Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies
Abstract
1. Introduction
2. Antimicrobial Resistance in Polymicrobial Biofilms
- Composition of EPS matrix: Matrix supports the microbial cells for adherence, immobilization, and protects from environmental stress and antimicrobial agents. EPS composition varies from species to species and also with the environment. P. aeruginosa polysaccharide (Psl) is reported to provide resistance against colistin, polymyxin B, tobramycin, and ciprofloxacin, and a similar effect is also observed in non-psl producers such as Escherichia coli and S. aureus, possibly via electrostatic forces [14]. Matrix composition differs in multispecies biofilm, which confers more resistance than mono-species biofilm. Candida albicans protects S. aureus from vancomycin treatment by secreting exopolysaccharide, β-1,3-glucan, while Streptococcus mutans produces glucans that protect the Candida from fluconazole in mixed biofilms [15].
- Commensal-like interactions: One member in the community provides a suitable condition for the survival of other members in an inhospitable environment. It was exemplified by Elias and Banin, 2012 [5], who found that the presence of aerobes provides a better condition for the survival of anaerobes when the oxygen concentration is high.
- Alteration of physiology by neighbouring species: It was reported that HQNO (4-hydroxy-2-heptylquinoline-N-oxide) produced by P. aeruginosa could be used by S. aureus to increase the tolerance to antibiotics (vancomycin & tobramycin). Prolonged exposure to HQNO or with P. aeruginosa makes the resistant small-colony variants (SCV) of S. aureus [16].
- Resistance to host immune response: The alpha toxin of S. aureus disrupts the host immunity and the barriers of epithelial cells, which leads to the co-infection with P. aeruginosa and eventually causes pulmonary dysfunction [17].
- Interspecies genetic exchange: Horizontal gene transfer (HGT) of resistance genes in multispecies biofilm results in the emergence of resistance in evolutionarily distant species. HGT facilitates a range of adaptations, such as changes in metabolic levels, antimicrobial resistance, and biofilm formation. It was reported that the conjugative plasmid induces biofilm development and stimulates biofilm formation [8]. It was found that plasmid having the carbapenemase resistance gene (blaNDM-1) was transferred from E. coli to either P. aeruginosa or Acinetobacter baumannii via conjugation in multispecies biofilms. Moreover, exchange of mobile genetic elements such as mecA cassette has also been reported [12].
- β-lactamases-producing strains: β-lactamases are the enzymes that hydrolyse β-lactam antibiotics (cell wall-targeting drugs). Inactivation of β-lactam antibiotics effectively protects the cell itself and other cells in the polymicrobial biofilm. For example, co-culturing of Haemophilus influenza (β-lactamase producer) with Streptococcus pneumoniae (β-lactamase non-producer) increases MIC/MBC of amoxicillin (β-lactam antibiotic) [12].
3. Polymicrobial Infections
4. Polymicrobial Interactions
4.1. Bacterial-Bacterial Biofilms
4.2. Bacterial-Fungal Biofilms
4.3. Fungal-Fungal Biofilms
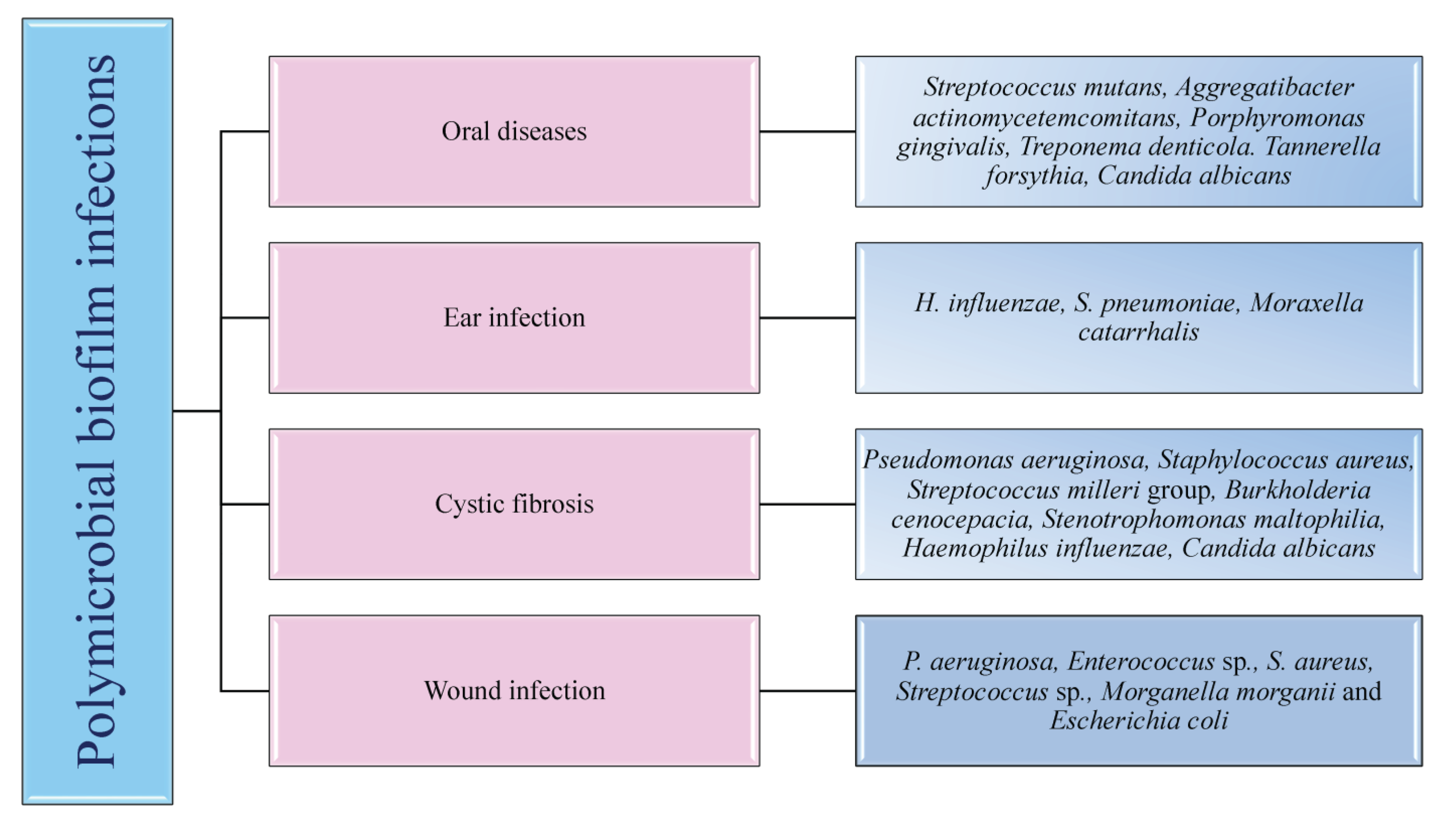
5. Clinically Relevant Human Polymicrobial Biofilm Infections
5.1. Oral Infections
