Abstract
The drug-resistance problem is widely spread and becoming more common in community-acquired and nosocomial strains of bacteria. Therefore, finding new antimicrobial agents remains an important drug target. From this perspective, new derivatives of benzothiazole were synthesized and evaluated for their antimicrobial activity and ability to inhibit the DHPS enzyme. The synthesis was carried out by the reaction of benzothiazole N-arylsulphonylhydrazone with N-aryl-2-cyano-3-(dimethylamino)acrylamide, N-aryl-3-(dimethylamino)prop-2-en-1-one, arylaldehydes or diazonium salt of arylamine derivatives, which led to the formation of N-arylsulfonylpyridones 6a–d (yield 60–70%) and 12a–c (yield 50–60%), N-(2-(benzo[d]thiazole-2-yl)-3-arylacryloyl-4-methylsulfonohydrazide 14a–c (yield 60–65%), 4-(benzo[d]thiazole-2-yl)-5-aryl-1H-pyrazol-3(2H)-one 16a–c (yield 65–75%), and N′-(2-(benzo[d]thiazol-2-yl)-2-(2-arylhydrazono)acetyl)-4-arylsulfonohydrazide 19a–e (yield 85–70%). The antimicrobial evaluations resulted into a variety of microbial activities against the tested strains. Most compounds showed antimicrobial activity against S. aureus with an MIC range of 0.025 to 2.609 mM. The most active compound, 16c, exhibited superior activity against the S. aureus strain with an of MIC 0.025 mM among all tested compounds, outperforming both standard drugs ampicillin and sulfadiazine. The physicochemical–pharmacokinetic properties of the synthesized compounds were studied, and it was discovered that some compounds do not violate rule of five and have good bioavailability and drug-likeness scores. The five antimicrobial potent compounds with good physicochemical–pharmacokinetic properties were then examined for their inhibition of DHPS enzyme. According to the finding, three compounds, 16a–c, had IC50 values comparable to the standard drug and revealed that compound 16b was the most active compound with an IC50 value of 7.85 μg/mL, which is comparable to that of sulfadiazine (standard drug) with an IC50 value of 7.13 μg/mL. A docking study was performed to better understand the interaction of potent compounds with the binding sites of the DHPS enzyme, which revealed that compounds 16a–c are linked by two arene-H interactions with Lys220 within the PABA pocket.
1. Introduction
Recently, designing and developing new therapeutic agents to treat microbial infections has emerged as one of the most important goals in the field of medicinal chemistry [1]. This is due to the fact that microbial infections kill over one million people each year. As micro-organisms develop resistance to antimicrobial drugs, the number of deaths rises, affecting people’s health worldwide [2]. Several drugs with heterocyclic rings, such as benzothiazole, have been discovered to have a wide range of pharmaceutical applications and biological activities [3,4,5], due to the extended π-delocalized systems which are capable of binding to DNA molecules via π–π interactions, including anti-inflammatory [6,7], antitumor [8,9,10,11,12], and antiviral [13,14]. In recent years, a considerable number of patents and scientific studies have demonstrated that benzothiazole scaffolds have remarkable antibacterial properties [15,16,17]. For example, pyridine substituted benzothiazoles have been shown to be particularly effective against a variety of bacterial species, including C. pneumonia, E. faecalis, and S. aureus [18]. In addition, benzamide-linked benzothiazole conjugates linked via an ether bond were found to have significant antibacterial activity and low MIC values [18]. Compounds containing an N-sulfonamide 2-pyridone derivative substituted with a benzothiazole moiety have shown considerable antibacterial activity against a wide range of bacterial species [19,20]. Furthermore, sulfonamide-containing compounds demonstrated a wide range of biological activities [21,22], including antiviral [22,23,24,25], anticancer [26], and anti-inflammatory activity [27]. Sulfonamide drugs were discovered to have broad antibacterial activity [19] against bacterial infections by inhibiting the dihydropteroate synthase (DHPS) enzyme [20] through competition with 4-aminobenzoic acid (PABA) [28]. Unfortunately, sulfonamide resistance has spread and is becoming more common in community-acquired and nosocomial strains of bacteria, including S. aureus, S. mutans, and P. aeruginosa. This resistance can occur by developing point mutations in the folP gene of an organism, which encodes a structural change in DHPS that results in an enzyme with a lower affinity for sulfonamide [29]. As a result, DHPS continues to be a significant drug target, motivating us to develop and synthesize new enzyme inhibitors. In pursuit of that goal, we synthesized new compounds containing benzothiazole and sulfonamide moiety and investigated their antimicrobial activities, DHPS enzyme inhibition, physicochemical properties, and protein interaction using molecular docking.
2. Results and Discussion
2.1. Chemistry
Derivatives of N-arylsulfonylpyridones substituted with benzothiazole moiety 6a–d were synthesized starting from the reaction of N-arylsulphonylhydrazones 3a–c, which prepared from the reaction of benzothiazolehydrazide with arylsulfonyl chloride [30], with N-aryl-2-cyano-3-(dimethylamino)acrylamide 4a–c [31] in a dry dioxane containing potassium hydroxide, Scheme 1. The reaction may have proceeded via Michael addition and elimination of NH(CH3)2, intermediate 5, which followed by the intramolecular cyclization caused by the addition of NH proton to the cyano group to furnish with the substituted N-arylsulfonylpyridones 6a–d. On the basis of elemental analysis and spectral data (IR, 1H, and 13C NMR), the structure of compounds 6a–d were determined. According to the IR spectral analysis of compounds 6a–d, the appearance of a broad absorption band at a range of 3426–3437 cm−1 confirmed the presence of NH group. In addition, the IR spectra showed a sharp band at a range of 1624–1629 cm−1 which corresponding to C=O group. As an illustration, the 1H NMR of compound 6a displayed two singlet signals that were assigned to the protons of the NH group and the CH group of pyidone ring at δ 8.94 and 10.21 ppm, respectively. Additionally, the 1H NMR of compound 6a showed two doublet of doublet but appear triplet signals at δ 7.08 and 7.26 as well as two doublets at δ 7.86 and 7.97 ppm. These four characteristic signals correspond to the four protons of the benzothiazole ring. The two C=O carbons at δ 164.5 and 166.5 ppm were detected in 6b 13C NMR spectrum.
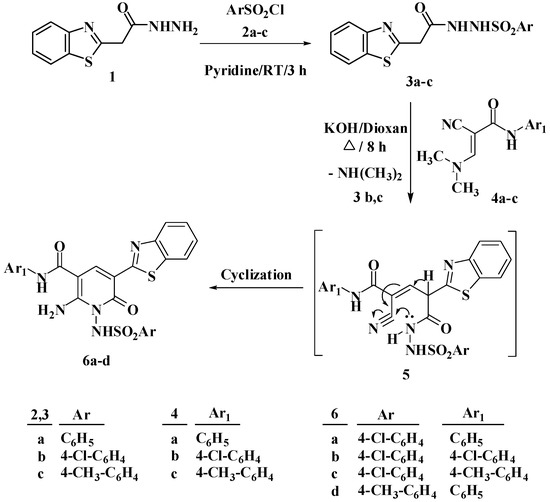
Scheme 1.
Reaction of N-arylsulphonylhydrazones 3a–c with N-aryl-2-cyano-3-(dimethylamino)acrylamide 4a–c in basic medium.
We attempted to synthesize derivatives of benzothiazole substituted with N-arylsulfonylpyridones through the reaction of N-arylsulphonylhydrazones 3a–c with N-aryl-3-(dimethylamino)prop-2-en-1-one 9a–c [32]. The reaction produced the 2-pyridone derivatives 12a–c through the removal of the NH-arylsulphonyl group, intermediate 11 and 11′, Scheme 2. The produced compounds 12a–c have a tautomerism structure 12′a–c as illustrated in Scheme 2. Both of spectral data (IR, 1H NMR and 13C NMR) and elemental analysis confirmed the structure of compounds 12a–c, Scheme 2. The IR spectrum of compounds 12a–c confirmed the presence of NH at a range of 3427–3433 cm−1 and C=O groups at a range of 1626–1637 cm−1. The 1H NMR revealed the absence of the signals corresponding to the arylsulfonyl group in all derivatives 12a–c. The 1H NMR of compound 12a, as an example, showed two doublet signals, which assigned for two pyridone-H at δ 6.81 and 8.41 ppm, respectively.
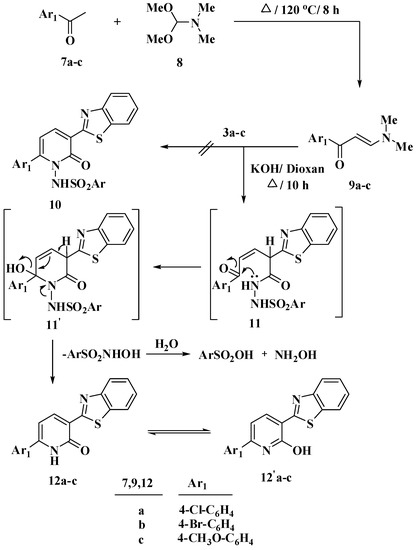
Scheme 2.
