A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Activity and Biofilm Inhibition by P. clarkii Extracts
2.2. MS Analysis of the Amino Acid Sequence of the Peptides Identified in Hemocytes and Hemolymph Extracts
2.3. AMP Predictions through In Silico Analysis
2.4. Antimicrobial Screening of Selected Natural Peptides
2.5. Optimization of In Vitro Natural Peptide Potency through Bioinformatic Analysis
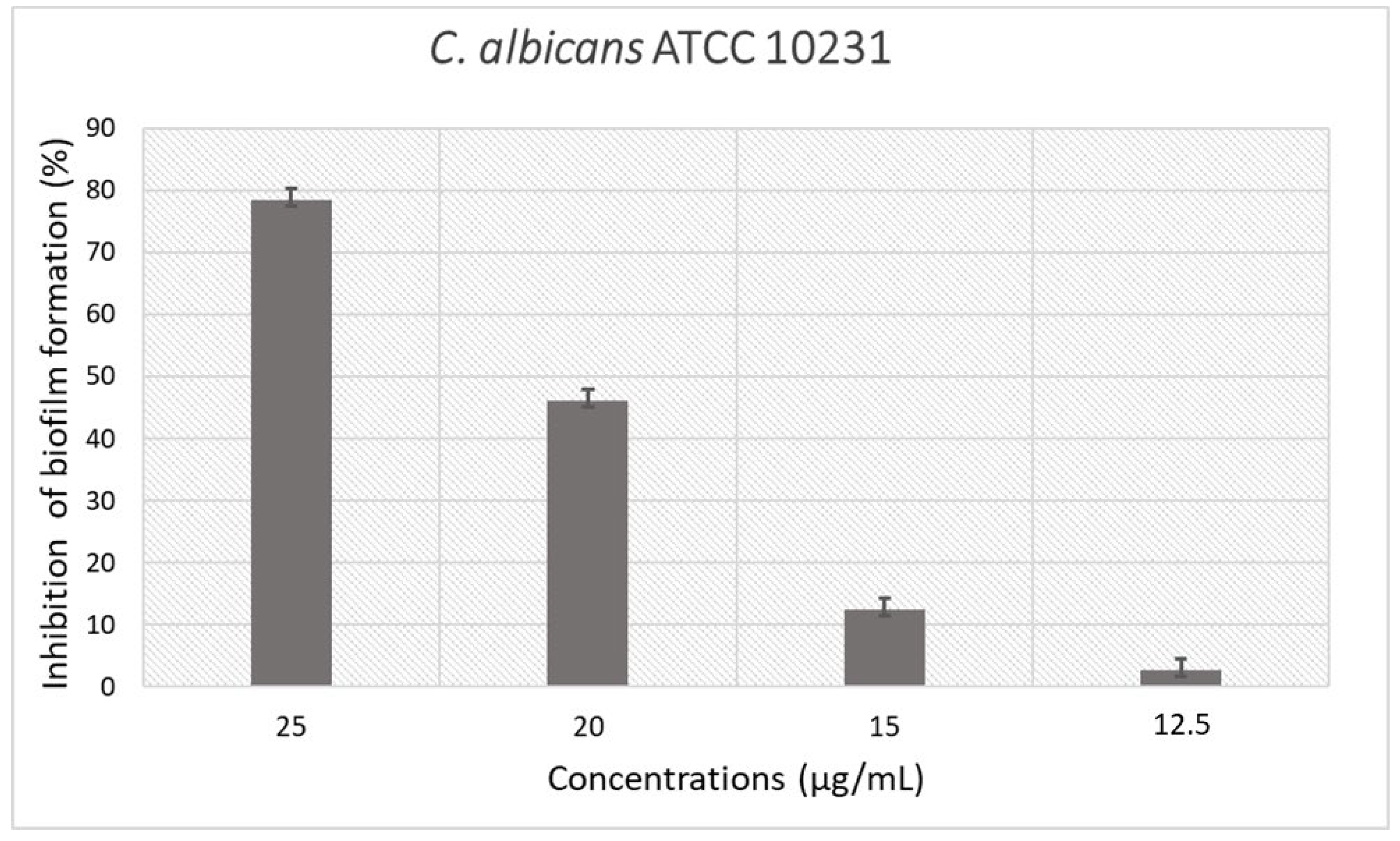
2.6. Antimicrobial and Antibiofilm Activity of Synthetic Derivative Peptides
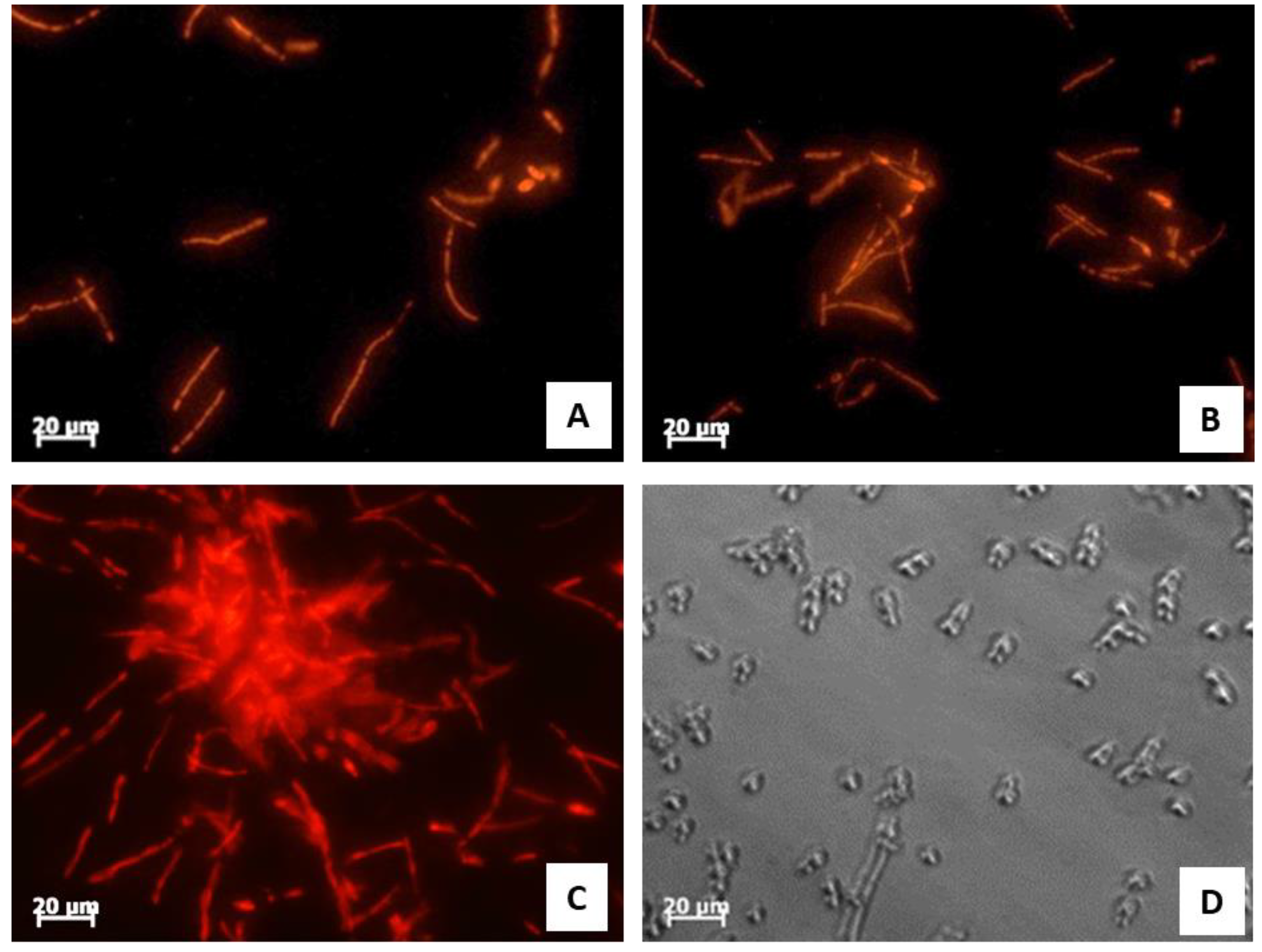
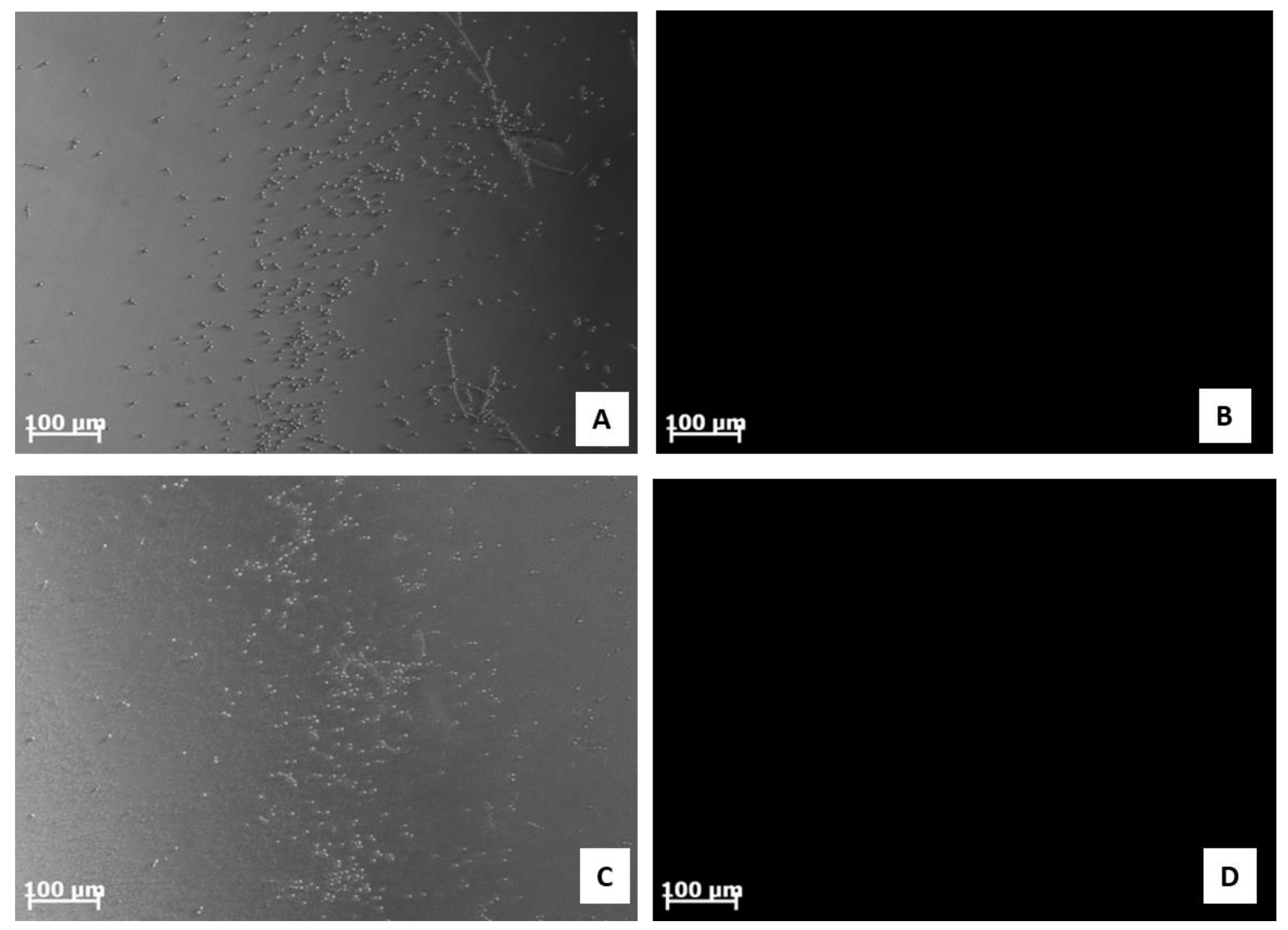
2.7. Analysis of C. albicans ATCC 10231 Morphology
2.8. Effect of Synthetic Peptide #14d on Membrane Integrity
2.9. Citotoxicity Assays on Synthetic Peptide #14d
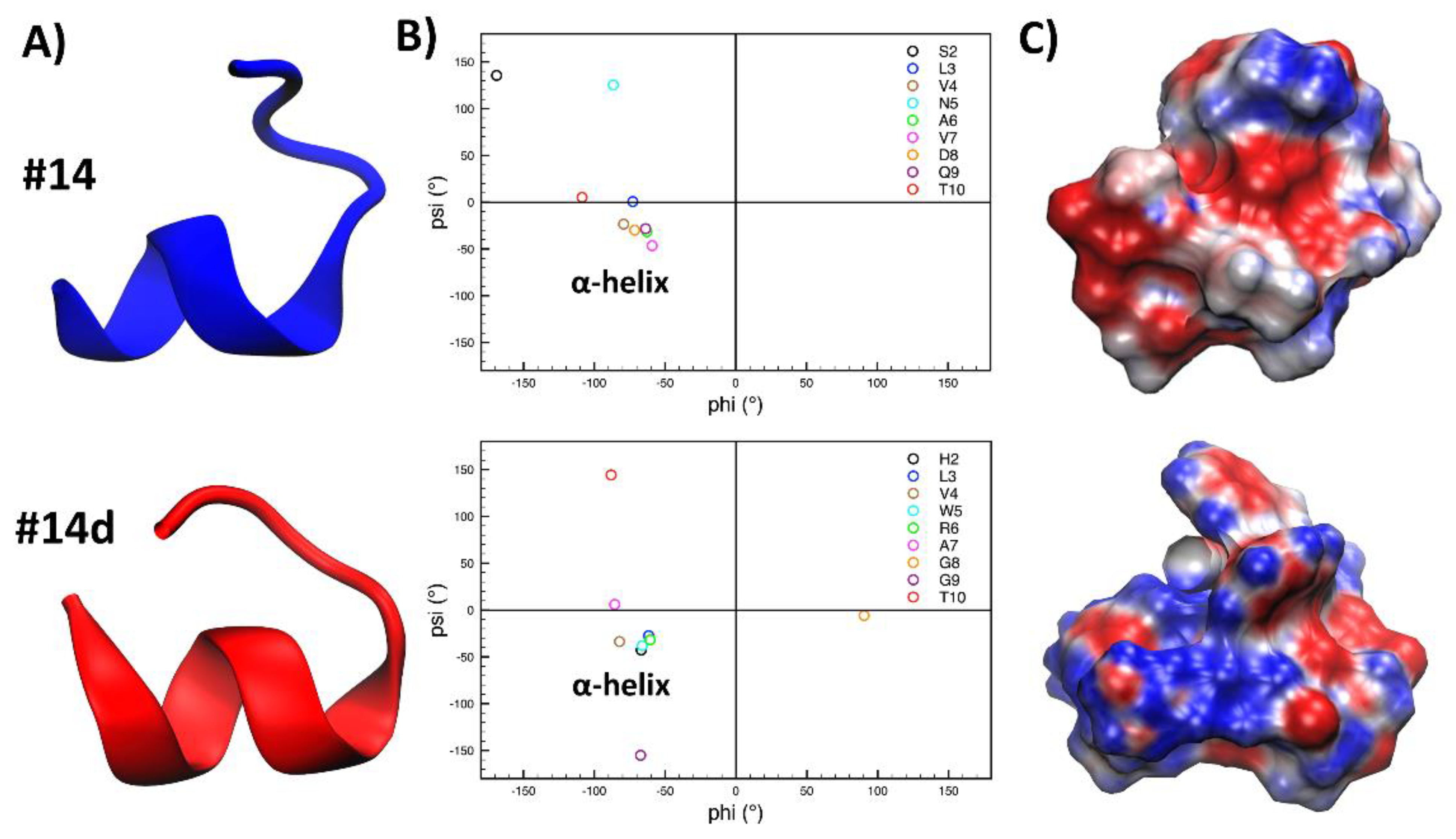
2.10. Molecular Dynamics Simulations
3. Discussion
4. Materials and Methods
4.1. P. clarkii Collection and Extract Preparation
4.2. Protein Concentration
4.3. Bacterial Strains
4.4. Determination of Minimal Inhibitory Concentrations (MICs)
4.5. Determination of EC50
4.6. Inhibition of Biofilm Formation (Crystal Violet Method)
4.7. Mass Spectrometry Analysis
4.8. Database Search
4.9. AMP Prediction and Bioinformatic Analysis
4.10. Peptide Synthesis
4.11. Molecular Dynamics (MD) Simulations
4.12. Cytocompatibility Assays
4.13. Cell Membrane Integrity Assay
4.14. Overproduction of Reactive Oxygen Species (ROS)
4.15. Scanning Electron Microscopy (SEM)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schrader, S.M.; Vaubourgeix, J.; Nathan, C. Biology of Antimicrobial Resistance and Approaches to Combat It. Sci. Transl. Med. 2020, 12, eaaz6992. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Sugár, S.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma. J. Mar. Sci. Eng. 2022, 10, 1292. [Google Scholar] [CrossRef]
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Drahos, L.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Cytotoxic capability and the associated proteomic profile of cell-free coelomic fluid extracts from the edible sea cucumber Holothuria tubulosa on HepG2 liver cancer cells. EXCLI J. 2022, 21, 722–743. [Google Scholar] [CrossRef] [PubMed]
- Punginelli, D.; Schillaci, D.; Mauro, M.; Deidun, A.; Barone, G.; Arizza, V.; Vazzana, M. The Potential of Antimicrobial Peptides Isolated from Freshwater Crayfish Species in New Drug Development: A Review. Dev. Comp. Immunol. 2022, 126, 104258. [Google Scholar] [CrossRef] [PubMed]
- Lazzara, V.; Arizza, V.; Luparello, C.; Mauro, M.; Vazzana, M. Bright Spots in the Darkness of Cancer: A Review of Starfishes-Derived Compounds and Their Anti-Tumor Action. Mar. Drugs 2019, 17, 617. [Google Scholar] [CrossRef] [PubMed]