5.2. Wound Infections
5.3. Diabetic Foot Ulcers
5.4. Respiratory Infections
5.5. Otitis Media
6. Model Systems to Study Polymicrobial Biofilms
7. Metagenomic Approaches in Detection, Prevention, and Inhibition of Polymicrobial Biofilms
Discovery of Novel Biofilm and Quorum Sensing Inhibitors through Metagenomics
8. Innovative Approaches to Mitigate Polymicrobial Biofilms
8.1. Nanoparticle and Nanoconjugate Mediated Therapy
8.2. Antimicrobial Photodynamic Therapy (aPDT)
8.3. Antimicrobial Peptides (AMPs)
8.4. Quorum Sensing Inhibitors/Natural Products Based Anti-Biofilm Agents
8.5. Phage Therapy
8.6. Probiotic Combinations
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zupančič, J.; Raghupathi, P.K.; Houf, K.; Burmølle, M.; Sørensen, S.J.; Gunde-Cimerman, N. Synergistic Interactions in Microbial Biofilms Facilitate the Establishment of Opportunistic Pathogenic Fungi in Household Dishwashers. Front. Microbiol. 2018, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, J.M.; Rai, A.K.; Sharma, M.; Tripathi, M.; Prasad, R. Microbial Biofilms: Recent Advances and Progress in Environmental Bioremediation. Sci. Total Environ. 2022, 824, 153843. [Google Scholar] [CrossRef] [PubMed]
- Jahid, I.K.; Ha, S. Do the Paradox of Mixed-Species Biofilms in the Context of Food Safety. Compr. Rev. Food Sci. Food Saf. 2014, 13, 990–1011. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Schulz, A.; Lang, R.; Behr, J.; Hertel, S.; Reich, M.; Kümmerer, K.; Hannig, M.; Hannig, C.; Hofmann, T. Targeted metabolomics of pellicle and saliva in children with different caries activity. Sci. Rep. 2020, 10, 697. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Liu, W.; Røder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sørensen, S.J.; Burmølle, M. Interspecific Bacterial Interactions are Reflected in Multispecies Biofilm Spatial Organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Wu, H.; Høiby, N.; Molin, S.; Song, Z. Current understanding of multi-species biofilms. Int. J. of Oral Sci. 2011, 3, 74–81. [Google Scholar] [CrossRef]
- Gabrilska, R.A.; Rumbaugh, K.P. Biofilm models of polymicrobial infection. Future Microbiol. 2015, 10, 1997–2015. [Google Scholar] [CrossRef]
- Estrela, S.; Brown, S.P. Metabolic and Demographic Feedbacks Shape the Emergent Spatial Structure and Function of Microbial Communities. PLoS Comput. Biol. 2013, 9, e1003398. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Alonso, V.P.P.; Harada, A.M.M.; Kabuki, D.Y. Competitive and/or Cooperative Interactions of Listeria monocytogenes With Bacillus cereus in Dual-Species Biofilm Formation. Front. Microbiol. 2020, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Jorge, P.; Magalhães, A.P.; Grainha, T.; Alves, D.; Sousa, A.M.; Lopes, S.P.; Pereira, M.O. Antimicrobial resistance three ways: Healthcare crisis, major concepts and the relevance of biofilms. FEMS Microbiol. Ecol. 2019, 95, fiz115. [Google Scholar] [CrossRef] [PubMed]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Cendra, M.d.M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef]
- Limoli, D.H.; Yang, J.; Khansaheb, M.K.; Helfman, B.; Peng, L.; Stecenko, A.A.; Goldberg, J.B. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 947–953. [Google Scholar] [CrossRef]
- Brogden, K.A.; Guthmiller, J.M.; Taylor, C.E. Human polymicrobial infections. Lancet. 2005, 365, 253–255. [Google Scholar] [CrossRef]