Reaction of N-arylsulphonylhydrazones 3a–c with N-aryl-3-(dimethylamino)prop-2-en-1-one 9a–c in basic medium.
Furthermore, the expected compounds N-(2-(benzo[d]thiazole-2-yl)-3-arylacryloyl-4-methylsulfonohydrazide 14a–c were produced when the N-tosylhydrazone derivative 3c was reacted with 4-substituted benzaldehydes 13a–c in ethanol containing a catalytic amount of piperidine at room temperature. Performing the same reaction under reflux resulted in the formation of pyrazolone derivatives 16a–c via cyclization by addition of NH group to the benzylidine group, intermediate 15, and removal of the tosyl group after hydrolysis, Scheme 3. When the 1H NMR spectra of 14a and 16b were compared, a singlet signal at δ 7.61 ppm was observed due to the presence of a proton of CH benzylidine in compound 14a, whereas this signal was absent in the 1H NMR spectrum of compound 16b. Furthermore, the 1H NMR spectra of compound 14a showed two doublet signals at δ 7.21 and 7.68 ppm, both of which represents two protons that corresponded to tosyl protons, whereas these signals were absent in the 1H NMR spectra of compound 16b. This observation indicates that the tosyl group has been removed from 14a–c after reflux. The produced compounds 16a–c have a tautomerism structure 16′a–c, as illustrated in Scheme 3. The structure of compound 16b was finally confirmed using LC-MS. The LC-MS of compound 16b showed a molecular ion peak M+1 at m/z = 328 and M+3 at m/z = 330 due to the presence of Cl atom, in the positive mode, as well as M-H at m/z = 326, in the negative mode. The IR spectra of compounds 14a–c showed the presence of NH and C=O groups at a range of 3434–344 cm−1 and 1629–1654 cm−1, respectively. 13C NMR spectrum of 14a confirmed the presence of the C=O carbons at δ 166.0 ppm.
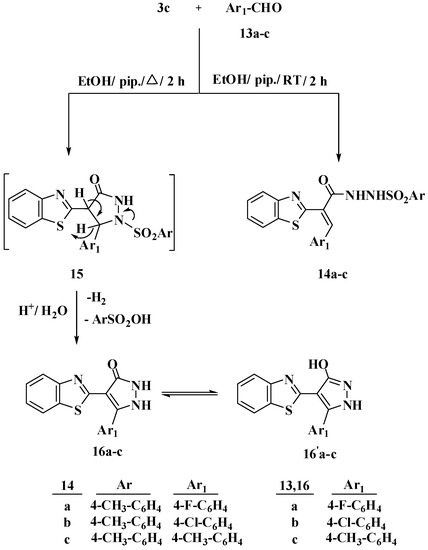
Scheme 3.
Reaction of N-tosylhydrazone derivative 3c with 4-substituted benzaldehydes 13a–c using a catalytic amount of piperidine at room temperature and under reflux.
Azodyes were synthesized to further study the reactivity of the methylene group by coupling N-arylsulphonylhydrazones 3a,b with the diazonium salt of arylamine derivatives 17a–c, Scheme 4. In ethanolic sodium acetate, a coupling reaction occurred between active methylene, which acts as a nucleophilic center, and the diazonium salt, which acts as an electrophilic center, yielding compounds 19a–e. To confirm the structure of 19a–e, the compound 19a was used as an example. The 1H NMR spectrum of this compound revealed two doublet peaks at δ 7.81 and 7.89 ppm, both of which represents two protons, confirming the presence of four aromatic amine’s protons. Furthermore, three broad signals appeared at δ 10.00, 10.50 and 14.86 ppm, confirming the presence of three NH group protons. The presence of C=O carbon was confirmed by the appearance of a signal at δ 163.6 ppm in the 13C NMR spectrum of 19a.
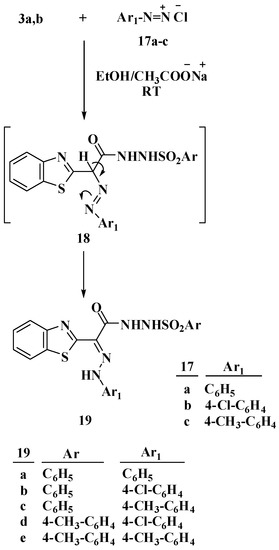
Scheme 4.
Reaction of N-arylsulphonylhydrazones 3a,c with the diazonium salt of arylamine derivatives 17a–c.
2.2. Antimicrobial Evaluation
The new synthetic compounds, 6a–c, 12a–c, 14a–c, 16a–c and 19a–e, were tested in vitro to evaluate antibacterial activities against Gram-negative and Gram-positive bacteria, such as E. coli (ATCC: 3008), K. pneumonia (ATCC: 4415) and P. aeruginosa (ATCC: 27853), as an example for Gram-negative bacteria, as well as S. aureus (ATCC: 6538) and S. mutans (ATCC: 25175) for Gram-positive bacteria. The antifungal activities were evaluated also for the novel compounds in vitro against C. albicans (ATCC: 10231). The antibacterial and antifungal activities were determined by the agar-diffusion method using gentamicin, ampicillin and sulfadiazine, example of sulfa drug, as standard drugs against Gram-negative and Gram-positive, while nystatin was used as standard drug against the fungal strain. Table 1 summarizes the results for all tested compounds as the average diameter of inhibition zones of microbial growth around the discs in mm ± SD. The minimum inhibitory concentration (MIC) for the most active compounds was also determined using a twofold serial dilution method, and the results are summarized in Table 2.

Table 1.
Antibacterial inhibition zone in mm ± standard deviation of synthesized compounds.

Table 2.
The minimum inhibitory concentration (MIC, mM) of the most active compounds.
Except for two compounds, 19a and 19e, all of the tested compounds showed antibacterial activity against the S. aureus strain with an MIC value lower than sulfadiazine. Surprisingly, none of the compounds tested were active against E. coli strain. It was also observed that only two compounds, 14b and 16a, exhibited antibacterial activities against four different bacterial strains, while six compounds showed activities against the fungal strain C. albicans strain. Compound 16c demonstrated superior activity against the S. aureus strain with the highest inhibition zone, 40.3 ± 0.6 mm, and the lowest MIC, 0.025 mM, among all tested compounds, outperforming both standard drugs ampicillin and sulfadiazine, which had inhibition zones of 22.0 ± 0.1 and 21.5 ± 0.6 mm and MIC values of 0.179 and 1.998 mM, respectively. Furthermore, the same compound, 16c, revealed the highest inhibition zone, 31.3 ± 0.6 mm, with low MIC value of 0.203 mM against the S. mutans strain among all tested compounds, which is similar to the value of the inhibition zone and MIC of ampicillin and higher than sulfadiazine. Among all tested compounds, the same compound, 16c, had the highest inhibition zone, 24.3 ± 0.6 mm, with a low MIC value of 0.813 mM against the K. pneumonia strain, which is similar to the value of the inhibition zone but with a higher MIC of ampicillin. Not only that, two additional compounds, 14b, and 14c, demonstrated inhibition zones against the S. aureus strain greater than ampicillin with lower MIC values, while two additional compounds, 16a and 16b, showed an inhibition zone lower than ampicillin against the S. mutans strain but with a close MIC value. Only one compound, 12a, exhibited an inhibition zone of 19.6 ± 0.6 mm, similar to sulfadiazine against the S. aureus strain but with a lower MIC value of 0.369 mM.
The structure–activity relationships revealed that compounds containing benzothiazole substituted with pyrazolone ring, 16a–c, have the highest antimicrobial activities followed by compounds containing benzylidine moiety of benzothiazole sulfonylhydrazide, 14a–c. Replacement of pyazolone moiety with pyridone or arylazo moieties led to less potent compounds, 6a–c, 12a–c and 19a–e.
Among the compounds with a pyrazolone ring, the presence of both 4-chloro and 4-methyphenyl groups bonded to the pyrazolone ring had a positive effect on the antimicrobial activities over the presence of 4-florophenyl group. The activity order can be presented as follows: 16c > 16b > 16a. On the other hand, the presence of both 4-chloro and 4-methylphenyl groups increased the activities of benzylidine moiety of benzothiazole sulfonylhydrazide compounds over the 4-florophenyl group. Therefore, compound activity is determined not only by the type of rings or groups bonded to the benzothiazole ring, but also by the type of substituted phenyl group.
2.3. Drug Likeness, and Physicochemical–Pharmacokinetic/ADMET Properties
To investigate the potential of the synthesized compounds as drug candidates, the computed drug likeness, violation of various rules, and ADMET properties were calculated using Molsoft software and the SwissADME program. Poor oral bioavailability of a molecule is more likely in drug discovery when there are more than five hydrogen bond donors, ten hydrogen bond acceptors, a molecular weight greater than 500 g/mol, and a calculated Log P greater than 5.