- Mauro, M.; Lazzara, V.; Punginelli, D.; Arizza, V.; Vazzana, M. Antitumoral compounds from vertebrate sister group: A review of Mediterranean ascidians. Dev. Comp. Immunol. 2020, 108, 103669. [Google Scholar] [CrossRef]
- Fredrick, W.S.; Ravichandran, S. Hemolymph Proteins in Marine Crustaceans. Asian Pac. J. Trop. Biomed. 2012, 2, 496–502. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, R.-R.; Fan, Z.-X.; Zhao, X.-F.; Wang, X.-W.; Wang, J.-X. Characterization of a type-I crustin with broad-spectrum antimicrobial activity from red swamp crayfish Procambarus clarkii. Dev. Comp. Immunol. 2016, 61, 145–153. [Google Scholar] [CrossRef]
- Vazquez, L.; Alpuche, J.; Maldonado, G.; Agundis, C.; Pereyra-Morales, A.; Zenteno, E. Review: Immunity Mechanisms in Crustaceans. Innate Immun. 2009, 15, 179–188. [Google Scholar] [CrossRef]
- Söderhäll, K.; Cerenius, L. Role of the Prophenoloxidase-Activating System in Invertebrate Immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Wu, B.; Zhang, C.; Qin, X.; Shi, L.; Zhao, M. Identification and Function of Penaeidin 3 and Penaeidin 5 in Fenneropenaeus Merguiensis. Fish Shellfish Immunol. 2019, 89, 623–631. [Google Scholar] [CrossRef]
- Shirdel, I.; Kalbassi, M.R.; Hosseinkhani, S.; Paknejad, H.; Wink, M. Cloning, characterization and tissue-specific expression of the antimicrobial peptide hepcidin from caspian trout (Salmo caspius) and the antibacterial activity of the synthetic peptide. Fish Shell. Immunol. 2019, 90, 288–296. [Google Scholar] [CrossRef]
- Nicolas, P. Multifunctional Host Defense Peptides: Intracellular-Targeting Antimicrobial Peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Sperstad, S.V.; Haug, T.; Vasskog, T.; Stensvåg, K. Hyastatin, a Glycine-Rich Multi-Domain Antimicrobial Peptide Isolated from the Spider Crab (Hyas araneus) Hemocytes. Mol. Immunol. 2009, 46, 2604–2612. [Google Scholar] [CrossRef]
- Donpudsa, S.; Rimphanitchayakit, V.; Tassanakajon, A.; Söderhäll, I.; Söderhäll, K. Characterization of Two Crustin Antimicrobial Peptides from the Freshwater Crayfish Pacifastacus leniusculus. J. Invertebr. Pathol. 2010, 104, 234–238. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Sharma, A.; Singla, D.; Rashid, M.; Raghava, G.P.S. Designing of peptides with desired half-life in intestine-like environment. BMC Bioinform. 2014, 15, 282. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Zaalishvili, G.; Karapetian, M.; Nasrashvili, T.; Kuljanishvili, N.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Grigolava, M.; et al. De Novo Design and In Vitro Testing of Antimicrobial Peptides against Gram-Negative Bacteria. Pharmaceuticals 2019, 12, 82. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, P.; Kumar, R.; Bhardwaj, A. dPABBs: A Novel in silico Approach for Predicting and Designing Anti-biofilm Peptides. Sci. Rep. 2016, 6, 21839. [Google Scholar] [CrossRef]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Raghava, G.P. Computer-Aided Virtual Screening and Designing of Cell-Penetrating Peptides. In Cell-Penetrating Peptides. Methods in Molecular Biology; Langel, Ü., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1324, pp. 59–69. [Google Scholar] [CrossRef]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Open source drug discovery consortium; Raghava, G.P.S. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open Source Drug Discovery Consortium; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. Peptide Toxicity Prediction. In Computational Peptidology. Methods in Molecular Biology; Zhou, P., Huang, J., Eds.; Humana Press: New York, NY, USA, 2015; Volume 1268, pp. 143–157. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols. Methods in Molecular Biology; Link, A.J., Ed.; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 531–552. [Google Scholar] [CrossRef]
- Meher, P.K.; Sahu, T.K.; Saini, V.; Rao, A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, 42362. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Hansen, I.K.Ø.; Lövdahl, T.; Simonovic, D.; Hansen, K.Ø.; Andersen, A.J.C.; Devold, H.; Richard, C.S.M.; Andersen, J.H.; Strøm, M.B.; Haug, T. Antimicrobial Activity of Small Synthetic Peptides Based on the Marine Peptide Turgencin A: Prediction of Antimicrobial Peptide Sequences in a Natural Peptide and Strategy for Optimization of Potency. Int. J. Mol. Sci. 2020, 21, 5460. [Google Scholar] [CrossRef]
- Pereira, R.; dos Santos Fontenelle, R.O.; de Brito, E.H.S.; de Morais, S.M. Biofilm of Candida albicans: Formation, regulation and resistance. J Appl Microbiol. 2021, 131, 11–22. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Grande, R.; Puca, V.; Muraro, R. Antibiotic resistance and bacterial biofilm. Expert Opin. Ther. Pat. 2020, 30, 897–900. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; pp. 348–365. [Google Scholar]
- Hathroubi, S.; Mekni, M.A.; Domenico, P.; Nguyen, D.; Jacques, M. Biofilms: Microbial Shelters Against Antibiotics. Microb. Drug Resist. 2017, 23, 147–156. [Google Scholar] [CrossRef]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Ann. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Lima, P.G.; Oliveira, J.T.A.; Amaral, J.L.; Freitas, C.D.T.; Souza, P.F.N. Synthetic antimicrobial peptides: Characteristics, design, and potential as alternative molecules to overcome microbial resistance. Life Sci. 2021, 278, 119647. [Google Scholar] [CrossRef]