- Espinosa, A.; Palencia, S.L.; García, A.; Palencia, M. Polymicrobial Biofilms: Fundamentals, diseases, and treatments—A review. J. Sci. Technol. Appl. 2020, 8, 4–34. [Google Scholar] [CrossRef]
- Tay, W.H.; Chong, K.K.L.; Kline, K.A. Polymicrobial–Host Interactions during Infection. J. Mol. Biol. 2016, 428, 3355–3371. [Google Scholar] [CrossRef]
- Pierce, E.C.; Dutton, R.J. Putting microbial interactions back into community contexts. Curr. Opin. Microbiol. 2022, 65, 56–63. [Google Scholar] [CrossRef]
- Kuramitsu, H.K.; He, X.; Lux, R.; Anderson, M.H.; Shi, W. Interspecies Interactions within Oral Microbial Communities. Microbiol. Mol. Biol. Rev. 2007, 71, 653–670. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Rhoads, D.D.; Dowd, S.E. Biofilms and chronic wound inflammation. J. Wound Care 2008, 17, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Smith, E.; Rhoads, D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS ONE 2008, 3, e3326. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.J., Jr.; Kazmerzak, K.; Hansen, M.C.; Kolenbrander, P.E. Mutualism versus independence: Strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001, 69, 5794–5804. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; Connell, J.L.; Stacy, A.; Turner, K.H.; Whitely, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014, 52, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. microbiol. 2000, 54, 413. [Google Scholar] [CrossRef]
- Williams, C.F.; Klinzman, D.; Yamashita, T.E.; Xiang, J.; Polgreen, P.M.; Rinaldo, C.; Liu, C.; Phair, J.; Margolick, J.B.; Zdunek, D.; et al. Persistent GB Virus C Infection and Survival in HIV-Infected Men. N. Engl. J. Med. 2004, 350, 981–990. [Google Scholar] [CrossRef]
- Keays, T.; Ferris, W.; Vandemheen, K.L.; Chan, F.; Yeung, S.W.; Mah, T.F.; Ramotar, K.; Saginur, R.; Aaron, S.D. A retrospective analysis of biofilm antibiotic susceptibility testing: A better predictor of clinical response in cystic fibrosis exacerbations. J. Cyst. Fibros. 2009, 8, 122–127. [Google Scholar] [CrossRef]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef]
- Monasta, L.; Ronfani, L.; Marchetti, F.; Montico, M.; Vecchi Brumatti, L.; Bavcar, A.; Grasso, D.; Barbiero, C.; Tamburlini, G. Burden of disease caused by otitis media: Systematic review and global estimates. PLoS ONE 2012, 7, e36226. [Google Scholar] [CrossRef] [PubMed]
- Bair, K.L.; Campagnari, A.A. Moraxella catarrhalis Promotes Stable Polymicrobial Biofilms with the Major Otopathogens. Front. Microbiol. 2020, 10, 3006. [Google Scholar] [CrossRef] [PubMed]
- Hotterbeekx, A.; Kumar-Singh, S.; Goossens, H.; Malhotra-Kumar, S. In vivo and In vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 2017, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Hajjeh, R.A.; Sofair, A.N.; Harrison, L.H.; Lyon, G.M.; Arthington-Skaggs, B.A.; Mirza, S.A.; Phelan, M.; Morgan, J.; Lee-Yang, W.; Ciblak, M.A.; et al. Incidence of Bloodstream Infections Due to Candida Species and In Vitro Susceptibilities of Isolates Collected from 1998 to 2000 in a Population-Based Active Surveillance Program. J. Clin. Microbiol. 2004, 42, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-S.; Chen, Y.-C.; Kuo, S.-F. The Impact of Biofilm Formation on the Persistence of Candidemia. Front. Microbiol. 2008, 9, 1196. [Google Scholar] [CrossRef]
