badoux, P.; Kracht-Kosten, L.; Herpers, B.; Euser, S. Method Comparison of the ImmuView L. pneumophila and L. longbeachae Urinary Antigen Test with the BinaxNOW Legionella Urinary Antigen Card for Detection of Legionella pneumophila Serogroup 1 Antigen in Urine. J. Clin. Microbiol. 2020, 58, 1429. [Google Scholar] [CrossRef] [PubMed]

| Virulence Factors | Encoding Genes | Roles | |
|---|---|---|---|
| Surface Proteins | Hsp60 | htpB | Attachment in host cells, modulation of invasion and cytokine expression in macrophages [36] |
| MOMP | momps | Activation of an alternative pathway of complement CR1 and CR3, phagocytosis [37] | |
| Mip | Mip | Penetration and replication in host cells [38] | |
| Secretion systems | Type I Lss | lssXYZABD locus | Secretion of rtxA, attachment and penetration in host cells [34,59] |
| Type II Lsp | PilEL | Secretion of other virulence factors [42] | |
| Type IV | Lvh | Conjunction, secretion of virulence factors, Legionella survival [43,60] | |
| IcM/Dot | Conjunction, transport and injection of DNA [48,49] | ||
| Two component system | CpxRA | cpxA, cpxR, cpxRA | Transcription of anti-stressor genes, regulation of IcM/Dot system effectors [57,58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gattuso, G.; Rizzo, R.; Lavoro, A.; Spoto, V.; Porciello, G.; Montagnese, C.; Cinà, D.; Cosentino, A.; Lombardo, C.; Mezzatesta, M.L.; et al. Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies. Antibiotics 2022, 11, 370. https://doi.org/10.3390/antibiotics11030370
Gattuso G, Rizzo R, Lavoro A, Spoto V, Porciello G, Montagnese C, Cinà D, Cosentino A, Lombardo C, Mezzatesta ML, et al. Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies. Antibiotics. 2022; 11(3):370. https://doi.org/10.3390/antibiotics11030370
Chicago/Turabian StyleGattuso, Giuseppe, Roberta Rizzo, Alessandro Lavoro, Vincenzoleo Spoto, Giuseppe Porciello, Concetta Montagnese, Diana Cinà, Alessia Cosentino, Cinzia Lombardo, Maria Lina Mezzatesta, and et al. 2022. "Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies" Antibiotics 11, no. 3: 370. https://doi.org/10.3390/antibiotics11030370
APA StyleGattuso, G., Rizzo, R., Lavoro, A., Spoto, V., Porciello, G., Montagnese, C., Cinà, D., Cosentino, A., Lombardo, C., Mezzatesta, M. L., & Salmeri, M. (2022). Overview of the Clinical and Molecular Features of Legionella Pneumophila: Focus on Novel Surveillance and Diagnostic Strategies. Antibiotics, 11(3), 370. https://doi.org/10.3390/antibiotics11030370