According to the results in Table 3, some compounds, such as 12a–c and 16a–c, have no violations, whereas others have one or more violations. For the number of hydrogen bond donors, number of hydrogen bond acceptors, and Log P, none of the compounds exceeded the normal range. The results also revealed that all compounds had a drug-likeness score ranging from -0.45 to 0.71. On the other hand, the molecule weight and topological polar surface area (TPSA) of compounds 6a–d have exceeded the normal range, with Mwt exceeding 500 g/mol and TPSA exceeding 140 Å2. Furthermore, compounds 19a–e have more than 140 Å2 of TPSA.

Table 3.
Predicted drug likeness, number of violation of various rules and bioavailability of the synthesized compounds.
The structure–activity relationships revealed that compounds having benzothiazole substituted with the pyrazolone ring, 16a–c, and the pyridone ring, 12a–c, are the most promising drug candidates since they have no violations, a low TPSA and Log Po, and a good drug-likeness score. Furthermore, compounds having the benzylidine moiety of benzothiazole sulfonylhydrazide, 14a–c, could be designated as good drug candidates because they have one violation with a low TPSA, Log Po and a good drug-likeness score.
Moreover, both of blood–brain barrier (BBB) permeant, gastrointestinal (GI) absorption, and bioavailability of the synthesized compounds have also been studied using SwissADME program, Table 4. As shown in Table 3, all of the synthesized compounds have no blood–brain barrier, indicating that they are not BBB permeant. On the other hand, some of the tested compounds, such as 6a–d, 14a–c, and 19a–e, have low GI absorption, indicating a good absorbance in the human intestine, whereas others, such as 12a–c and 14a–c, have a high GI absorption. Furthermore, all of the tested compounds have a bioavailability score of 0.55, except 6b, indicating that they have good pharmacokinetic properties.

Table 4.
Predicted ADMET properties of the tested compounds.
2.4. Enzymatic Assay
Dihydropteroate synthase (DHPS) is an enzyme contributed in the Bacillus anthracis folate synthesis pathway via catalyzing PABA to convert into dihydropteroate [33]. Sulfonamides, a class of synthetic molecules that act as competitive inhibitors, are the most commonly used DHPS inhibitors [33]. Other DHPS inhibitors capable of inhibiting DHPS’s pterin binding site can also be utilized [34].
Based on the antimicrobial evaluation and predicted physicochemical properties of the tested compounds, we chose the most potent compounds, 12a, 14b, 14c, 16a, 16b, and 16c, to be tested for their ability to inhibit DHPS enzyme. Table 5 shows the inhibitory percent against the DHPS enzyme, and Figure 1 shows the IC50 of the tested compounds. Sulfadiazine was used as a positive control in this study. The data showed that compound 16b with 4-chlorophenyl group at C5 position of pyrazolone ring has the lowest IC50 value of 7.85 μg/mL, while compound 12a with 4-chlorophenyl group at C6 position of 3-pyridone ring has the highest IC50 value of 28.31 μg/mL. Surprisingly, the IC50 value of the most potent compound, 16b, is comparable to that of sulfadiazine with IC50 value 7.13 μg/mL. Compounds with benzylidine moiety, 14b and 14c, showed IC50 values of 16.76 μg/mL and 26.14 μg/mL, respectively. Additionally, compounds with pyrazolone ring, 16a–c, showed IC50 values of 11.17, 7.85 and 11.03 μg/mL, respectively. According to this finding, the presence of a pyrzolone ring bonded to benzothiazole inhibited the DHPS enzyme more than the presence of a pyridone ring or benzylidine moiety bonded to benzothiazole. Furthermore, the reactivity depended on the type of substituent in the para position of the benzene ring. The presence of chlorine atom at the para position of the benzene ring increased the potency of the compounds more than the methyl group.

Table 5.
Inhibition % values of compounds 12a, 14b, 14c, 16a, 16b, 16c and sulfadiazine against DHPS enzyme.
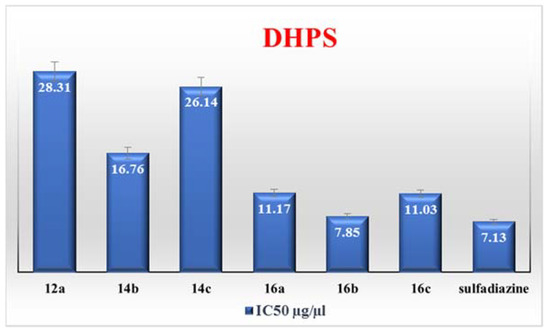
Figure 1.
IC50 values for the synthesized compounds 12a, 14b, 14c, 16a, 16b and 16c against dihydropteroate synthase enzyme.
Based on the enzyme assay study, we can conclude that three compounds, 16a–c, are potentially effective DHPS inhibitors.
2.5. Docking Study
In vitro evaluations of the most potent synthesized compounds revealed that benzothiazole compounds bearing pyrazolone ring, 16a–c, have the potential to inhibit the DHPS enzyme. Therefore, docking studies were conducted using a crystal structure of DHPS (PDB ID: 3TYE) obtained from the Protein Data Bank server to better understand the interaction of these compounds with the binding sites of DHPS enzyme. The DHPS enzyme has two binding pockets, one binds dihydropterin pyrophosphate (DHPP) and the other binds p-aminobenzoic acid (PABA). The key amino acid residues that recognized the pterin substrate via hydrogen bonding are Asn120, Asp184, Lys220, and a water molecule, while the amino acid residues that recognized the PABA binding site via arene-H interactions are Lys220 and Phe189 and Ser221 through amino group hydrogen bonds [35].
Docking cocrystallized ligand (XTZ) inside the active site after extraction from the respective receptor was used to validate the docking study, as shown in Figure 2. The cocrystallized ligand XTZ docking resulted in a root mean square deviation value of −6.9784 kcal/mol, Table 6. The docking results revealed that XTZ formed four H-bond acceptors with Ser221, Asp184, and Lys220, three H-bond donors with Asp184 and Asn120, and one arene-H interaction with Lys220. On the other hand, Figure 2 depicts the various types of interactions between the ligand XTZ and DHPS pockets.
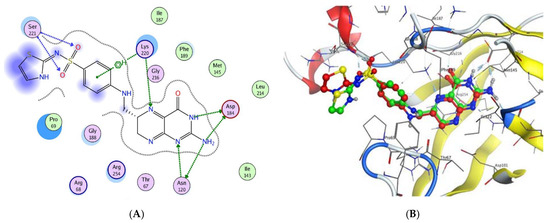
Figure 2.
Docking poses of TXZ ligand inside DHPS. (A) 2D interaction of TXZ ligand with DHPS pockets. (B) 3D Docking of TXZ ligand for validation.

Table 6.
Molecular docking free binding energy estimates to DHPS.
The top ranked poses of the most active compounds 16a–c within the active site of DHPS were summarized in Figure 3A–F. Surprisingly, the docking analysis of the evaluated compounds revealed that all three compounds fit inside the PABA pocket. Compound 16a exhibited three arene-H interactions, two between the pyrazolone and the benzene ring of benzothiazole with Lys220 and one between the thiazole ring with Arg234 in addition to one H-bond donor with bond length 2.22 Å between Gly188 and NH group in the pyrazolone ring, as in Figure 3A,B. Figure 3C–F showed similar interactions in compounds 16b and 16c in the active site of DHPS, as well as one extra arene-H interaction between the NH group in the pyrazolone ring and Phe189. Based on these findings, all three compounds are linked not only by one but two arene-H interactions with Lys220 within the PABA pocket. This suggests that these new compounds could be used as alternatives to sulfonamide drugs to combat bacterial resistance.

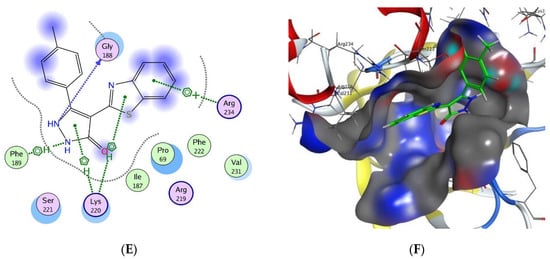
Figure 3.
Docking poses of compounds 16a-c inside DHPS (A) 2D interaction of 16a with DHPS pockets (B) 3D interaction of 16a with DHPS pockets (C) 2D interaction of 16b with DHPS pockets (D) 3D interaction of 16b with DHPS pockets (E) 2D interaction of 16c with DHPS pockets (F) 3D interaction of 16c with DHPS pockets.
3. Experimental
3.1. Chemistry
Melting Points were measured on an SMP3 melting point apparatus. The 1H and 13C NMR Spectra were recorded on a Bruker advance (III)-400 Spectrometer (400 and 100 MHz, respectively) in DMSO-d6 using Si(CH3)4 as an internal standard (See Supplementary Materials) at the Ain Shams University, Cairo, Egypt. Chemical shifts are expressed in δ values (ppm). All coupling constants (J) values are given in Hertz. The abbreviations used are as follows: s, singlet; d, doublet and m, multiplet. Reaction courses and product mixtures were routinely monitored by thin layer chromatography (TLC) on silica gel pre-coated F254 plates Merck, and using UV lamp.