- Delaney, C.; Kean, R.; Short, B.; Tumelty, M.; McLean, W.; Nile, C.J.; Ramage, G. Fungi at the Scene of the Crime: Innocent Bystanders or Accomplices in Oral Infections? Curr. Clin. Microbiol. Rep. 2018, 5, 190–200. [Google Scholar] [CrossRef]
- Ponde, N.O.; Lortal, L.; Ramage, G. Candida albicans biofilms and polymicrobial interactions. Crit. Rev. Microbiol. 2021, 47, 91–111. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; De Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal biofilms and polymicrobial diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Coco, B.J.; Bagg, J.; Cross, L.J.; Jose, A.; Ramage, G. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol. Immunol. 2008, 23, 377–383. [Google Scholar] [CrossRef]
- Kirkpatrick, W.R.; Lopez-Ribot, J.L.; Mcatee, R.K.; Patterson, T.F. Growth competition between Candida dubliniensis and Candida albicans under broth and biofilm growing conditions. J. Clin. Microbiol. 2000, 38, 902–904. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.H.G.; Pires, R.H.; Cunha, A.O.; Pereira, C.A.M.; de Lacorte Singulani, J.; Abrão, F.; de Moraes, T.; Mendes-Giannini, M.J.S. Candida/Candida biofilms. First description of dual-species Candida albicans/C. rugosa biofilm. Fungal Biol. 2016, 120, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N. Grand Challenges in Oral Infections and Microbes. Front. Oral. Health 2020, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Kang, M.; Zhang, M.J. Polymicrobial periodontal disease triggers a wide radius of effect and unique virome. npj Biofilms Microbiomes. 2020, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the Normal Bacterial Flora of the Oral Cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2005, 75, 1704–1712. [Google Scholar] [CrossRef]
- Shaikh, H.F.M.; Patil, S.H.; Pangam, T.S.; Rathod, K.V. Polymicrobial synergy and dysbiosis: An overview. J. Indian Soc. Periodontol. 2018, 22, 101. [Google Scholar] [CrossRef]
- Mohanty, R.; Asopa, S.J.; Joseph, M.D.; Singh, B.; Rajguru, J.P.; Saidath, K.; Sharma, U. Red complex: Polymicrobial conglomerate in oral flora: A review. J. Fam. Med. Prim. Care. 2019, 8, 3480. [Google Scholar]
- Sukhvinder Singh, O.; Shabina, S.; Shibani, G. Various complexes of the oral microbial flora in periodontal disease. J. Dent. Probl. Solut. 2021, 8, 032–033. [Google Scholar] [CrossRef]
- Havsed, K.; Stensson, M.; Jansson, H.; Carda-Dieguez, M.; Pedersen, A.; Neilands, J.; Svensäter, G.; Mira, A. Bacterial Composition and Metabolomics of Dental Plaque from Adolescents. Front. Cell Infect. Microbiol. 2021, 11, 716493. [Google Scholar] [CrossRef]
- Mayumi, S.; Kuboniwa, M.; Sakanaka, A.; Hashino, E.; Ishikawa, A.; Ijima, Y.; and Amano, A. Potential of Prebiotic D-Tagatose for Prevention of Oral Disease. Front. Cell. Infect. Microbiol. 2021, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Barraza, J.P.; Arthur, R.A.; Hara, A.; Lewis, K.; Liu, Y.; Scisci, E.L.; Hajishengallis, E.; Whiteley, M.; Koo, H. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl. Acad. Sci. USA 2020, 117, 12375–12386. [Google Scholar] [CrossRef]
- Morse, D.J.; Smith, A.; Wilson, M.J.; Marsh, L.; White, L.; Posso, R.; Bradshaw, D.J.; Wei, X.; Lewis, M.A.; Williams, D.W. Molecular community profiling of the bacterial microbiota associated with denture-related stomatitis. Sci Rep. 2019, 9, 10228. [Google Scholar] [CrossRef] [PubMed]
