Abstract
Anthropogenic pressure is known to be a key driver of antimicrobial resistance (AMR) dissemination in the environment. Especially in lower income countries, with poor infrastructure, the level of AMR dissemination is high. Therefore, we assessed the levels and diversity of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in Lebanese rivers at estuaries’ sites (n = 72) of the Mediterranean Sea in spring 2017 and winter 2018. Methods: A combined approach using culture techniques and high throughput qPCR were applied to identify ARB and ARGs in rivers along the Lebanese coast. Results: Multidrug-resistant Gram-negative (Enterobacterales and Pseudomonas spp.) and Gram-positive bacterial pathogens were isolated. Levels of ARGs were highest in the winter campaign and areas with high anthropogenic activities and population growth with an influx of refugees. Conclusion: Qualitative analysis of ARB and the analysis of the Lebanese estuaries’ resistome revealed critical levels of contamination with pathogenic bacteria and provided significant information about the spread of ARGs in anthropogenically impacted estuaries.
1. Introduction
Antimicrobial resistance (AMR) is a One Health crisis aggravated by the lack of water and pollution management on a global scale [1,2]. Anthropogenic activities are demonstrated to be the key drivers of AMR dissemination in the environment [3,4,5,6], subsequently altering its ecosystems [7,8,9]. Through discharge of treated or untreated wastewater (WW) effluents into surface water, a high abundance of antibiotic resistance genes (ARGs), antibiotic-resistant bacteria (ARB), and mobile genetic elements (MGEs) mixed with a cocktail of micropollutants and drug residues are continuously disseminated into the environment [10,11,12,13]. Additionally, agricultural practices such as soil fertilization with manure and sludge, or irrigation with WW effluents, further expand the environmental background levels of pollutants associated with the dissemination of antimicrobial resistance [9,14]. In Lebanon, rules to control the overuse and misuse of antibiotics for treatment, growth promotion, and prophylaxis in agriculture and animal husbandry are not strictly implemented [15,16], which would likely contribute to increasing AMR in the Lebanese and connected environments [17,18].
The surveillance of AMR in the environment is often assessed through ARG quantification [19,20]. High abundances of ARGs were found to be associated with fecal contamination [21,22,23].
Estuaries are transitional zones between rivers and sea bodies [24,25], exhibiting properties of marine and freshwater and underlining continental–oceanic interactions [26]. Estuaries are also considered as ecosystems that are broadly anthropized [24,27]. Contaminants can reside for prolonged periods in estuarian ecosystems due to the tidal streams [28,29]. The levels of ARB and ARGs and their spatial–temporal dissemination in Lebanese estuaries remains understudied [26]. The Mediterranean coastline is densely populated with a large anthropic footprint, i.e., intensive fishing, shipping, recreational, and industrial activities [30]. Our study aimed to assess the level of AMR dissemination in the Mediterranean Sea through the Lebanese estuaries. The objectives of this study were to monitor in the Lebanese river estuaries, the dissemination of i) ARB, notably extended-spectrum β-Lactamase-producing-Enterobacterales (ESBL-E), multidrug-resistant Pseudomonas aeruginosa, and methicillin-resistant Staphylococcus aureus (MRSA), and ii) the resistome through the detection of ARGs and MGEs.
2. Results
2.1. Bacterial Culture
In total, we isolated 19 different bacterial species in different quantities in spring and winter from the estuarine water samples along the Lebanon coastline (see the Materials and Methods section) (Table 1). From 1 mL of the water samples cultivated on selective media, we obtained 50 and 10,064 CFUs (colony forming units) resistant Enterobacterales and 41 and 43 CFUs resistant Pseudomonas spp. in spring and winter, respectively. However, among the 10,064 CFUs in winter, 104 corresponded to the same bacterial species (Hafnia alvei) in the Beirut estuary. A more diverse panel of Enterobacterales species was isolated in spring. Enterobacterales and/or Pseudomonas spp. strains were isolated from all samples, except the Bared River. Moreover, Enterobacterales were not detected in the Kaleb River and Pseudomonas spp. were not found in the Zahnari river. MRSA isolates were detected only in spring.

Table 1.
Gram-negative bacterial species and Gram-positive MRSA isolated in the rivers’ estuaries.
2.2. Susceptibility Profiles
We performed susceptibility testing for a panel of Enterobacterales (33) and Pseudomonas (39) strains. The results are shown in Table 2. We found 16 out of the 33 Enterobacterales and 6 out of the 39 Pseudomonas strains tested that expressed an ESBL phenotype detected with the double-disk synergy test according to the European Committee on Antimicrobial Susceptibility (EUCAST) v8 guidelines.

Table 2.
Resistance of the tested Enterobacterales and Pseudomonas species: numbers indicate the number of strains resistant out of the strains tested (%).
Resistance towards the tested antibiotics was broadly disseminated. All tested strains of Pseudomonas in spring were susceptible to gentamicin. In winter, we detected two Pseudomonas aeruginosa strains with an ESBL synergy test, one in Janoubi and one in Beirut. We detected five strains resistant to imipenem in spring, three ESBL-positive Pseudomonas luteola strains (two in Aarqa and one in Beirut), and two Enterobacterales strains (one ESBL Klebsiella oxytoca in Aarqa and one Serratia marcescens in Qadicha). No resistant strains to imipenem were detected in winter.