- General procedure for the synthesis of 2-amino-N-aryl-5-(benzo[d]thiazole-2-yl)-1-(4-chlorophenylsulfonamido)-6-oxo-1,6-dihydropyridine-3-carboxamide (6a–d):
N-(2-(benzo[d]thiazole-2-yl)acetyl)arylsulfonohydrazide (3b,c) (10 mmol) was added to a stirred solution of the N-aryl-2-cyano-3-(dimethyl amino) acrylamide (5 mmol) in a dry dioxane (30 mL) containing potassium hydroxide (10 mmol). The reaction mixture was refluxed for 8 h and the precipitate was collected by filtration while hot, washed with hot ethanol, dried and crystallized from ethanol.
- 2-Amino-5-(benzo[d]thiazole-2-yl)-1-(4-chlorophenylsulfonamido)-6-oxo-N-phenyl-1,6-dihydropyridine-3–carboxamide (6a)
Brown solid (yield 65%), m.p. over 350 °C; IR (KBr, cm−1): υ 3428 (NH-NH2), 2925 (Ar-CH), 1624 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.08 (t, J = 7.6 Hz, 1H, benzothiazole-H), 7.26 (t, J = 8.0 Hz, 1H, benzothiazole-H), 7.32–7.44 (m, 5H, Ar-H), 7.69 (d, J = 8.8 Hz, 2H, Ar-H), 7.78 (d, J = 9.2 Hz, 2H, Ar-H), 7.86 (d, J = 8.0 Hz, 1H, benzothiazole H), 7.97 (d, J = 8.4 Hz, 1H, benzothiazole H), 8.94 (s, 1H, CH-pyridine), 10.21 (br, 1H, NH); Anal.Calcd.for C25H18ClN5O4S2 (552.02): calc. C% 54.39, H% 3.29, N% 12.69, found C% 54.41, H% 3.27, N% 12.71.
- 2-Amino-5-(benzo[d]thiazole-2-yl)-N-(4-chlorophenyl)-1-(4-chlorophenylsulfonamido)-6-oxo-1,6-dihydropyridine-3–carboxamide (6b)
Buff solid (yield 70%), m.p. 334–336 °C; IR (KBr, cm−1): υ 3426 (NH-NH2), 2926 (Ar-CH), 1624 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.26 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.39–7.44 (m, 5H, 4Ar-H, 1benzothiazole-H), 7.72–7.78 (m, 4H, Ar-H), 7.87 (d, J = 10.0 Hz, 1H, benzothiazole-H), 7.97 (d, J = 8.4 Hz, 1H, benzothiazole-H), 8.94 (s, H, CH-pyridine), 10.31 (br, 1H, NH); 13C NMR (100 MHz, DMSO- d6): δ 93.4, 105.6, 121.0, 122.0, 122.9, 123.7, 126.1, 127.4, 127.9, 128.5, 128.8, 134.2, 135.1, 135.5, 138.5, 147.2, 152.2, 157.6, 160.1 (Ar-C), 164.5, 166.5 (2C=O); Anal.Calcd.for C25H17Cl2N5O4S2 (586.47): calc. C% 51.20, H% 2.92, N% 11.94, found C% 51.28, H% 2.85, N% 11.90.
- 2-Amino-5-(benzo[d]thiazole-2-yl)-1-(4-chlorophenylsulfonamido)-6-oxo-N-(p-tolyl)-1,6-dihydropyridine-3–carboxamide (6c)
Buff solid (yield 75%), m.p. 302–305 °C; IR (KBr, cm−1): υ 3427 (NH-NH2), 2925 (Ar-CH), 1628 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.29 (s, 3H, CH3), 7.15 (d, J =9.6 Hz, 2H, Ar-H), 7.25 (t, J = 7.8 Hz, 1H, benzothiazole-H), 7.38–7.44 (m, 3H, 2Ar-H, 1benzothiazole-H), 7.56 (d, J = 11.2 Hz, 2H, Ar-H), 7.77 (d, J = 11.6 Hz, 2H, Ar-H), 7.86 (d, J = 10.4 Hz, 1H, benzothiazole-H), 7.97 (d, J =12.8 Hz, 1H, benzothiazole-H), 8.92 (s, 1H, CH-pyridine), 10.13 (br, 1H, NH); 13C NMR (100 MHz, DMSO- d6): δ 21.9 (CH3), 114.5, 121.0, 121.3, 122.0, 123,4, 125.9, 126.8, 127.9, 128.5, 129.3, 129.7, 132.5, 134.0, 135.4, 135.5, 137.2, 147.8, 152.5, 157.6 (Ar-C), 164.7, 166.5 (2C=O); Anal.Calcd.for C26H20ClN5O4S2 (566.05): calc. C% 55.17, H% 3.56, N% 12.37, found C% 55.19, H% 3.58, N% 12.39.
- 2-Amino-5-(benzo[d]thiazole-2-yl)-1-(4-methylphenylsulfonamido)-6-oxo-N-phenyl-1,6-dihydropyridine-3–carboxamide (6d)
Brown solid (yield 60%), m.p. over 350 °C; IR (KBr, cm−1): υ 3414 (NH-NH2), 2921 (Ar-CH), 1618 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.36 (s, 3H, CH3), 7.13 (d, J = 7.6 Hz, 2H, Ar-H), 7.27 (t, J = 8.4 Hz, 1H, benzothiazole-H), 7.32–7.36 (m, 3H, Ar-H), 7.42 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.67–7.71 (m, 4H, Ar-H), 7.87 (d, J = 10.4 Hz, 1H, benzothiazole-H), 7.97 (d, J = 8.8 Hz, 1H, benzothiazole-H), 8.94 (s, 1H, CH-pyridine), 10.20 (br, 1H, NH); 13C NMR (100 MHz, DMSO- d6): δ 21.4 (CH3), 121.0, 121.3, 121.9, 123.6, 126.0, 126.6, 127.6, 128.5, 128.6, 128.9, 135.4, 135.6, 139.1, 139.9, 145.9, 152.5, 157.8, 160.3 (Ar-C), 164.5, 166.5 (2C=O); Anal.Calcd.for C26H21N5O4S2 (531.61): calc. C% 58.74, H% 3.98, N% 13.17, found C% 58.77, H% 3.99, N% 13.19.
- General procedure for the synthesis of 3-(benzo[d]thiazol-2-yl)-6-arylpyridin-2(1H)-one (12a–c):
To a stirred solution of the N-aryl-3-(dimethylamino)prop-2-en-1-one (9a–c) (5 mmol) in a dry dioxane (30 mL) containing potassium hydroxide (10 mmol), N-(2-(benzo[d]thiazole-2-yl)arylsulfonohydrazide (3a–c) (10 mmol) was added and the reaction mixture was refluxed for 10 h. The precipitate was collected by filtration while hot, washed with hot ethanol, dried and crystallized from ethanol.
- 3-(Benzo[d]thiazol-2-yl)-6-(4-chlorophenyl)pyridin-2(1H)-one (12a)
Yellow solid (yield 60%), m.p. over 350 °C; IR (KBr, cm−1): υ 3427 (NH), 2924 (Ar-CH), 1637 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 6.81 (d, J = 8.0, 1H, CH-pyridine), 7.26 (t, J = 9.4 Hz, 1H, benzothiazole-H), 7.40 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.47 (d, J = 10.0 Hz, 2H, Ar-H), 7.85 (d, J = 8.0 Hz, 1H, benzothiazole-H), 7.97 (d, J = 5.6 Hz, 1H, benzothiazole-H), 8.04 (d, J =6.4 Hz, 2H, Ar-H), 8.41 (d, J = 8.4 Hz, 1H, CH-pyridine); Anal.Calcd.for C18H11ClN2OS (338.81): calc. C% 63.81, H% 3.27, N% 8.27, found C% 63.83, H% 3.26, N% 8.28
- 3-(Benzo[d]thiazol-2-yl)-6-(4-bromophenyl)pyridin-2(1H)-one (12b)
Yellow solid (yield 50%), m.p. 345–346 °C; IR (KBr, cm−1): υ 3429 (NH), 2926 (Ar-CH), 1631 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 6.78 (d, J = 10.4 Hz, 1H, CH-pyridine), 7.39–7.46 (m, 2H, benzothiazole-H), 7.60 (d, J = 8.8 Hz, 2H, Ar-H), 7.82 (d, J = 5.6 Hz, 1H, benzothiazole-H), 7.88 (d, J = 9.6 Hz, 1H, benzothiazole-H), 8.10 (d, J = 6.4 Hz, 2H, Ar-H), 8.36 (d, J = 7.2 Hz, 1H, CH-pyridine); Anal.Calcd.for C18H11BrN2OS (383.26): calc. C% 56.41, H% 2.89, N% 7.31, found C% 56.42, H% 2.88, N% 7.34.