- Rouabhia, M.; Chmielewski, W. Diseases Associated with Oral Polymicrobial Biofilms. Open Mycol. J. 2012, 6, 27–32. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.E.; Millhouse, E.; Sherry, L.; Kean, R.; Malcolm, J.; Nile, C.J.; and Ramage, G. Polymicrobial Candida biofilms: Friends and foe in the oral cavity. FEMS Yeast Res. 2015, 15, fov077. [Google Scholar] [CrossRef]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 2011, 6, e27317. [Google Scholar] [CrossRef]
- Andersen, B.M. Prevention of Postoperative Wound Infections. In Prevention and Control of Infections in Hospitals; Springer International Publishing: Cham, Switerland, 2019; pp. 377–437. [Google Scholar]
- Falanga, V. The chronic wound: Impaired healing and solutions in the context of wound bed preparation. Blood Cells Mol. Dis. 2004, 32, 88–94. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Rhoads, D.D. A study of biofilm-based wound management in subjects with critical limb ischaemia. J. Wound Care. 2008, 17, 145–155. [Google Scholar] [CrossRef]
- Metcalf, D.G.; Bowler, P.G. Biofilm delays wound healing: A review of the evidence. Burns Trauma. 2013, 1, 2321–3868. [Google Scholar] [CrossRef]
- Bessa, L.J.; Fazii, P.; Di Giulio, M.; Cellini, L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: Some remarks about wound infection. Int. Wound J. 2015, 12, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom Distribution of Pseudomonas aeruginosa and Staphylococcus aureus in Chronic Wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 2018, 76, fty003. [Google Scholar] [CrossRef]
- Fleming, D.; Chahin, L.; Rumbaugh, K. Glycoside Hydrolases Degrade Polymicrobial Bacterial Biofilms in Wounds. Antimicrob. Agents Chemother. 2017, 61, e01998-16. [Google Scholar] [CrossRef]
- Rondas, A.A.; Schols, J.M.; Stobberingh, E.E.; Halfens, R.J. Prevalence of chronic wounds and structural quality indicators of chronic wound care in Dutch nursing homes. Int. Wound J. 2015, 12, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Driver, V.R.; Blume, P.A. Evaluation of wound care and health-care use costs in patients with diabetic foot ulcers treated with negative pressure wound therapy versus advanced moist wound therapy. J. Am. Podiatr. Med. Assoc. 2015, 104, 147–153. [Google Scholar] [CrossRef]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. Apmis 2013, 121, 1–58. [Google Scholar] [CrossRef]
- Clinton, A.; Carter, T. Chronic wound biofilms: Pathogenesis and potential therapies. Lab. Med. 2015, 46, 277–284. [Google Scholar] [CrossRef]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef]
- Bertesteanu, S.; Triaridis, S.; Stankovic, M.; Lazar, V.; Chifiriuc, M.C.; Vlad, M.; Grigore, R. Polymicrobial wound infections: Pathophysiology and current therapeutic approaches. Int. J. Pharm. 2014, 463, 119–126. [Google Scholar] [CrossRef]
- Hrynyshyn, A.; Simões, M.; Borges, A. Biofilms in Surgical Site Infections: Recent Advances and Novel Prevention and Eradication Strategies. Antibiotics 2022, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, G.; Zavahir, J.S.; Bandara, W.M.M.S.; Weerasekara, M.L.M.A.W. Fungal-bacterial biofilms: Their development for novel biotechnological applications. World J. Microbiol. Biotechnol. 2008, 24, 739–743. [Google Scholar] [CrossRef]