2.3. Targeted Resistome Assessment by High-Throughput qPCR
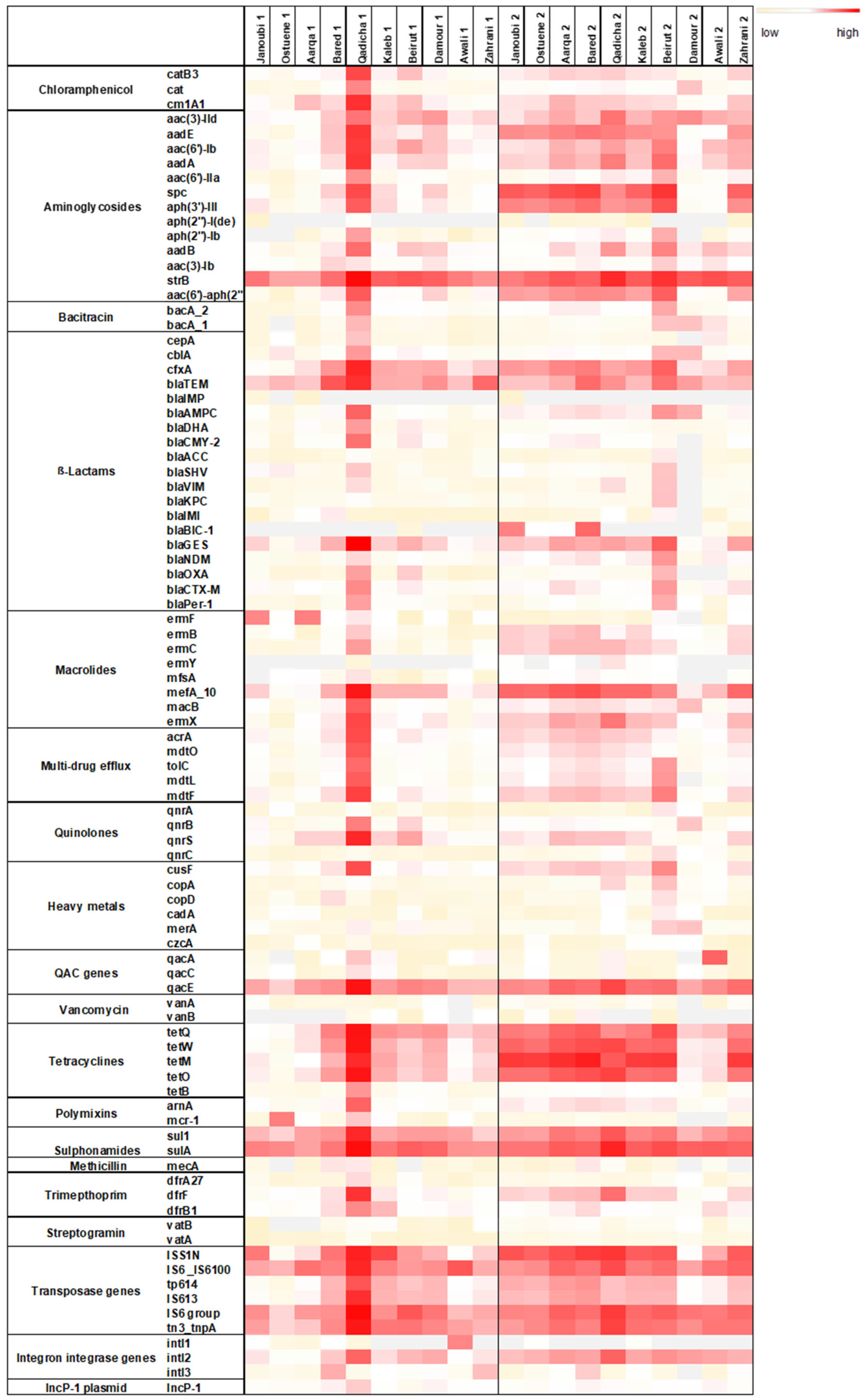
Total DNA was extracted from triplicate river samples collected at the 12 estuaries of the major Lebanese rivers. The resistome analysis could only be performed on 10 rivers during both sampling campaigns since for 2 rivers DNA concentration was not sufficient. Figure 1 depicts the abundance of each targeted gene normalized to the 16S rRNA encoding gene.
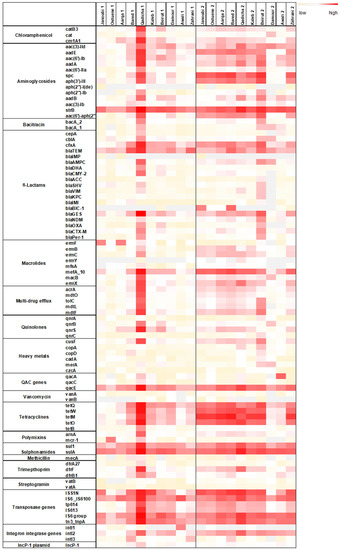
Figure 1.
Heat map depicting the normalized abundances to the 16S rRNA gene of each targeted gene in spring (1) and in winter (2) in the 10 estuarine Lebanese river samples. Three-color legend with red: high normalized abundance, white: medium normalized abundance, and light yellow: low normalized abundance. Grey fields: gene undetectable. qac genes: genes conferring resistance to quaternary ammonium compounds.
Overall, a higher normalized abundance of the targeted resistome was detected during the winter sampling campaign (Figure 1). Normalized abundance of individual ARGs was markedly high for the Qadicha estuarine during the spring sampling campaign, compared to all other estuaries.
The marker IncP-1 was detected in all river estuaries. IncP-1 plasmids are highly promiscuous and considered as anthropogenic markers in the environment. They frequently carry multiple ARGs and are suggested to be important vectors of ARGs and integrons [31].
Noticeably, transposase genes that are associated with the dissemination of ARGs amongst bacteria were detected in high normalized abundance in most river samples, especially during the winter sampling campaign. Individual resistance genes such as the strB, spc (conferring resistance to streptomycin, spectinomycin), the mefA gene (conferring resistance to macrolides), the tetracycline resistance gene tetM, and the resistance gene sulA conferring resistance to sulphonamides, were also detected in higher normalized abundance in different rivers during the winter sampling campaign.
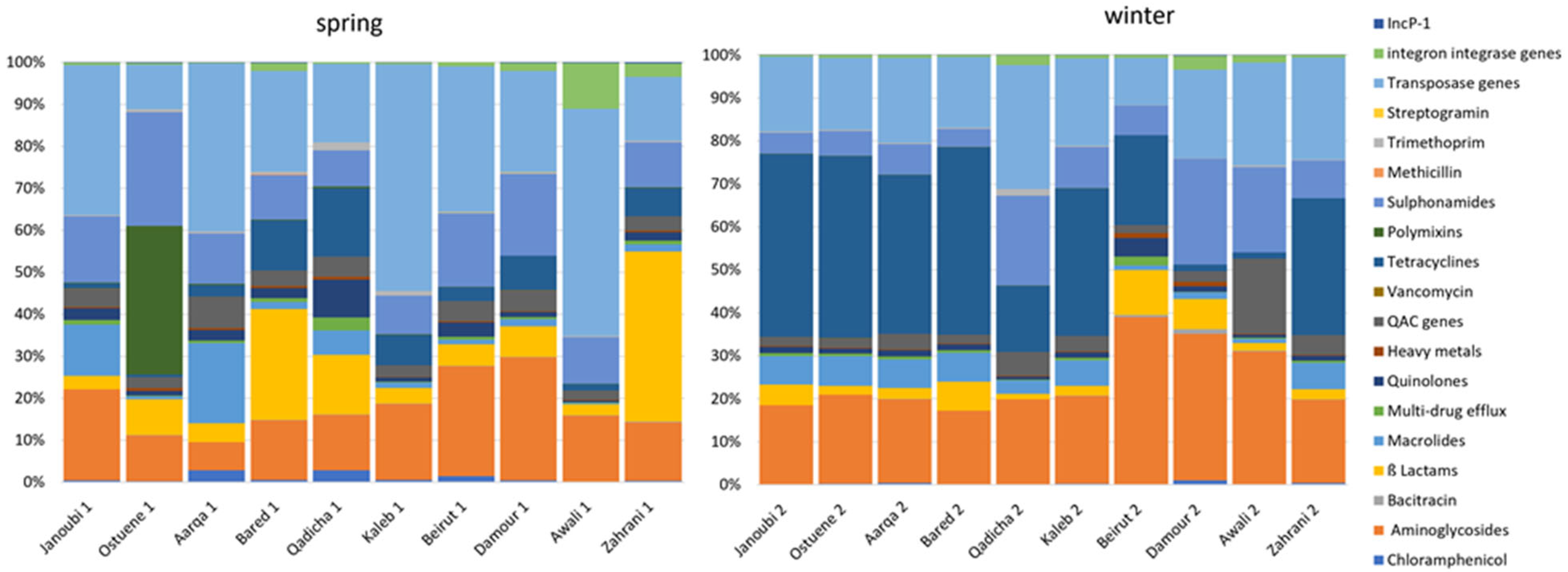
Figure 2 exhibits different resistome signatures between the two sampling campaigns, with a higher diversity between individual rivers in spring than in winter. The resistome signature is less variable and remarkably similar between the northern sampled estuaries (Janoubi, Ostuene, Aarqa, and Bared) during the winter campaign.