- 3-(Benzo[d]thiazol-2-yl)-6-(4-methoxyphenyl)pyridin-2(1H)-one (12c)
Yellow solid (yield 60%), m.p. over 350 °C; IR (KBr, cm−1): υ 3428 (NH), 2926 (Ar-CH), 1626 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.50 (s, 3H, OCH3), 7.14–7.98 (m, 10H, 4Ar-H, 4benzothiazole-H, 2CH-pyridine); Anal.Calcd.for C19H14N2O2S (334.39): calc. C% 68.24, H% 4.22, N% 8.38, found C% 68.27, H% 4.23, N% 8.39.
- General procedure for the synthesis of N-(2-(benzo[d]thiazole-2-yl)-3-arylacryloyl-4-methylbenzenesulfonohydrazide (14a–c):
To a stirred solution of the para derivatives of benzaldehyde (13a–c) (10 mmol) in ethanol (30 mL) containing piperidine (1 ml), N-(2-(benzo[d]thiazole-2-yl)acetyl) arylsulfonohydrazide (3c) (10 mmol) was added and the reaction mixture was stirred at room temperature for 4 h. The solid precipitate was filtered off and then recrystallized from ethanol.
- N-(2-(Benzo[d]thiazole-2-yl)-3-(4-florophenylacryloyl)-4-methylbenzene-sulfonohydrazide (14a)
White solid (yield 65%), m.p. 201–202 °C; IR (KBr, cm−1): ʋ 3440 (NH), 2925 (Ar-CH), 1629 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.40 (s, 3H, CH3), 7.21–7.26 (m, 2H, Ar-H), 7.36 (d, J = 8.4 Hz, 2H, Ar-H), 7.47 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.56 (t, J = 8.4 Hz, 1H, benzothiazole-H), 7.61 (s, 1H, CH), 7.68–7.72 (m, 2H, Ar-H), 7.78 (d, J = 8.0 Hz, 2H, Ar-H), 8.00 (d, J = 7.6 Hz, 1H, benzothiazole-H), 8.12 (d, J = 7.6 Hz, 1H, benzothiazole-H), 10.20 (br, 1H, NH), 10.71 (br, 1H, NH); 13C NMR (100 MHz, DMSO-d6): δ 21.4 (CH3), 116.1 (d, J = 28 Hz, C-F), 116.3, 122.6, 123.1, 126.2, 127.2, 128.2, 129.7, 130.1 (d, J = 10 Hz, C-F), 130.5, 132.5, 132.6, 134.6, 137.1, 143.7, 153.4 (Ar-C), 165.7 (d, J = 230 Hz, C-F), 166.0 (C=O); Anal.Calcd.for C23H18FN3O3S2 (467.54): calc. C% 50.09, H% 3.88, N% 8.99, found C% 50.11, H% 3.89, N% 8.92.
- N-(2-(Benzo[d]thiazole-2-yl)-3-(4-chlorophenylacryloyl)-4-methylbenzene-sulfonohydrazide (14b)
White solid (yield 60%), m.p. 188–189 °C; IR (KBr, cm−1): ʋ 3444 (NH), 2926 (Ar-CH), 1630 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.37 (s, 3H, CH3), 7.36–7.77 (m, 13H, 8Ar-H, 4benzothiazole-H, 1CH), 10.23 (br, 1H, NH), 10.71 (br, 1H, NH); 13C NMR (100 MHz, DMSO-d6): δ 21.5 (CH3), 1227, 123.2, 126.3, 127.9, 128.1, 128.5, 129.2, 129.7, 131.0, 131.8, 132.8, 134.5, 134.7, 137.1, 143.7, 153.5, 165.6 (Ar-C), 165.8 (C=O); Anal.Calcd.for C23H18ClN3O3S2 (483.99): calc. C, 57.08, H, 3.75, N, 8.68, found C% 57.09, H% 3.76, N% 8.69.
- N-(2-(Benzo[d]thiazole-2-yl)-3-(p-tolyl)acryloyl)-4-methylbenzene-sulfonohydrazide (14c)
White solid (yield 60%), m.p. 195–196.5 °C; IR (KBr, cm−1): ʋ 3439 (NH), 2926 (Ar-CH), 1654 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.37 (s, 3H, CH3), 2.38 (s, 3H, CH3), 7.22 (d, J = 8.8 Hz, 2H, Ar-H), 7.35–7.42 (m, 3H, 2Ar-H, benzothiazole-H), 7.46 (t, J = 8.2 Hz, 1H, benzothiazole-H), 7.53–7.57 (m, 3H, 2Ar-H, 1CH), 7.80 (d, J = 9 Hz, 2H, Ar-H), 8.01 (d, J = 8.0 Hz, 1H, benzothiazole-H), 8.10 (d, J = 8.0 Hz, 1H, benzothiazole-H), 10.15 (br, 1H, NH), 10.70 (br, 1H, NH); Anal.Calcd.for C24H21N3O3S2 (463.57): calc. C% 62.18, H% 4.57, N% 9.06, found C% 62.19, H% 4.56, N% 9.09.
- General procedure for the synthesis of 5-aryl-4-(benzo[d]thiazole-2-yl)-1H-pyrazol-3(2H)-one (16a–c):
To a stirred solution of the para derivatives of benzaldehyde (13a–c) (10 mmol) in ethanol (30 mL) containing piperidine (1 mL), N-(2-(benzo[d]thiazole-2-yl)acetyl)-4-methylsulfonohydrazide (3c) (10 mmol) was added and the reaction mixture was refluxed for 4 h. After cooling, the reaction mixture poured in water and neutralized by HCl. The precipitate formed was collected by filtration, washed with hot ethanol, dried and crystallized from ethanol.
- 4-(Benzo[d]thiazole-2-yl)-5-(4-florophenyl)-1H-pyrazol-3(2H)-one (16a)
Buff solid (yield 65%), m.p. 238–240 °C; IR (KBr, cm−1): υ 3437 (NH), 2929 (Ar-CH), 1631 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.31–7.37 (m, 3H, 2Ar-H, 1benzothiazole-H), 7.42 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.79–7.85 (m, 3H, 2Ar-H, 1benzothiazole-H), 8.01 (d, J = 7.6 Hz, 1H, benzothiazole-H), 12.53 (br, 1H, NH); 13C NMR (100 MHz, DMSO-d6): δ 115.6 (d, J = 28 Hz, C-F), 121.7, 122.0, 124.5, 126.4, 126.7, 131.7 (d, J = 8 Hz, C-F), 133.8, 152.4, 160.9 (Ar-C), 161.5 (d, J = 245 Hz, C-F), 164.2 (C=O); Anal.Calcd.for C16H10FN3OS (311.33): calc. C% 61.73, H% 3.24, N% 13.50, found C% 61.75, H% 3.25, N% 13.52.
- 4-(Benzo[d]thiazole-2-yl)-5-(4-chlorophenyl)-1H-pyrazol-3(2H)-one (16b)
Buff solid (yield 75%), m.p. 236–237.5 °C; IR (KBr, cm−1): υ 3439 (NH), 2924 (Ar-CH), 1631 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.32 (t, J = 8.8 Hz, 1H, benzothiazole-H), 7.43 (t, J = 8.4 Hz, 1H, benzothiazole-H), 7.57 (d, J = 6.8 Hz, 2H, Ar-H), 7.80- 7.82 (m, 3H, 2Ar-H, 1benzothiazole-H), 8.02 (d, J = 8.0 Hz, 1H, benzothiazole-H), 10.69 (br, 1H, NH); LC-MS: m/z 328; Anal.Calcd.for C16H10ClN3OS (327.79): calc. C% 58.63, H% 3.07, N% 12.82, found C% 58.65, H% 3.08, N% 12.84.
- 4-(Benzo[d]thiazole-2-yl)-5-(p-tolyl)-1H-pyrazol-3(2H)-one (16c)
Buff solid (yield 70%), m.p. 239–241 °C; IR (KBr, cm−1): ʋ 3432 (NH), 2929 (Ar-CH), 1629 (C=O); 1H NMR (400 MHz, DMSO- d6): δ 2.35 (s, 3H, CH3), 7.05 (t, J = 7.2 Hz, 1H, benzothiazole-H), 7.15 (d, J = 8.0 Hz, 2H, Ar-H), 7.21 (t, J = 7.0 Hz, 1H, benzothiazole-H), 7.46 (d, J = 7.6 Hz, 1H, benzothiazole-H), 7.77 (d, J = 6.4 Hz, 1H, benzothiazole-H), 7.94 (d, J = 7.2 Hz, 2H, Ar-H); 13C NMR (100 MHz, DMSO-d6): δ 21.4 (CH3), 119.6, 119.9, 121.1, 121.6, 125.0, 128.1, 129.1, 133.5, 136.1, 153.4, 163.6 (Ar-C), 165.8 (C=O); Anal.Calcd.for C17H13N3OS (307.37): calc. C% 66.43, H% 4.26, N% 13.67, found C% 66.46, H% 4.27, N% 13.68.