- Souza, P.F.N.; Marques, L.S.M.; Oliveira, J.T.A.; Lima, P.G.; Dias, L.P.; Neto, N.A.S.; Lopes, F.E.S.; Sousa, J.S.; Silva, A.F.B.; Caneiro, R.F.; et al. Synthetic antimicrobial peptides: From choice of the best sequences to action mechanisms. Biochimie 2020, 175, 132–145. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, J.; Gao, L. Catalytic antimicrobial therapy using nanozymes. WIREs Nanomed. Nanobiotechnology 2022, 14, e1769. [Google Scholar] [CrossRef]
- Yang, S.; Veerana, M.; Yu, N.; Ketya, W.; Park, G.; Kim, S.; Kim, Y. Copper(II)-MOF Containing Glutarate and 4,4′-Azopyridine and Its Antifungal Activity. Appl. Sci. 2022, 12, 260. [Google Scholar] [CrossRef]
- Struyfs, C.; Cammue, B.P.A.; Thevissen, K. Membrane-interacting antifungal peptides. Front. Cell Dev. Biol. 2021, 9, 649875. [Google Scholar] [CrossRef]
- Oliveira, J.T.A.; Souza, P.F.N.; Vasconcelos, I.M.; Dias, L.P.; Martins, T.F.; Van Tilburg, M.F.; Guedes, M.I.F.; Sousa, D.O.B. Mo-CBP3-PepI, Mo-CBP3-PepII, and Mo-CBP3-PepIII are synthetic antimicrobial peptides active against human pathogens by stimulating ROS generation and increasing plasma membrane permeability. Biochimie 2019, 157, 10–21. [Google Scholar] [CrossRef]
- Lima, P.G.; Souza, P.F.N.; Freitas, C.D.T.; Oliveira, J.T.A.; Dias, L.P.; Neto, J.X.S.; Vasconcelos, I.M.; Lopes, J.L.S.; Sousa, D.O.B. Anticandidal activity of synthetic peptides: Mechanism of action revealed by scanning electron and fluorescence microscopies and synergism effect with nystatin. J. Pept. Sci. 2020, 26, e3249. [Google Scholar] [CrossRef]
- Bezerra, L.P.; Freitas, C.D.T.; Silva, A.F.B.; Amaral, J.L.; Neto, N.A.S.; Silva, R.G.G.; Parra, A.L.C.; Goldman, G.H.; Oliveira, J.T.A.; Mesquita, F.P.; et al. Synergistic Antifungal Activity of Synthetic Peptides and Antifungal Drugs against Candida albicans and C. parapsilosis Biofilms. Antibiotics 2022, 11, 553. [Google Scholar] [CrossRef]
- Belmadani, A.; Semlali, A.; Rouabhia, M. Dermaseptin-S1 decreases Candida albicans growth, biofilm formation and the expression of hyphal wall protein 1 and aspartic protease genes. J. Appl. Microbiol. 2018, 125, 72–83. [Google Scholar] [CrossRef]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xu, W.-T.; Zhang, H.-W.; Dong, L.-P.; Zhang, T.; Zhao, X.-F.; Wang, J.-X. An anti-lipopolysaccharide factor from red swamp crayfish, Procambarus clarkii, exhibited antimicrobial activities in vitro and in vivo. Fish Shell. Immunol. 