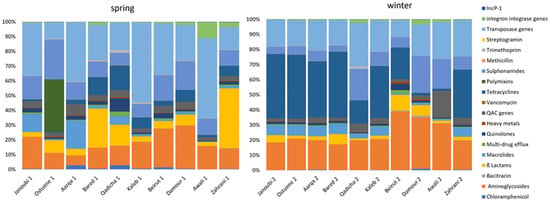
Figure 2.
Proportional abundances of targeted genes grouped into gene families according to their function per river in spring and in winter in 10 estuarine Lebanese rivers samples. qac genes: genes conferring resistance to quaternary ammonium compounds.
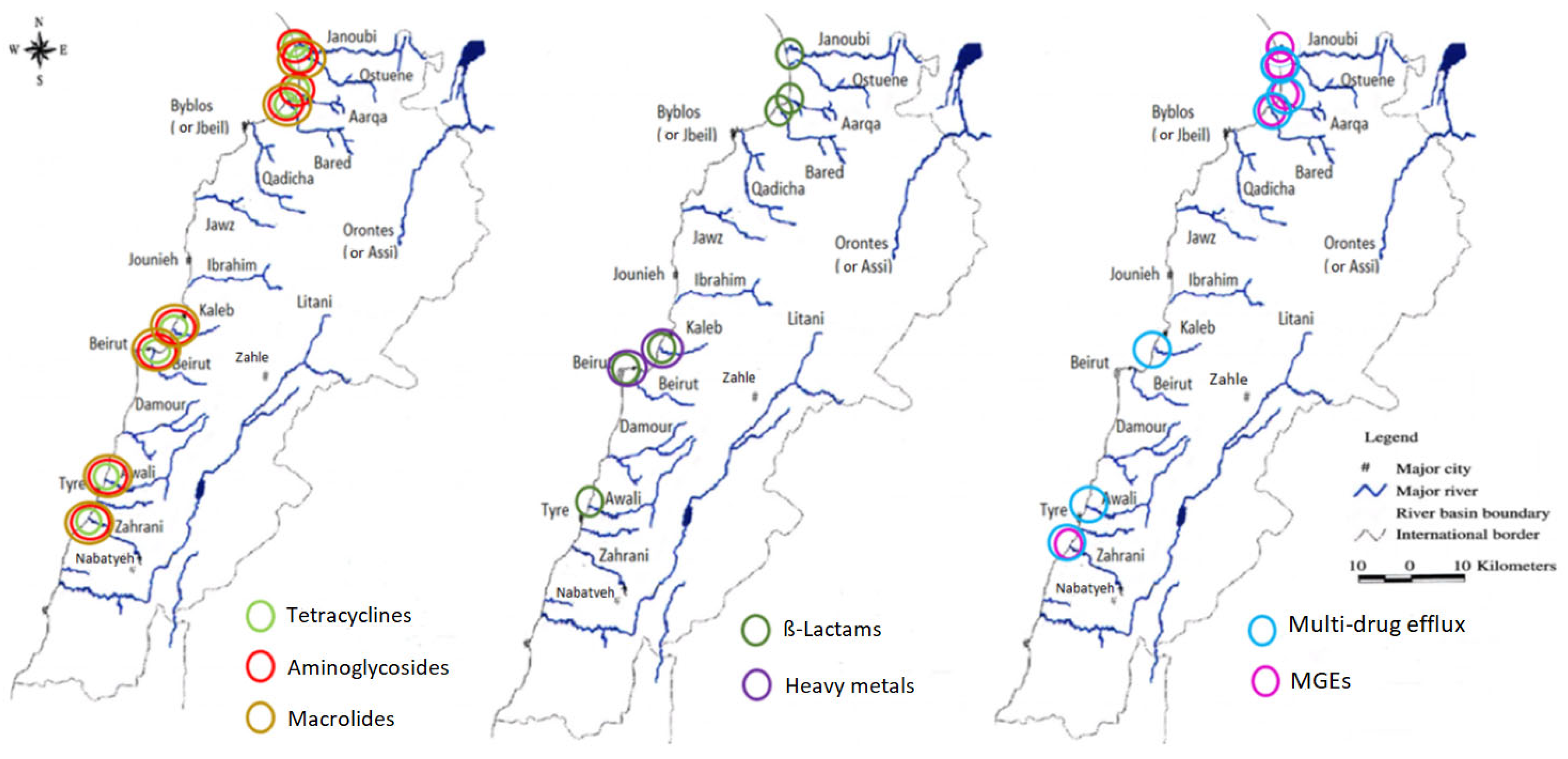
Variation in normalized abundance for the different gene families in the estuaries during the two sampling campaigns in winter and spring was assessed using the non-parametric Wilcoxon’s test. Figure 3 depicts the estuaries where a significant difference in the mean normalized abundance was observed between spring and winter. The p values corresponding to all the significant variations are available in Supplementary Table S2.
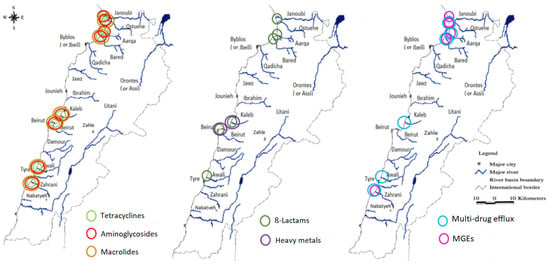
Figure 3.
Graphical representation showing the estuaries where an increase in the mean normalized abundances of the ARGs families and MGEs (transposase genes, integron integrase genes, and the IncP-1 gene were grouped together as mobile genetic elements) from spring (2017) to winter (2018) was significant (p < 0.05).
Although we observed a significant increase for most ARG families in all samples from spring to winter, the mean normalized abundances of tetracyclines and β-Lactams, decreased from spring to winter in the Qadicha and Damour estuaries. In Qadicha, the aminoglycoside, macrolide, tetracycline, multidrug efflux, and transposase genes decreased significantly from spring to winter.
3. Discussion
Here we show that high levels of ARB and ARGs could be detected in all studied Lebanese rivers during all time points. Previous studies underlined the contamination of Lebanese rivers with fecal bacteria, especially Escherichia coli and coliforms, reaching 70.4% of the rivers in the North and 60% in the Bekaa region, which was above the acceptable threshold according to the French SEQ-EAU-2003 recommendations for irrigation water [32].
In our study, we showed that the levels of ARB and ARGs varied according to the location.