- General procedure for the synthesis of N′-(2-(benzo[d]thiazol-2-yl)-2-(2-arylhydrazono)acetyl)-4-arylsulfonohydrazide (19a–e):
Aniline derivatives (10 mmol) were dissolved in a concentrated hydrochloric acid (2 mL) in an ice bath. Sodium nitrite (10 mmol) was dissolved in cold water (0 °C) and added dropwise to the reaction mixture under vigorous mechanical stirring. The diazonium salts (17a–c) were obtained and used for the next coupling reaction. N-(2-(Benzo[d]thiazole-2-yl)acetyl)arylsulfonohydrazide (3a,c) (5 mmol) was dissolved in ethanol. Freshly prepared diazonium salt was added dropwise to the reaction mixture under vigorous mechanical stirring (0 °C). After addition, the mixture was neutralized with sodium acetate anhydrous to pH 7, and the precipitate was filtered, washed with ethanol and dried in a vacuum. The product was recrystallized from ethanol.
- N′-(2-(Benzo[d]thiazol-2-yl)-2-(2-phenylhydrazono)acetyl)benzenesulfono-hydrazide (19a)
Yellow solid (yield 85%) m.p. 246–248.6 °C; IR (KBr, cm−1): ʋ 3438 (NH), 2925 (Ar-CH), 1633 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.17 (t, J = 7.4 Hz, 1H, benzothiazole-H), 7.44–7.52 (m, 3H, Ar-H), 7.67 (t, J = 7.0 Hz, 1H, benzothiazole-H), 7.81 (d, J = 7.6 Hz, 2H, Ar-H), 7.89 (d, J = 6.4 Hz, 2H, Ar-H), 8.13 (d, J = 8.4 Hz, 1H, benzothiazole-H), 8.28 (d, J = 7.6 Hz, 1H, benzothiazole-H), 10.02 (br, 1H, NH), 10.58 (br, 1H, NH), 14.84 (br, 1H, NH); 13 C NMR (100 MHz, DMSO-d6): δ 116.5, 121.8, 122.3, 123.1, 124.7, 126.6, 127.0, 128.1, 129.3, 129.8, 133.4, 134.2, 139.8, 142.4, 150.6, 158.4 (Ar-C), 163.6 (C=O); Anal.Calcd.For C21H17N5O3S2 (451.52): calc. C% 55.86, H% 3.79, N% 15.51, found C% 55.89, H% 3.78, N% 15.53.
- N′-(2-(Benzo[d]thiazol-2-yl)-2-(2-(4-chlorophenyl)hydrazono)acetyl)benzene-sulfonohydrazide (19b)
Yellow solid (yield 85%,) m.p. 265–267 °C; IR (KBr, cm−1): ʋ 3445 (NH), 2925 (Ar-CH), 1632 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 7.27–7.67 (m, 7H, 5Ar-H, 2benzothiazole-H), 7.86–7.88 (m, 4H, Ar-H), 8.13 (d, J = 8.4 Hz, 1H, benzothiazole-H), 8.28 (d, 1H, J = 7.6 Hz, benzothiazole-H), 10.02 (br, 1H, NH), 10.58 (br, 1H, NH), 14.84 (br, 1H, NH); Anal.Calcd.For C21H16ClN5O3S2 (485.97): calc. C% 51.90, H% 3.32, N% 14.41, found C% 51.93, H% 3.33, N% 14.43.
- N′-(2-(Benzo[d]thiazol-2-yl)-2-(2-(4-methylphenyl)hydrazono)acetyl)benzene-sulfonohydrazide (19c)
Yellow solid (yield 85%,) m.p. 262–263 °C; IR (KBr, cm−1): ʋ 3440 (NH), 2925 (Ar-CH), 1648 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.29 (s, 3H, CH3), 7.26 (d, J = 6.8 Hz, 2H, Ar-H), 7.48 (t, J = 7.6 Hz, 1H, benzothiazole-H), 7.57–7.59 (m, 3H, 2Ar-H, 1benzothiazole-H), 7.65–7.70 (m, 3H, 2Ar-H, 1benzothiazole-H), 7.88 (d, J = 9.6 Hz, 2H, Ar-H), 8.12 (d, J = 8.0 Hz, 1H, benzothiazole-H), 8.25 (d, J = 10.4 Hz, 1H, benzothiazole-H), 9.98 (br, 1H, NH), 10.47 (br, 1H, NH), 14.87 (br, 1H, NH); 13 C NMR (100 MHz, DMSO-d6): δ 21.0 (CH3), 116.5, 121.2, 122.3, 123.0, 126.5, 126.9, 128.1, 129.3, 130.3, 133.4, 134.0, 134.2, 139.9, 140.1, 150.6, 158.5 (Ar-C), 163.7 (C=O); Anal.Calcd.For C22H19N5O3S2 (465.55): calc. C% 56.76, H% 4.11, N% 15.04, found C% 56.78, H% 4.12, N% 15.06.
- N′-(2-(Benzo[d]thiazol-2-yl)-2-(2-(4-chlorophenyl)hydrazono)acetyl)-4-methylbenzenesulfono-hydrazide (19d)
Yellow solid (yield 70%,) m.p. 280–281 °C; IR (KBr, ʋ cm−1): 3445 (NH), 2925 (Ar-CH), 1632 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.36 (s, 3H, CH3), 7.35–7.57 (m, 6H, 4 Ar-H, 2benzothiazole-H), 7.77 (d, J = 7.8 Hz, 2H, Ar-H), 7.84 (d, J = 7.2 Hz, 2H, Ar-H), 8.11 (d, J = 5.6 Hz, 1H, benzothiazole-H), 8.25 (d, J = 8.4 Hz, 1H, benzothiazole-H), 9.85 (br, 1H, NH), 10.52 (br, 1H, NH), 14.81 (br, 1H, NH); Anal.Calcd.For C22H18ClN5O3S2 (499.99): calc. C% 52.85, H% 3.63, N% 14.01, found C% 52.88, H% 3.64, N% 14.0.
- N′-(2-(Benzo[d]thiazol-2-yl)-2-(2-(4-methylphenyl)hydrazono)acetyl)-4-methylbenzenesulfono-hydrazide (19e)
Yellow solid (yield 85%) m.p. 285–286 °C; IR (KBr, cm−1): ʋ 3442 (NH), 2924 (Ar-CH), 1636 (C=O); 1H NMR (400 MHz, DMSO-d6): δ 2.88 (s, 3H, CH3), 2.41 (s, 3H, CH3), 7.25 (d, J = 7.2 Hz, 2H, Ar-H), 7.36 (d, J = 8.0 Hz, 2H, Ar-H), 7.48 (t, J = 7.2 Hz, 1H, benzothiazole-H), 7.57 (t, J = 7.8 Hz, 1H, benzothiazole-H), 7.70 (d, J = 7.6 Hz, 2H, Ar-H), 7.76 (d, J = 8.4 Hz, 2H, Ar-H), 8.12 (d, J = 8.8 Hz, 1H, benzothiazole-H), 8.25 (d, J = 7.6 Hz, 1H, benzothiazole-H), 9.91 (br, 1H, NH), 10.41 (br, 1H, NH), 14.83 (br, 1H, NH); Anal.Calcd.For C23H21N5O3S2 (479.57): calc. C% 57.60, H% 4.41, N% 14.60, found C% 57.62, H% 4.42, N% 14.62.
3.2. Antimicrobial Activity
The antimicrobial activity of synthesized compounds was determined using agar well diffusion method [36]. All the compounds were tested in vitro for their antibacterial activity against S. aureus and S. mutans (Gram-positive bacteria), E. coli, P. aeruginosa and K. pneumonia (Gram-negative bacteria) using nutrient agar medium. Ampicillin and gentamicin were used as standard drugs for Gram-positive and Gram-negative, respectively. DMSO was used as solvent control. The compounds were tested at a concentration of 15 mg/mL against both bacterial and fungal strains. The sterilized media was poured onto the sterilized Petri dishes (20–25 mL, each petri dish) and allowed to solidify at room temperature. Microbial suspension was prepared in sterilized saline equivalent to McFarland 0.5 standard solution (1.5 × 105 CFU mL−1) and its turbidity was adjusted to OD= 0.13 using spectrophotometer at 625 nm. Optimally, within 15 min after adjusting the turbidity of the inoculum suspension, a sterile cotton swab was dipped into the adjusted suspension and was flooded on the dried agar surface, then allowed to dry for 15 min with lid in place. Wells of 6 mm diameter was made in the solidified media with the help of sterile borer. A total of 100 μL of the solution of the tested compound was added to each well with the help of micropipette. The plates were incubated at 37 °C for 24 h in case of antibacterial activity. This experiment was carried out in triplicate and zones of inhibition were measured in mm scale.