2011, 30, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Lee, D.G. Antifungal activity and pore-forming mechanism of astacidin 1 against Candida albicans. Biochimie 2015, 105, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Petit, V.W.; Rolland, J.-L.; Blond, A.; Cazevieille, C.; Djediat, C.; Peduzzi, J.; Goulard, C.; Bachère, E.; Dupont, J.; Destoumieux-Garzón, D.; et al. A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2016, 1860, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-Q.; Shi, Y.-H.; Wang, Q. Characterisation of a novel Type I crustin involved in antibacterial and antifungal responses in the red claw crayfish, Cherax quadricarinatus. Fish Shell.Immunol. 2016, 48, 30–38. [Google Scholar] [CrossRef]
- Li, S.; Jin, X.-K.; Guo, X.-N.; Yu, A.-Q.; Wu, M.-H.; Tan, S.-J.; Zhu, Y.-T.; Li, W.-W.; Wang, Q. A double WAP domain-containing protein Es-DWD1 from Eriocheir sinensis exhibits antimicrobial and proteinase inhibitory activities. PLoS ONE 2013, 8, e73563. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Romero, S.M.; Cardillo, A.B.; Martínez Ceron, M.C.; Camperi, S.A.; Giudicessi, S.L. Temporins: An Approach of Potential Pharmaceutic Candidates. Surg. Infect. 2020, 21, 309–322. [Google Scholar] [CrossRef]
- Cunsolo, V.; Schicchi, R.; Chiaramonte, M.; Inguglia, L.; Arizza, V.; Cusimano, M.G.; Schillaci, D.; Di Francesco, A.; Saletti, R.; Lo Celso, F.; et al. Identification of New Antimicrobial Peptides from Mediterranean Medical Plant Charybdis pancration (Steinh.) Speta. Antibiotics 2020, 9, 747. [Google Scholar] [CrossRef]
- Cusimano, M.G.; Spinello, A.; Barone, G.; Schillaci, D.; Cascioferro, S.; Magistrato, A.; Parrino, B.; Arizza, V.; Vitale, M. A Synthetic Derivative of Antimicrobial Peptide Holothuroidin 2 from Mediterranean Sea Cucumber (Holothuria tubulosa) in the Control of Listeria monocytogenes. Mar. Drugs 2019, 17, 159. [Google Scholar] [CrossRef]
- Girasolo, M.A.; Schillaci, D.; Di Salvo, C.; Barone, G.; Silvestri, A.; Ruisi, G. Synthesis, spectroscopic characterization and in vitro antimicrobial activity of diorganotin(IV) dichloride adducts with [1,2,4] triazolo-[1,5-a] pyrimidine and 5,7-dimethyl-[1,2,4] triazolo-[1,5-a] pyrimidine. J. Organomet. Chem. 2006, 691, 693–701. [Google Scholar] [CrossRef]
- Carbone, A.; Cascioferro, S.; Parrino, B.; Carbone, D.; Pecoraro, C.; Schillaci, D.; Cusimano, M.G.; Cirrincione, G.; Diana, P. Thiazole Analogues of the Marine Alkaloid Nortopsentin as Inhibitors of Bacterial Biofilm Formation. Molecules 2020, 26, 81. [Google Scholar] [CrossRef]
- Merlani, M.; Scheibel, D.M.; Barbakadze, V.; Gogilashvili, L.; Amiranashvili, L.; Geronikaki, A.; Catania, V.; Schillaci, D.; Gallo, G.; Gitsov, I. Enzymatic Synthesis and Antimicrobial Activity of Oligomer Analogues of Medicinal Biopolymers from Comfrey and Other Species of the Boraginaceae Family. Pharmaceutics 2022, 14, 115. [Google Scholar] [CrossRef]