The Lebanese coastal belt from Tripoli to Tyre is an urbanized zone with significant agricultural activities [33,34]. The highest bacterial counts, (Enterobacterales (Hafnia alvei (104)) in winter and Pseudomonas luteola (104) in spring) were detected in the estuary of the capital Beirut. The highest normalized abundance of ARGs was observed in the Qadicha estuary in spring. The Qadicha river is a small river impacted by population growth, high industrial activities, and a chronic default of wastewater treatment. It has been shown that the estuary of this river was highly contaminated with pollutants [35], which may explain the high levels of ARGs and ARB detected here. The highest diversity of Enterobacterales was isolated from the Aarqa estuary in spring 2017, while a higher bacterial count was observed in winter (Klebsiella pneumoniae 50 CFU). The Aarqa estuary is located on the northern rural coast, inhabited by local and refugee communities with high agricultural activities.
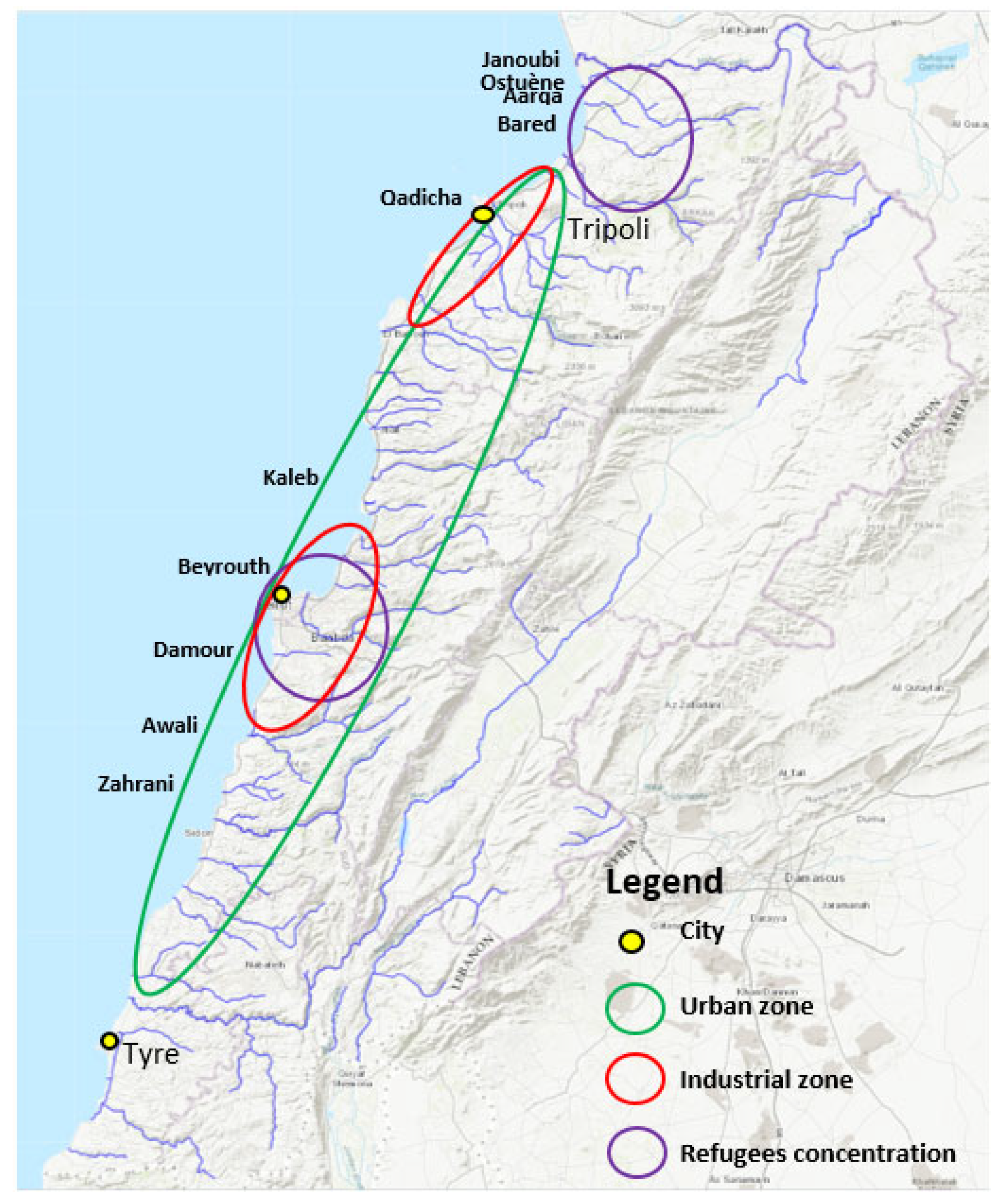
Interestingly, we observed particularly high levels of ARGs in the North (Figure 3), which corresponds to the area with the highest number of inhabitants and with a high density of refugees’ camps (Figure 4). Lebanon hosts the largest number of refugees per capita worldwide. The Syrian crisis in the year 2011 has escalated river water pollution, particularly on the northern coast, following the migratory inflow of displaced persons searching for water points and settling to find refuge while infrastructure is lacking [36]. This inflow has contributed to sustaining extreme hot spots of water stress in urban areas in informal settlements (the Northern coast or in the Bekaa region), while water networks and governance are already insufficient [32]. In addition to the efforts from humanitarian organizations such as the UNHCR and the UNICEF to improve the sanitary of a fraction of the displaced people by the implementation of water and wastewater facilities, local initiatives to strengthen and/or rehabilitate existing infrastructure also exist and must be continued to alleviate the vulnerability of displaced communities and to reduce the water stress in the country [37,38].
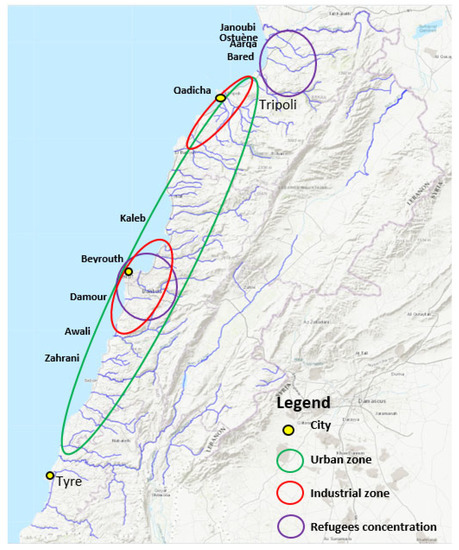
Figure 4.
High anthropogenic impact locations. A map from “Rivers, Lebanon, 2012. https://maps.princeton.edu/catalog/stanford-wn533df2039” (accessed on 10 December 2021) representing the Lebanese rivers, modified to show locations on the Lebanese cost of the largest industrial zones (Beirut and Tripoli), the highest urbanized area, and the location of refugees, according to the UNHCR, UN-habitat 2014, and the UNRWA 2021 organizations.
River water levels in Lebanon increase during spring due to the rise of underground water and the snowmelt [39]. The difference in water levels between seasons may contribute to the observed lower normalized abundances of the targeted resistome in spring compared with the winter season (Figure 1). Notably, we observed a higher diversity of ARB and ARGs in the spring sampling campaign (Figure 2). The observed diversity for the targeted resistome and ARB in spring may also reflect an impact due to higher recreational activities in spring compared with winter.