3.3. Methodology of MIC
For each strain, three to five isolated colonies were selected from the fresh agar plate and were transferred into a tube containing 3–4 mL of sterile broth medium. The bacterial suspension was mixed well and incubated at 35–37 °C for 2–6 h. The turbidity of the bacterial suspension should be equal to or greater than the turbidity of a McFarland Standard 0.5. After that, 1 mg of the tested compound (antimicrobial agent) was dissolved in 1 mL DMSO and two-fold serial dilution was done using broth medium. A fixed volume of the prepared bacterial inoculum was added to each tube and incubated for at 37 °C 16–20 h. The MIC is defined as the lowest concentration of the antimicrobial agent that inhibits visible growth of the tested isolate as observed with the unaided eye [37].
3.4. Drug Likeness Predictions and Physicochemical–Pharmacokinetic/ADMET Properties
A qualitative concept used in drug design to predict a drug-like property is drug-likeness. Drug-like properties, such as solubility, permeability, transporter effects, and metabolic stability are critical for drug candidates’ success. They have an impact on oral bioavailability, toxicity, metabolism, clearance, and in vitro pharmacology. Five separate filters were used to evaluate the drug-likeness of the synthesized compounds, Lipinski [38], Veber [39], Muegge [40], Ghose [41] and Egan [42] rules in addition to bioavailability, and the drug-likeness scores using the Molsoft software and SwissADME program.
3.5. Enzyme Assay
DHPS catalytic assay DHPS was assayed for the most potent compounds. Assay tubes contained 5 mM MgCl2, 5 m MDTT, 10AM H2PtCH20PP, 1 mm [3H] PABA (finalspact, 2Ci/mmol), and 40 mm Tris/HCI, pH8.3 in a total reaction volume of 100 mL. Various concentrations of inhibitors were added when applicable. Reactions were initiated by the addition of 0.5 mU of enzyme (1U= 1 nmol of product formed/min). After a 30-min incubation at 37 °C the reaction tubes were placed on ice to terminate the reactions and then spotted onto 3×30 cm strips of 3 mm chromatography paper (Whatman Laboratory Products Inc.). The strips were developed in a descending chromatography tank using an elution buffer of 0.1 M KH2PO4, pH 7.0. The origins containing the labeled products of the reaction were cut from the strips, placed in scintillation vials, and counted in a liquid scintillation counter (Packard Tri-Carb; Packard Instrument Co., Inc., Downers Grove, IL, USA) 24 h after the addition of 9.5 mL of counting cocktail (3A70b; Research Products International Corp., Mt.Pros-pect, IL, USA). A total of 100 uL of lysate (100 μg of protein) was combined with 14C-labeled PABA, diphosphoric acid, and mono[(2-amino1,4,7,8-tetrahydro-4-oxo-6-pteridinyl)-methyl]ester in Tris buffer, pH 8.3, containing dithiothreitol and MgCl2 in a 200-μL reaction volume, and the mixture was held at 37 °C for 15 min. One hundred microliters of each reaction volume was spotted onto 3 mm paper (Whatman International, Ltd., Maidstone, England), and ascending chromatography was performed in 0.01 M phosphate buffer. Under these conditions, free substrate (14C-labeled PABA) migrates with the solvent front and radiolabeled product (14C-labeled dihydropteroate) remains at the origin. The chromatogram was dried, the area (1 cm2) representing the origin was placed into scintillation fluid, and radioactivity was measured using an LS 6000IC Liquid Scintillation System (Beckman Instruments, Fullerton, Calif.). Results were expressed as the mean and standard deviation of triplicate samples in picomoles of product formed per milligram of total protein.
3.6. Molecular Docking Study
The Molecular Operating Environment (MOE 2014) was used for the molecular studies. The ligand molecules were drawn by builder molecule and their energy was minimized. All of the minimizations were done until an rmsd gradient of 0.01 kcal/mol was reached with the MMFF94X force field, and the partial charges were calculated automatically. Docking simulations were run using the Protein Data Bank’s crystal structure of the DHPS enzyme in complex with XTZ (PDB ID: 3TYE). Water molecules and ligands that were bound were removed. The MOE protonate 3D application was used to add the missing hydrogens and to correctly assign the ionization states. The MOE-Alpha site finder was used to generate the active site. The obtained alpha spheres were used to create dummy atoms. Ligands were then docked within the active sites using the MOE-Dock. The GBVI/WSA DG free-energy estimates were used to rank the optimized poses, and docking poses were visually examined. The interactions with binding pocket residues were finally investigated.
4. Conclusions
Eighteen new benzothiazole derivatives were synthesized and tested for their antimicrobial activity against six microbial strains. Most compounds showed antimicrobial activity against S. aureus with MIC range in 0.025 to 2.609 mM. The most active compound 16c exhibited superior activity against the S. aureus strain with an MIC value of 0.025 mM among all tested compounds, outperforming both standard drugs, ampicillin and sulfadiazine, as well as having very good activity against S. mutans and K. pneumonia with MIC values of 0.203 and 0.813 mM, respectively. The predicted physicochemical properties, bioavailability and drug-likeness scores indicated that six compounds 12a–c and 16a–c have not violated any rule with TPSA range of 73.99–89.78, and three compounds violate only one rule with TPSA 124.78. The bioavailability score of all compounds was approximately 0.55. The enzyme assay of potent compounds revealed that benzothiazole derivative 16b, having a pyrzolone ring and 4-chlorophenyl moiety, was the most active compound with an IC50 value of 7.85 μg/mL, which is comparable to that of sulfadiazine (standard drug) with IC50 value of 7.13 μg/mL. Finally, the docking study reveals that compounds 16a–c are linked by two arene-H interactions with Lys220 within the PABA pocket.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics11121799/s1, Supplementary Materials: IR, 1H NMR, and 13C NMR spectra of new compounds, 6a–d, 12a–c, 14a–c, 16a–c, and 19a–e.
Author Contributions
Conceptualization, G.H.E. and R.A.A.; methodology, G.H.E., R.A.A. and H.A.E.; validation, G.H.E. and R.A.A.; formal analysis, G.H.E.; investigation, G.H.E. and R.A.A.; resources, G.H.E.; data curation, H.A.E.; writing—original draft preparation, H.A.E.; writing—review and editing, G.H.E. and R.A.A.; visualization, R.A.A.; supervision, G.H.E. and R.A.A.; project administration, G.H.E.; funding acquisition, G.H.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Desai, N.C.; Rajpara, K.M.; Joshi, V.V. Synthesis of pyrazole encompassing 2-pyridone derivatives as antibacterial agents. Bioorg. Med. Chem. Lett. 2013, 23, 2714–2717. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Mitscher, L.A.; Shen, L.L. The 2-pyridone antibacterial agents: Bacterial topoisomerase inhibitors. Med. Res. Rev. 2000, 20, 231–293. [Google Scholar] [CrossRef] [PubMed]
- Azzam, R.A.; Osman, R.R.; Elgemeie, G.H. Efficient synthesis and docking studies of novel benzothiazole-based pyrimidinesulfonamide scaffolds as new antiviral agents and hsp90α inhibitors. ACS Omega 2020, 5, 1640–1655. [Google Scholar] [CrossRef]
- Azzam, R.A.; Elgemeie, G.H.; Osman, R.R. Synthesis of novel pyrido[2,1-b]benzothiazole and N-substituted 2-pyridylbenzothiazole derivatives showing remarkable fluorescence and biological activities. J. Mol. Struct. 2020, 1201, 127194. [Google Scholar] [CrossRef]
- Gill, R.K.; Rawal, R.K.; Bariwal, J. Recent advances in the chemistry and biology of benzothiazoles. Arch. Pharm. 2015, 348, 155–178. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Wyllie, S.; Wells, G.; Stevens, M.F.; Wyatt, P.G. Antitumor quinol PMX464 is a cytocidal anti-trypanosomal inhibitor targeting trypanothione metabolism. J. Biol. Chem. 2011, 286, 8523–8533. [Google Scholar] [CrossRef]
- Lee, Y.R.; Jin, G.H.; Lee, S.M.; Park, J.W.; Ryu, J.H.; Jeon, R.; Park, B.H. Inhibition of TNF-α-mediated inflammatory responses by a benzodioxolylacetylamino-linked benzothiazole analog in human fibroblast-like synoviocytes. Biochem. Biophys. Res. Commun. 2011, 408, 625–629. [Google Scholar] [CrossRef]