- Federico, S.; Catania, V.; Palumbo, F.S.; Fiorica, C.; Schillaci, D.; Pitarresi, G.; Giammona, G. Photothermal nanofibrillar membrane based on hyaluronic acid and graphene oxide to treat Staphylococcus aureus and Pseudomonas aeruginosa infected wounds. Int. J. Biol. Macromol. 2022, 214, 470–479. [Google Scholar] [CrossRef]
- Martorana, A.; Pitarresi, G.; Palumbo, F.S.; Catania, V.; Schillaci, D.; Mauro, N.; Fiorica, C.; Giammona, G. Fabrication of silver nanoparticles by a diethylene triamine-hyaluronic acid derivative and use as antibacterial coating. Carbohydr. Polym. 2022, 295, 119861. [Google Scholar] [CrossRef]
- Plescia, F.; Venturella, F.; Lauricella, M.; Catania, V.; Polito, G.; Schillaci, D.; Piccionello, A.P.; Daidone, G.; D’Anneo, A.; Raffa, D. Chemical composition, cytotoxic effects, antimicrobial and antibiofilm activity of Artemisia arborescens (Vaill.) L. growing wild in the province of Agrigento, Sicily, Italy. Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2022, 1–10. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Schillaci, D.; Cusimano, M.G.; Spinello, A.; Barone, G.; Russo, D.; Vitale, M.; Parrinello, D.; Arizza, V. Paracentrin 1, a synthetic antimicrobial peptide from the sea-urchin Paracentrotus lividus, interferes with staphylococcal and Pseudomonas aeruginosa biofilm formation. AMB Express 2014, 4, 78. [Google Scholar] [CrossRef]
- Schillaci, D.; Spinello, A.; Cusimano, M.G.; Cascioferro, S.; Barone, G.; Vitale, M.; Arizza, V. A peptide from human β thymosin as a platform for the development of new anti-biofilm agents for Staphylococcus spp. and Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2016, 32, 124. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982, 76, 2662–2666. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chang, W.; Zhang, M.; Li, X.; Jiao, Y.; Lou, H. Diorcinol D Exerts Fungicidal Action against Candida albicans through Cytoplasm Membrane Destruction and ROS Accumulation. PLoS ONE 2015, 10, e0128693. [Google Scholar] [CrossRef]


| MIC | ||
|---|---|---|
| Hemocytes | Hemolymph | |
| S. aureus ATCC 25923 | 50% v/v (11 μg/mL) | 50% v/v (700 μg/mL) |
| P. aeruginosa ATCC 15442 | 50% v/v (11 μg/mL) | 50% v/v (700 μg/mL) |
| E. coli ATCC 25922 | 50% v/v (11 μg/mL) | 50% v/v (700 μg/mL) |
| E. faecalis ATCC 29212 | 50% v/v (11 μg/mL) | 50% v/v (700 μg/mL) |
| C. albicans ATCC 10231 | 50% v/v (11 μg/mL) | 12.5% v/v (175 μg/mL) |
| BIC50 | ||
|---|---|---|
| Hemocytes | Hemolymph | |
| S. aureus ATCC 25923 | >1 | >70 |