Most bacteria that were isolated from the river estuaries were highly resistant to β Lactams, with 16/33 Enterobacterales strains and 6/39 Pseudomonas strains expressing an ESBL phenotype. The targeted resistome analysis of the river water samples detected a panel of ESBL-encoding genes, including a high abundance of ESBL-encoding genes that were specifically detected in the Mediterranean Sea [40,41], such as blaTEM and blaGES in most river samples during both seasons.
The high normalized abundance detected for transposase genes in the estuaries in both sampling campaigns might be indicative of an increased dissemination of ARGs among the bacteria in Lebanese estuaries. Transposons in rivers have been suggested to drive the dissemination of ARGs [42]. It was recently shown that transfer between resistance plasmids and bacterial chromosomes is mediated by insertion sequences, mainly those belonging to the IS6 family, IS26, and IS6100 [43]. The clinically relevant IS6-group and IS6100 genes were detected in high abundance in the estuaries studied.
The high prevalence of the tetracycline resistance genes (tetQ, tetW, tetM, and tetO) might be the result of antibiotic selective pressure, considering that tetracyclines persist for a long time in the environment and are used frequently in human and veterinary medicine and aquacultural and agricultural practices [44,45,46].
Studies in lower income countries with lack of sanitation measures and operational wastewater treatment systems have shown high levels of ARB and ARGs that were significantly correlated with human gut bacteria and pathogens [23,47,48]. Inversely, resistant Enterobacterales in the urbanized San Francisco Bay were absent from near-shore sediments, suggesting that urbanized estuaries may not constitute a major human exposure hazard when secondary and tertiary treatment operations and control measures for all wastewaters that drain into the studied environment are implemented [49]. Altogether, the presence of ESBL and carbapenemase-producing pathogens and corresponding ARGs in the Lebanon estuaries raises concerns and warrants further attention [50].
The presence of MRSA in Aarqa, Ostuene, and Awali estuaries detected in spring should be closely monitored since studies have clearly shown that Staphylococcus aureus and MRSA persist in anthropized river samples, presenting a potential source of the dissemination and transmission of resistant bacteria [51,52,53].
Currently, wastewater treatment plants (WWTP) in Lebanon mainly use secondary systems, although their status of operationality is undetermined [36].
Here, we confirmed the estuaries of Lebanon as putative host-spots for the dissemination of ARGs and resistant bacteria into the Mediterranean Sea. Recreational waters such as rivers and beaches have gained increased attention as having a central role in the persistence, dissemination, and emergence of antibiotic resistance [54]. Specific surveillance systems should be put in place that trace urban movements, contact with recreational waters, pollution levels, and WW treatment levels and management to trace the actual dissemination of antimicrobial resistance through recreational waters [55].
4. Materials and Methods
Lebanon is located in the eastern Mediterranean area along a coastal length of 220 km [30] and is characterized by a short, cold, and wet winter season from January to March, with annual rainfall ranging between 850 mm and 1800 mm/year, and by a dry summer [56,57]. The surface waters in Lebanon are under increased pressure from anthropic activities, i.e., urban, industrial, and recreational. Agriculture in the coastal zone in Lebanon requires irrigation with surface and groundwater, causing the depletion of water resources while increasing pollution according to the Food and Agriculture organization report of the year 2016 [36].
Figure 4 marks anthropogenic impact on the different geographic locations of Lebanon by indicating important urban zones, industrial zones, and activity, as well as zones with high population densities due to the influx of refugees.
Fifteen Lebanese rivers spread through or at the extension of the Mount of Lebanon for a dozen of km eastward before discharging in small catchment areas leading to the Mediterranean Sea [58]. The rivers spread over the territory, making it a dense network of watercourses at a less than 10 km distance from each other and sharing similar basin characteristics [58]. To evaluate the seasonal impact on the presence and abundance of ARB and ARGs, sampling campaigns were performed at two different periods.
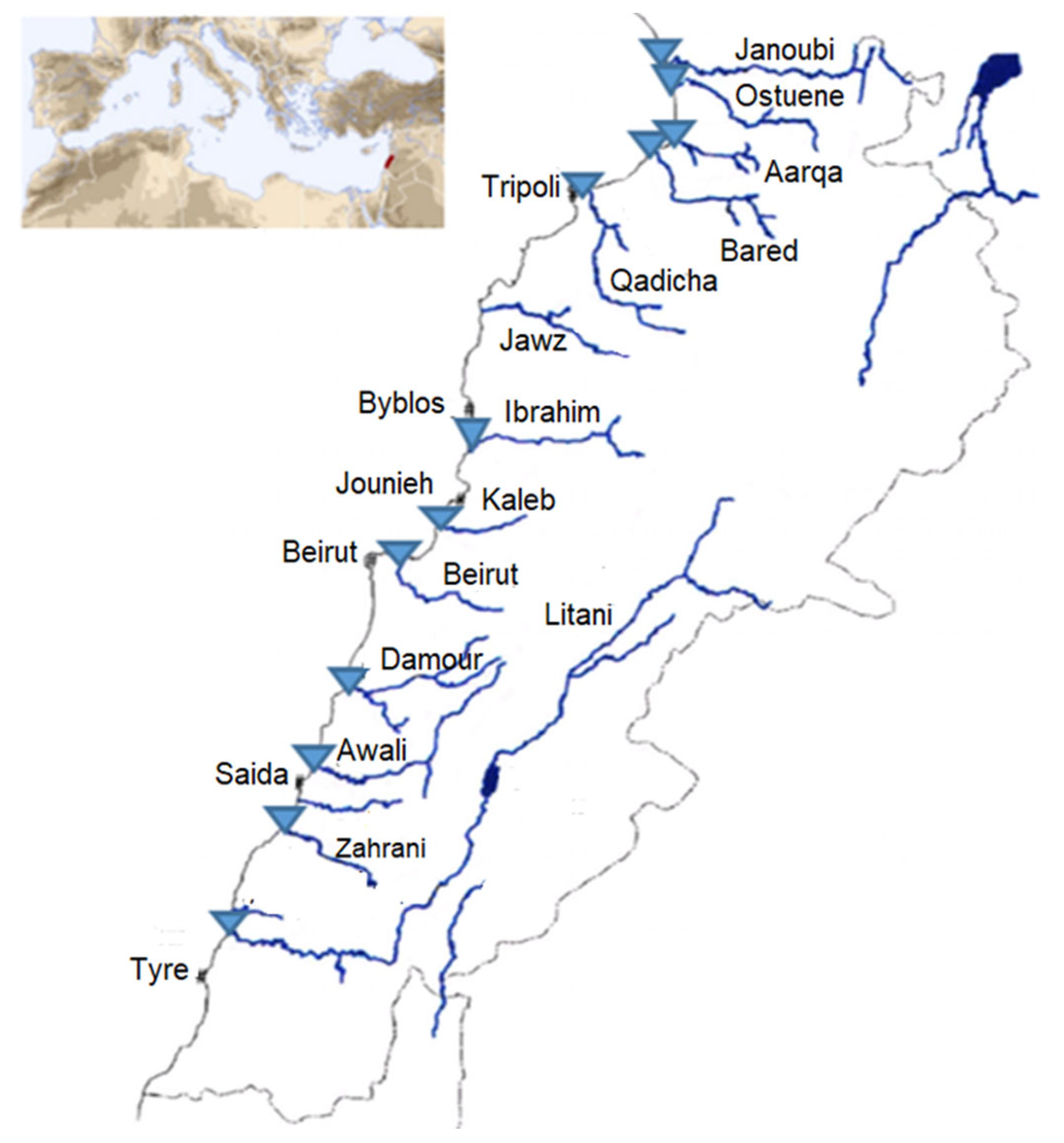
Among the rivers’ estuaries, twelve estuaries or mouths were sampled in triplicate in sterile cups in April 2017 and January 2018, resulting in n = 72 river water samples: 36 in spring and 36 in winter (3 samples of 60 mL each taken one after the other per river) (Figure 5). The samples from two rivers (Litani and Ibrahim) did not contain sufficient biomass and were excluded from the study. Weather conditions in terms of temperature with slight or no precipitation were similar, along with the month duration for each of the two sampling campaigns. Samples were transported on ice directly to our Lebanese laboratory for further analysis within 2 h. The exact coordinates registered for each sampling location as well as the temperatures are available in Supplementary Table S1.