- Wang, X.; Sarris, K.; Kage, K.; Zhang, D.; Brown, S.P.; Kolasa, T.; Surowy, C.; El Kouhen, O.F.; Muchmore, S.W.; Brioni, J.D.; et al. Synthesis and evaluation of benzothiazole-based analogues as novel, potent, and selective fatty acid amide hydrolase inhibitors. J. Med. Chem. 2009, 52, 170–180. [Google Scholar] [CrossRef]
- Kamal, A.; Srikanth, Y.V.V.; Naseer Ahmed Khan, M.; Ashraf, M.; Kashi Reddy, M.; Sultana, F.; Kaur, T.; Chashoo, G.; Suri, N.; Sehar, I.; et al. 2-Anilinonicotinyl linked 2-aminobenzothiazoles and [1,2,4]triazolo[1,5-b] [1,2,4]benzothiadiazine conjugates as potential mitochondrial apoptotic inducers. Bioorganic Med. Chem. 2011, 19, 7136–7150. [Google Scholar] [CrossRef]
- Mohammed, A.K.; Wafaa, A.Z.; Gihad, E.E.; Rasha, A.A.; Galal, H.E. Purine analogs: Synthesis, evaluation and molecular dynamics of pyrazolopyrimidines based benzothiazole as anticancer and antimicrobial CDK inhibitors. Nucleosides Nucleotides Nucleic Acids 2022, 1–28. [Google Scholar] [CrossRef]
- Tasler, S.; Muller, O.; Wieder, T. Substituted 2-arylbenzothiazole as kinase inhibitor: Hit-to-lead optimization. Bioorganic Med. Chem. 2009, 17, 6728–6737. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, C.G.; Wells, G.; Crochard, J.P. Antitumour benzothiazoles. 26. 2-(3,4-dimethoxyphenyl)-5-fluorobenzothiazole (GW 610, NSC 721648), a simple fluorinated 2-arylbenzothiazole, shows potent and selective inhibitory activity against lung, colon, and breast cancer cell lines. J. Med. Chem. 2006, 49, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Jin, K.J.; Brewer, S.C.; Peng, L.; Platz, M.S.; Novaak, M. Indirect and direct detection of the 4-(benzothiazol-2-yl)phenylnitrenium ion from a putative metabolite of a model anti-tumor drug. Org. Lett. 2009, 11, 4862–48665. [Google Scholar] [CrossRef]
- Ke, S.; Wei, Y.; Yang, Z.; Wang, K.; Liang, Y.; Shi, L. Novel cycloalkylthiophene–imine derivatives bearing benzothiazole scaffold: Synthesis, characterization and antiviral activity evaluation. Bioorganic Med. Chem. Lett. 2013, 23, 5131–5134. [Google Scholar] [CrossRef]
- Catalano, A.; Rosato, A.; Salvagno, L.; Iacopetta, D.; Ceramella, J.; Fracchiolla, G.; Sinicropi, M.S.; Franchini, C. Benzothiazole-containing analogues of triclocarban with potent antibacterial activity. Antibiotics 2021, 10, 803. [Google Scholar] [CrossRef]
- Al-Tel, T.H.; Al-Qawasmeh, R.A.; Zaarour, R. Design, synthesis and in vitro antimicrobial evaluation of novel Imidazo[1,2-a]pyridine and imidazo[2,1-b][1,3]benzothiazole motifs. Eur. J. Med. Chem. 2011, 46, 1874–1881. [Google Scholar] [CrossRef] [PubMed]
- Bondock, S.; Fadaly, W.; Metwally, M.A. Synthesis and antimicrobial activity of some new thiazole, thiophene and pyrazole derivatives containing benzothiazole moiety. Eur. J. Med. Chem. 2010, 45, 3692–3701. [Google Scholar] [CrossRef]
- Kamal, A.; Syed, M.A.H.; Mohammed, S.M. Therapeutic potential of benzothiazoles: A patent review (2010–2014). Expert. Opin. Ther. Pat. 2015, 25, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Azzam, R.A.; Elsayed, R.E.; Elgemeie, G.H. Design, synthesis, and antimicrobial evaluation of a new series of N-sulfonamide 2-pyridones as dual inhibitors of DHPS and DHFR enzymes. ACS Omega 2020, 5, 10401–10414. [Google Scholar] [CrossRef]
- Azzam, R.A.; Elsayed, R.E.; Elgemeie, G.H. Design and synthesis of a new class of pyridine-based N-sulfonamides exhibiting antiviral, antimicrobial, and enzyme inhibition characteristics. ACS Omega 2020, 5, 26182–26194. [Google Scholar] [CrossRef]
- Elgemeie, G.H.; Azzam, R.A.; Elsayed, R.E. Sulfa drug analogs: New classes of N-sulfonyl aminated azines and their biological and preclinical importance in medicinal chemistry (2000–2018). Med. Chem. Res. 2019, 28, 1099–1131. [Google Scholar] [CrossRef]
- Elgemeie, G.; Azzam, R.; Zaghary, W.; Aly, A.; Metwally, N.; Sarhan, M.; Abdelhafez, E.; Elsayed, R. N-Sulfonated-N-Heterocycles: Synthesis, Chemistry, And Biological Applications, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Azzam, R.A.; Elboshi, H.A.; Elgemeie, G.H. Novel synthesis and antiviral evaluation of new benzothiazole-bearing N-sulfonamide 2-pyridone derivatives as USP7 enzyme inhibitors. ACS Omega 2020, 5, 30023–30036. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, R.E.; Madkour, T.M.; Azzam, R.A. Tailored-design of electrospun nanofiber cellulose acetate/poly(lactic acid) dressing mats loaded with a newly synthesized sulfonamide analog exhibiting superior wound healing. Int. J. Biol. Macromol. 2020, 164, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wolkenberg, S.E.; Lu, M.; Munshi, V.; Moyer, G.; Feng, M.; Carella, A.V.; Ecto, L.T.; Gabryelski, L.J.; Lai, M.T.; et al. Novel indole-3-sulfonamides as potent HIV non-nucleoside reverse transcriptase inhibitors (NNRTIs). Bioorganic Med. Chem. Lett. 2008, 18, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Dastagiri, D.; Janaki Ramaiah, M.; Surendranadha Reddy, J.; Vijaya Bharathi, E.; Kashi Reddy, M.; Pal-Bhadra, M. Synthesis and apoptosis inducing ability of new anilino substituted pyrimidine sulfonamides as potential anticancer agents. Eur. J. Med. Chem. 2011, 46, 5817–5824. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Javed, K.; Ahmad, S.; Rathish, I.G.; Singh, S.; Alam, M.S. Synthesis and biological evaluation of some new 2-pyrazolines bearing benzene sulfonamide moiety as potential anti-inflammatory and anti-cancer agents. Eur. J. Med. Chem. 2011, 46, 5763–5768. [Google Scholar] [CrossRef]
- Potshangbam, A.M.; Rathore, R.S.; Nongdam, P. Discovery of sulfone-resistant dihydropteroate synthase (DHPS) as a target enzyme for kaempferol, a natural flavanoid. Heliyon 2020, 6, 03378. [Google Scholar] [CrossRef]
- Nurjadi, D.; Zizmann, E.; Chanthalangsy, Q.; Heeg, K.; Boutin, S. Integrative analysis of whole genome sequencing and phenotypic resistance toward prediction of trimethoprim-sulfamethoxazole resistance in Staphylococcus aureus. Front. Microbiol. 2021, 11, 607842. [Google Scholar] [CrossRef]
- Azzam, R.A.; Elgemeie, G.H.; Elsayed, R.E.; Jones, P.G. Crystal structure of N′-[2-(benzo[d]thiazol-2-yl)acetyl]-4-methylbenzenesulfonohydrazide. Acta Crystallogr. E Crystallogr. Commun. 2017, 73, 1041–1043. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Nakatani, T.; Tanaka, R.; Okada, M.; Torii, E.; Harayama, T.; Kimachi, T. α-Dimethylaminomethylenation-induced Houben–Hoesch-type cyclization of cyanoacetanilides: A practical synthesis of 3-formyl-4-hydroxyquinolin-2 (1H)-ones. Tetrahedron 2011, 67, 3457–3463. [Google Scholar] [CrossRef]
- Lin, Y.I.; Lang, S.A., Jr. New synthesis of isoxazoles and isothiazoles. A convenient synthesis of thioenaminones from enaminones. J. Org. Chem. 1980, 45, 4857–4860. [Google Scholar] [CrossRef]
- Vinnicombe, H.G.; Derrick, J.P. Dihydropteroate synthase from streptococcus pneumoniae: Characterization of substrate binding order and sulfonamide inhibition. Biochem. Biophy. Res. Commun. 1999, 258, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Hevener, K.E.; Yun, M.K.; Qi, J.; Kerr, I.D.; Babaoglu, K.; Hurdle, J.G.; Balakrishna, K.; White, S.W.; Lee, R.E. Structural studies of pterin-based inhibitors of dihydropteroate synthase. J. Med. Chem. 2010, 53, 166–177. [Google Scholar] [CrossRef]
- Yun, M.K.; Wu, Y.; Li, Z.; Zhao, Y.; Waddell, M.B.; Ferreira, A.M.; Lee, R.E.; Bashford, D.; White, S.W. Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 2012, 335, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.C. Laboratory control of antimicrobial therapy. In Mackie and McCartney Practical Medical Microbiology, 13th ed.; Collee, J.G., Duguid, J.P., Fraser, A.G., Marmion, B.P., Eds.; Churchill Livingstone: Edinburgh, UK, 1989; Volume 2, pp. 161–182. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Robert, E.W. Hancock. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Wars, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Lagorce, D.; Sperandio, O.; Galons, H.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs2: Free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinform. 2008, 9, 396. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).