| P. aeruginosa ATCC 15442 | 1 | >70 |
| Polypeptide-Enriched Extracts | #No. | Identified Peptides | Better Chance to Be an AMP Predicted Ability to Interact with Membranes | Percentages of Similarity with Already Described AMPs |
|---|---|---|---|---|
| Hemocytes | #1 | MFLHGHAV a | - | 44.44% plicatamide (marine tunicate, invertebrates) |
| #2 | EGLDDDERL b | - | 44.44% SAAP fraction (mammals) | |
| #3 | SSGYGGYGGRF | yes | 50% Crinicepsin I (insects) | |
| #4 | LNVQAQMLLQ | yes | 38.46% Temporin–1Ee (frogs, amphibians) | |
| #5 | NNWTGADCKAATLK | yes | 42.85% Panurgirn I (insects) | |
| #6 | SHGDSALSSTF | - | 42.86% Temporin-1DYa (frogs, amphibians) | |
| #7 | YGGYFGNR | yes | 50% Crinicepsin I (insects) | |
| #8 | ETEASLTAALPRW | - | 38.46% RP9 (reptiles) | |
| #9 | LVDSNGALLDELPVAR | - | 41.18% Peptide #4 (frogs, amphibians) | |
| #10 | KLLLDNSAEDLEELASHK | yes | 45% Hb 98–114 (mammals) | |
| Hemolymph | #11 | AADSFGETFAATL | yes | 38.46% Urechistakynin II (marine worms, invertebrates) |
| #12 | FDTLSSHLVATD | yes | 42.86 Temporin-SN4 (frogs, amphibians) | |
| #13 | ETAPLSGVCF | - | 41.67% Peptide 7 (molluscs, marine invertebrates) | |
| #14 | FSLVNAVDQTT | yes | 38.46% VmCT2 (scorpions, arthropds) |
| Peptide #3 | Peptide #5 | Peptide #7 | Peptide #14 | |
|---|---|---|---|---|
| Peptide sequence | SSGYGGYGGGRF | NNWTGADCKAATLK | YGGYFGNR | FSLVNAVDQTT |
| Monoisotopic Theoretical mass (Da) | 1163.499 | 1491.714 | 932.414 | 1193.592 |
| Net charge | +1 | +1 | +1 | −1 |
| Isoelectric point | 6.228 | 6.091 | 6.360 | 5.474 |
| Wimley–White whole–residue (kcal/mol) | −1.88 kcal/mol | 2.2 kcal/mol | −1.75 kcal/mol | 1.26 kcal/mol |
| Hydrophobic ratio (%) | 8 | 40 | 13 | 45 |
| Hydrophobic moment (µH) | 0.336 | 0.044 | - | 0.315 |
| Protein–binding potential Boman index (kcal/mol) | 1.11 kcal/mol | 1.66 kcal/mol | 2 kcal/mol | 1.05 kcal/mol |
| Half-life (s) | 1.098 | 0.733 | 2.198 | 0.304 |
| Stability in a biological proteolytic environment | high | normal | high | normal |
| Properties | Peptide #3 | Peptide #5 | Peptide #7 | Peptide #14 |
|---|---|---|---|---|
| Sequences | SSGYGGYGGRF | NNWTGADCKAATLK | YGGYFGNR | FSLVNADQTT |
| CPP (cell penetrating peptides) | Not CPP | Not CPP | Not CPP | Not CPP |
| Antibacterial activity | 68.9% | 86.4% | 49,3% | 59% |
| Antifungal activity | 71.5% | 88.2% | 65.2% | 48% |
| Antibiofilm activity | Not predicted | Not predicted | Not predicted | Not predicted |
| Allergic potential | No | Yes | No | No |
| Hemolytic potential (probability) | 0.49 | 0.50 | 0.49 | 0.49 |
| Toxicity | No | No | No | No |
| Degradation by trypsin | Yes | Yes | Yes | No |