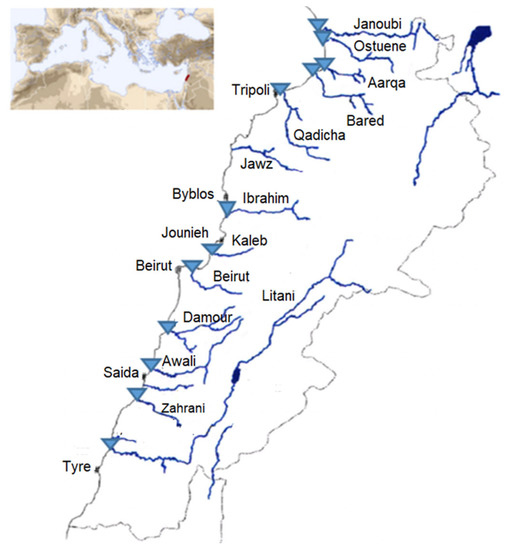
Figure 5.
Sampling sites are represented as blue triangles on the map of the Lebanese rivers. Figure adapted from Houri and El Jeblawi (2007).
The water samples were analyzed by the spread-plate method, culturing a V = 1 mL volume, using sterile rakes on MacConkey and Mannitol Salt agar selective of Gram-negative bacteria and Staphylococcus spp., respectively, with and without antibiotics. For Gram-negative enteric bacteria, we used ceftriaxone (2 mg/L), cefepime (4 mg/L), or ertapenem (0.5 mg/L). For Staphylococcus aureus, we used oxacillin (4 mg/L). All media were incubated at 37 °C for 48 h. For Gram-negative bacteria, species identification was performed with API® 20 NE (for Pseudomonas) or API® 20 E (for Enterobacterales) galleries (Biomérieux). For S. aureus, we used three phenotypic tests: catalase, DNAse, and coagulase.
The susceptibility testing was performed on Mueller–Hinton agar according to the European Committee on Antimicrobial Susceptibility (EUCAST) v8 guidelines. Antibiotic Bio-Rad® discs used were: amoxicillin–clavulanic acid (20 µg–10 µg), cefepim 30 µg, ceftazidim (10 µg), aztreonam (30 µg), gentamicin (10 µg), amikacin (30 µg), piperacillin–tazobactam (30 µg–6 µg), and imipenem (10 µg) for Gram-negative bacteria, and fusidic acid (10 µg), cefoxitin (30 µg), trimethoprim–sulfamethoxazole (1.25–23.75 µg), gentamicin (10 µg), and ciprofloxacin (5 µg) for S. aureus.
For molecular analysis, the three water samples per river were filtered (total volume of water filtered = 180 mL), using a filtration ramp (Sartorius, Göttingen, Germany) on a sterile 47 mm diameter filter with a pore size of 0.45 µm (Sartorius, Göttingen, Germany). Microorganisms were recovered from filters and subject to DNA extraction for downstream analysis using the DNeasy PowerWater® (Qiagen) adapted to water samples. All DNA samples were diluted or concentrated to a final concentration of 10 ng/µL for downstream qPCR and 16S rRNA analysis.
The Litani and Ibrahim rivers estuaries were excluded from the analysis, as the respective water samples contained insufficient biomass even after an additional 2 L water volume sampling and DNA extraction.
We targeted 71 ARGs, 6 heavy metal resistance genes, and 3 genes encoding resistance to quaternary ammonium compounds, 9 MGEs (transposases ISSW1, ISS1N, IS6100, IS613, IS6 group, Tn3, ISCEc9, tp614), and integron integrase genes (intI1, intI2, and intI3) [10]. The list of all the primers is published [10]. The 16S rRNA encoding genes were targeted to allow normalizing the abundance of individual resistance genes.
The targeted genes are grouped according to their function [10] with, in addition, the added environmental resistance marker: IncP-1 [31].
High throughput real-time PCR was performed using the Biomark microfluidic system from Fluidigm, in which every sample–gene combination is quantified using a 96.96 Dynamic Array™ IFCs (BMK-M-96.96, Fluidigm), as described previously [59]. A mean normalized abundance for each ARGs’ family and each estuary in spring and winter was calculated by dividing the cumulated abundance by the number of ARGs constituting the ARG family. The mean abundance of the ARGs families was then compared between spring and winter to assess a possible variation between the seasons. The estuaries where a significant variation was observed are listed in Supplementary Table S2.
5. Conclusions
To date, this is the first study to provide an accurate assessment and comparison of the targeted resistome in ten estuaries on the Lebanese coast. A combined approach using culture-based techniques and high throughput qPCR for the detection of ARB and ARGs identified Lebanese river estuaries as hotspots for antimicrobial resistance. This study highlights the need to implement regular antimicrobial resistance surveillance and improvement in wastewater management, in addition to the enforcement of regulations and guidelines stringency for sewage sludge or wastewater reuse, as per the new EU 2020/741 legislation of the European parliament.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics11030306/s1, Table S1: The exact coordinates of the sampling locations and the temperature recorded at the time of sampling, Table S2: Significant variations (p < 0.05) in ARGs families in the rivers between spring and winter.