| Degradation by pepsin (pH = 1.3) | Yes | Yes | Yes | Yes |
| Degradation by pepsin (pH > 2) | Yes | Yes | Yes | Yes |
| Starting Natural Peptide Sequence | Derivative Synthetic Peptide | Derivative Synthetic Peptide Sequence | Monoisotopic Theoretical Mass (Da) | Net Charge | Boman Index (kcal/mol) | Hydrophobic Ratio (%) |
|---|---|---|---|---|---|---|
| #3 SSGYGGYGGRF | Pep #3d | IIIRKGRW | 1041.31 | +3 | 2.17 | 50 |
| #5 NWTGADCKAATLK | Pep #5d | NWWTGARCKAATLK | 1634.87 | +3 | 1.46 | 50 |
| #14 FSLVNADQTT | Pep #14d | FHLVWRAGGTF | 1290.49 | +1.25 | 0.11 | 55 |
| Peptide | Antibiofilm Activity 1 | CPP 2 | Antimicrobial Activity 3 | Antifungal Activity 3 | Hemolytic Potential 4 | Toxic Potential 5 | Trypsin Cleavage Site | Pepsin Cleavage Site pH 1.3–pH > 2 |
|---|---|---|---|---|---|---|---|---|
| Peptide #3d | Yes | No | 82% | 69% | 0.49 | No | Yes | No-Yes |
| Peptide #5d | Yes | Yes | 91% | 61% | 0.48 | No | Yes | Yes-Yes |
| Peptide #14d | Yes | No | 41% | 14% | 0.48 | No | Yes | Yes-Yes |
| Peptide | Source | Target Fungi | In Vitro MIC (μg/mL) | References |
|---|---|---|---|---|
| PcALF1 | Procambarus clarkii | C. albicans | 20.0 | [46] |
| Astacidin 1 | Pacifastacus leniusculus | C. albicans T. biegelii Malassezia furfur Trichophyton rubrum | 6.3 6.3 12.5 25.0 | [47,48] |
| Crustin I | Cheraxquadricarinatus | C. albicans | 20.0 | [49] |
| Es-DWD1 | Eriocheir sinensis | P. pastoris | 30.0 | [50] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Punginelli, D.; Catania, V.; Vazzana, M.; Mauro, M.; Spinello, A.; Barone, G.; Barberi, G.; Fiorica, C.; Vitale, M.; Cunsolo, V.; et al. A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii. Antibiotics 2022, 11, 1792. https://doi.org/10.3390/antibiotics11121792
Punginelli D, Catania V, Vazzana M, Mauro M, Spinello A, Barone G, Barberi G, Fiorica C, Vitale M, Cunsolo V, et al. A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii. Antibiotics. 2022; 11(12):1792. https://doi.org/10.3390/antibiotics11121792
Chicago/Turabian StylePunginelli, Diletta, Valentina Catania, Mirella Vazzana, Manuela Mauro, Angelo Spinello, Giampaolo Barone, Giuseppe Barberi, Calogero Fiorica, Maria Vitale, Vincenzo Cunsolo, and et al. 2022. "A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii" Antibiotics 11, no. 12: 1792. https://doi.org/10.3390/antibiotics11121792
APA StylePunginelli, D., Catania, V., Vazzana, M., Mauro, M., Spinello, A., Barone, G., Barberi, G., Fiorica, C., Vitale, M., Cunsolo, V., Saletti, R., Di Francesco, A., Arizza, V., & Schillaci, D. (2022). A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii. Antibiotics, 11(12), 1792. https://doi.org/10.3390/antibiotics11121792