Author Contributions
Conceptualization, W.H., D.K.S. and C.D.; methodology, W.H., M.G. and E.B.; software, W.H. and E.B.; validation, E.B., D.K.S. and C.D.; formal analysis, W.H., E.B. and M.-C.P.; investigation, W.H.; resources, D.K.S. and M.-C.P.; data curation, E.B. and M.-C.P.; writing—original draft preparation, W.H.; writing—review and editing, W.H., E.B., C.D. and M.-C.P.; visualization, E.B. and M.-C.P.; supervision, E.B. and C.D.; project administration, D.K.S. and C.D.; funding acquisition, D.K.S., C.D. and M.-C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by The National Council for Scientific Research Lebanon CNRS-L, the Inserm Research Institute and French Ministry of Research, and the Research Council of the Saint-Joseph University of Beirut.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Collignon, P.; Beggs, J.J.; Walsh, T.; Gandra, S.; Laxminarayan, R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef]
- Pieri, A.; Aschbacher, R.; Fasani, G.; Mariella, J.; Brusetti, L.; Pagani, E.; Sartelli, M.; Pagani, L. Country income is only one of the tiles: The global journey of antimicrobial resistance among humans, animals, and environment. Antibiotics 2020, 9, 473. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Catena, F.; Coccolini, F.; Hardcastle, T.C.; Roques, C.; Salameh, P. Drivers of Antibiotic Resistance Transmission in Low- and Middle-Income Countries from a “One Health” Perspective—A Review. Antibiotics 2020, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Goulas, A.; Belhadi, D.; Descamps, A.; Andremont, A.; Benoit, P.; Courtois, S.; Dagot, C.; Grall, N.; Makowski, D.; Nazaret, S.; et al. How effective are strategies to control the dissemination of antibiotic resistance in the environment? A systematic review. Environ. Evid. 2020, 9, 4. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Chatterjee, A.; Modarai, M.; Naylor, N.; Boyd, S.E.; Atun, R.; Barlow, J.; Holmes, A.H.; Johnson, A.; Robotham, J. Quantifying drivers of antibiotic resistance in humans: A systematic review. Lancet Infect. Dis. 2018, 18, e368–e378. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, A.; Power, A.; Hansen, M.; Brandt, K.; Piliposian, G.; Appleby, P.; O’Neill, P.; Jones, R.; Sierocinski, P.; Koskella, B.; et al. Heavy metal pollution and co-selection for antibiotic resistance: A microbial palaeontology approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef]
- Xiang, Q.; Chen, Q.-L.; Zhu, D.; An, X.-L.; Yang, X.-R.; Su, J.-Q.; Qiao, M.; Zhu, Y.-G. Spatial and temporal distribution of antibiotic resistomes in a peri-urban area is associated significantly with anthropogenic activities. Environ. Pollut. 2018, 235, 525–533. [Google Scholar] [CrossRef]
- Buelow, E.; Rico, A.; Gaschet, M.; Lourenço, J.; Kennedy, S.P.; Wiest, L.; Ploy, M.-C.; Dagot, C. Hospital discharges in urban sanitation systems: Long-term monitoring of wastewater resistome and microbiota in relationship to their eco-exposome. Water Res. X 2020, 7, 100045. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Aubertheau, E.; Stalder, T.; Mondamert, L.; Ploy, M.-C.; Dagot, C.; Labanowski, J. Impact of wastewater treatment plant discharge on the contamination of river biofilms by pharmaceuticals and antibiotic resistance. Sci. Total Environ. 2017, 579, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.M.; Narciso-Da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buelow, E.; Ploy, M.-C.; Dagot, C. Role of pollution on the selection of antibiotic resistance and bacterial pathogens in the environment. Curr. Opin. Microbiol. 2021, 64, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Chaddad, A.; Hanna, J.; Matta, J.; Daoud, Z. Understanding the Epidemiology of Multi-Drug Resistant Gram-Negative Bacilli in the Middle East Using a One Health Approach. Front. Microbiol. 2019, 10, 1941. [Google Scholar] [CrossRef] [Green Version]
- Jammoul, A.; El Darra, N. Evaluation of Antibiotics Residues in Chicken Meat Samples in Lebanon. Antibiotics 2019, 8, 69. [Google Scholar] [CrossRef] [Green Version]
- Vaz-Moreira, I.; Ferreira, C.; Nunes, O.C.; Manaia, C.M. Sources of Antibiotic Resistance: Zoonotic, Human, Environment. Antibiot. Drug Resist. 2019, 10, 211–238. [Google Scholar]
- Lundborg, C.S.; Tamhankar, A.J. Antibiotic residues in the environment of South East Asia. BMJ 2017, 358, j2440. [Google Scholar] [CrossRef] [Green Version]
- Triggiano, F.; Calia, C.; Diella, G.; Montagna, M.T.; De Giglio, O.; Caggiano, G. The Role of Urban Wastewater in the Environmental Transmission of Antimicrobial Resistance: The Current Situation in Italy (2010–2019). Microorganisms 2020, 8, 1567. [Google Scholar] [CrossRef]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Flach, C.-F.; Larsson, D.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef] [PubMed]
- An, X.-L.; Wang, J.-Y.; Pu, Q.; Li, H.; Pan, T.; Li, H.-Q.; Pan, F.-X.; Su, J.-Q. High-throughput diagnosis of human pathogens and fecal contamination in marine recreational water. Environ. Res. 2020, 190, 109982. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Guo, X.; Feng, C.; Gu, E.; Tian, C.; Shen, Z. Spatial distribution, source apportionment and risk assessment of antibiotics in the surface water and sediments of the Yangtze Estuary. Sci. Total Environ. 2019, 671, 548–557. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, A.; Dai, T.; Li, F.; Xie, H.; Chen, L.; Wen, D. Cell-free DNA: A Neglected Source for Antibiotic Resistance Genes Spreading from WWTPs. Environ. Sci. Technol. 2017, 52, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-P.; Liu, X.; Niu, Z.-S.; Lu, D.-P.; Zhao, S.; Sun, X.-L.; Wu, J.-Y.; Chen, Y.-R.; Tou, F.-Y.; Hou, L.; et al. Seasonal and spatial distribution of antibiotic resistance genes in the sediments along the Yangtze Estuary, China. Environ. Pollut. 2018, 242, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lau, H.K.; Lee, T.; Lau, D.K.; Payne, J. In Silico Serotyping Based on Whole-Genome Sequencing Improves the Accuracy of Shigella Identification. Appl. Environ. Microbiol. 2019, 85, e00165-19. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Su, Y.; Xi, H.; Chen, X.; Xie, B. Urban and agriculturally influenced water contribute differently to the spread of antibiotic resistance genes in a mega-city river network. Water Res. 2019, 158, 11–21. [Google Scholar] [CrossRef]
- Wang, S.; Xue, N.; Li, W.; Zhang, D.; Pan, X.; Luo, Y. Selectively enrichment of antibiotics and ARGs by microplastics in river, estuary and marine waters. Sci. Total Environ. 2020, 708, 134594. [Google Scholar] [CrossRef]
- Kazour, M.; Jemaa, S.; Issa, C.; Khalaf, G.; Amara, R. Microplastics pollution along the Lebanese coast (Eastern Mediterranean Basin): Occurrence in surface water, sediments and biota samples. Sci. Total Environ. 2019, 696, 133933. [Google Scholar] [CrossRef]
- Plaza, J.J.G.; Blau, K.; Milaković, M.; Jurina, T.; Smalla, K.; Udiković-Kolić, N. Antibiotic-manufacturing sites are hot-spots for the release and spread of antibiotic resistance genes and mobile genetic elements in receiving aquatic environments. Environ. Int. 2019, 130, 104735. [Google Scholar] [CrossRef] [PubMed]
- Dagher, L.A.; Hassan, J.; Kharroubi, S.; Jaafar, H.; Kassem, I.I. Nationwide Assessment of Water Quality in Rivers across Lebanon by Quantifying Fecal Indicators Densities and Profiling Antibiotic Resistance of Escherichia coli. Antibiotics 2021, 10, 883. [Google Scholar] [CrossRef] [PubMed]
- Verner, D.; Ashwill, M.; Christensen, J.; McDonnell, R.; Redwood, J.; Jomaa, I.; Saade, M.; Massad, R.; Chehade, A.; Bitar, A.; et al. Droughts and Agriculture in Lebanon; World Bank: Washington, DC, USA, 2018. [Google Scholar] [CrossRef] [Green Version]
- Council for Development & Reconstruction. National Physical Master Plan of the Lebanese Territory; Council for Development & Reconstruction: Beirut, Lebanon, 2005.
- Merhabi, F.; Gomez, E.; Amine, H.; Rosain, D.; Halwani, J.; Fenet, H. Occurrence, distribution, and ecological risk assessment of emerging and legacy contaminants in the Kadicha river in Lebanon. Environ. Sci. Pollut. Res. 2021, 28, 62499–62518. [Google Scholar] [CrossRef] [PubMed]
- Salman, M.; Abukhalaf, M.; del Lungo, A. Assessment of Treated Wastewater for Agriculture in Lebanon; FAO: Rome, Italy, 2016. [Google Scholar]
- WHO. Lebanon Water Quality Survey; WHO/UNICEF: Geneva, Switzerland, 2019. [Google Scholar]
- Yassin, N. 101Facts & Figures, Issam Fares Institute for Public Policy and International Affairs; American University of Beirut: Beirut, Lebanon, 2018. [Google Scholar]
- Fayad, A. Evaluation of the Snow Water Resources in Mount Lebanon Using Observations and Modelling; Université Paul Sabatier-Toulouse III: Toulouse, France, 2017. [Google Scholar]
- Aujoulat, F.; Ragot, R.; Toubiana, M.; Bancon-Montigny, C.; Monfort, P.; Salles, C.; Masnou, A.; Delpoux, S.; Rio, M.; Tournoud, M.; et al. Environmental antimicrobial resistance in a small urban Mediterranean river: A focus on endemic beta-lactamases in clinically relevant bacteria. Water 2021, 13, 2010. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic resistance genes in phage particles from antarctic and mediterranean seawater ecosystems. Microorganisms 2020, 8, 1293. [Google Scholar] [CrossRef]
- Zhang, B.; Qin, S.; Guan, X.; Jiang, K.; Jiang, M.; Liu, F. Distribution of Antibiotic Resistance Genes in Karst River and Its Ecological Risk. Water Res. 2021, 203, 117507. [Google Scholar] [CrossRef]
- Che, Y.; Yang, Y.; Xu, X.; Břinda, K.; Polz, M.F.; Hanage, W.P.; Zhang, T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2008731118. [Google Scholar] [CrossRef]
- Yoo, K.; Lee, G. Investigation of the Prevalence of Antibiotic Resistance Genes According to the Wastewater Treatment Scale Using Metagenomic Analysis. Antibiotics 2021, 10, 188. [Google Scholar] [CrossRef]
- Sciencedirect. Tetracycline. 2021. Available online: https://www.sciencedirect.com/topics/chemistry/tetracycline (accessed on 19 February 2022).
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in food and feeding stuffs: From regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017, 2017, 1315497. [Google Scholar] [CrossRef]
- Nadimpalli, M.; Delarocque-Astagneau, E.; Love, D.C.; Price, L.B.; Huynh, B.-T.; Collard, J.-M.; Lay, K.S.; Borand, L.; Ndir, A.; Walsh, T.; et al. Combating Global Antibiotic Resistance: Emerging One Health Concerns in Lower- and Middle-Income Countries. Clin. Infect. Dis. 2018, 66, 963–969. [Google Scholar] [CrossRef]
- McInnes, R.S.; Uz-Zaman, H.; Alam, I.T.; Ho, S.F.S.; Moran, R.A.; Clemens, J.D.; Islam, S.; van Schaik, W. Metagenome-Wide Analysis of Rural and Urban Surface Waters and Sediments in Bangladesh Identifies Human Waste as a Driver of Antibiotic Resistance. mSystems 2021, 6, e0013721. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.F.; Snyder, R.E.; Riley, L.W.; Immke, D.W.; Greenfield, B.K. Antimicrobial Drug-Resistant Gram-Negative Saprophytic Bacteria Isolated from Ambient, Near-Shore Sediments of an Urbanized Estuary: Absence of β-Lactamase Drug-Resistance Genes. Antibiotics 2020, 9, 400. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.; Cerceo, E. Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics 2020, 9, 196. [Google Scholar] [CrossRef] [Green Version]
- Silva, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Diversity and genetic lineages of environmental staphylococci: A surface water overview. FEMS Microbiol. Ecol. 2020, 96, fiaa191. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.-C.; Tao, C.-W.; Hsu, B.-M.; Yang, Y.-Y.; Tseng, Y.-C.; Huang, T.-Y.; Huang, S.-W.; Kuo, Y.-J.; Chen, J.-S. Multidrug-resistance in methicillin-resistant Staphylococcus aureus (MRSA) isolated from a subtropical river contaminated by nearby livestock industries. Ecotoxicol. Environ. Saf. 2020, 200, 110724. [Google Scholar] [CrossRef]
- Ramessar, K.; Olaniran, A.O. Antibiogram and molecular characterization of methicillin-resistant Staphylococcus aureus recovered from treated wastewater effluent and receiving surface water in Durban, South Africa. World J. Microbiol. Biotechnol. 2019, 35, 142. [Google Scholar] [CrossRef]
- Hammer-Dedet, F.; Jumas-Bilak, E.; Licznar-Fajardo, P. The Hydric Environment: A Hub for Clinically Relevant Carbapenemase Encoding Genes. Antibiotics 2020, 9, 699. [Google Scholar] [CrossRef]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.-H. Antibiotic resistance in major rivers in the world: A systematic review on occurrence, emergence, and management strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Andraos, C.; Najem, W. Multi-model Approach for Reducing Uncertainties in Rainfall-Runoff Models. In Advances in Hydroinformatics; Springer: Singapore, 2020; pp. 545–557. [Google Scholar] [CrossRef]
- Baalbaki, R.; Nassar, J.; Salloum, S.; Shihadeh, A.L.; Lakkis, I.; Saliba, N.A. Comparison of atmospheric polycyclic aromatic hydrocarbon levels in three urban areas in Lebanon. Atmospheric Environ. 2018, 179, 260–267. [Google Scholar] [CrossRef]
- Shaban, A. Water Resources of Lebanon; Springer Science Publisher: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Buelow, E.; Bayjanov, J.R.; Majoor, E.; Willems, R.; Bonten, M.J.M.; Schmitt, H.; van Schaik, W. Limited influence of hospital wastewater on the microbiome and resistome of wastewater in a community sewerage system. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).