- Dowd, S.E.; Delton Hanson, J.; Rees, E. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J. Wound Care. 2011, 20, 40–47. [Google Scholar] [CrossRef]
- Tkaczyk, C.; Jones-Nelson, O.; Shi, Y.Y.; Tabor, D.E.; Cheng, L.; Zhang, T.; Sellman, B.R. Neutralizing Staphylococcus aureus Virulence with AZD6389, a Three mAb Combination, Accelerates Closure of a Diabetic Polymicrobial Wound. mSphere 2022, 7, e00130-22. [Google Scholar] [CrossRef]
- Afonso, A.C.; Oliveira, D.; Saavedra, M.J.; Borges, A.; Simões, M. Biofilms in diabetic foot ulcers: Impact, risk factors and control strategies. Int. J. Mol. Sci. 2021, 22, 8278. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Veiga, A.S.; Tavares, L.; Castanho, M.; Oliveira, M. Bacterial Biofilms in Diabetic Foot Ulcers: Potential Alternative Therapeutics. In Microbial Biofilms—Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; InTechOpen: London, UK, 2016. [Google Scholar]
- Noor, S.; Zubair, M.; Ahmad, J. Diabetic foot ulcer—A review on pathophysiology, classification and microbial etiology. Diabetes Metab. Syndr. Clin. Res. Rev. 2015, 9, 192–199. [Google Scholar] [CrossRef]
- Lecube, A.; Pachón, G.; Petriz, J.; Hernández, C.; Simó, R. Phagocytic activity is impaired in type 2 diabetes mellitus and increases after metabolic improvement. PLoS ONE 2011, 6, e23366. [Google Scholar] [CrossRef]
- Gonzalez, M.R.; Ducret, V.; Leoni, S.; Fleuchot, B.; Jafari, P.; Raffoul, W.; Applegate, L.A.; Que, Y.A.; Perron, K. Transcriptome analysis of Pseudomonas aeruginosa cultured in human burn wound exudates. Front. Cell Infect. Microbiol. 2018, 8, 39. [Google Scholar] [CrossRef]
- Seth, A.K.; Geringer, M.R.; Hong, S.J.; Leung, K.P.; Galiano, R.D.; Mustoe, T.A. Comparative analysis of single-species and polybacterial wound biofilms using a quantitative, in vivo, rabbit ear model. PLoS ONE 2012, 7, e42897. [Google Scholar] [CrossRef]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of Methicillin Resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in Polymicrobial Wound Infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef]
- Plichta, J.K.; Gao, X.; Lin, H.; Dong, Q.; Toh, E.; Nelson, D.E.; Gamelli, R.L.; Grice, E.A.; Radek, K.A. Cutaneous Burn Injury Promotes Shifts in the Bacterial Microbiome in Autologous Donor Skin. Shock 2017, 48, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Elgharably, H.; Sinha, M.; Ganesh, K.; Chaney, S.; Mann, E.; Miller, C.; Khanna, S.; Bergdall, V.K.; Powell, H.M.; et al. Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J. Pathol. 2014, 233, 331–343. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.J.; Welch, M. Recapitulation of polymicrobial communities associated with cystic fibrosis airway infections: A perspective. Future Microbiol. 2019, 14, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Ibberson, C.B.; Stacy, A.; Fleming, D.; Dees, J.L.; Rumbaugh, K.; Gilmore, M.S.; Whiteley, M. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol. 2017, 2, 17079. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, K. Cystic fibrosis in the year 2020: A disease with a new face. Acta Paediatrica. 2020, 109, 893–899. [Google Scholar] [CrossRef]
- Van den Bossche, S.; De Broe, E.; Coenye, T.; Van Braeckel, E.; Crabbé, A. The cystic fibrosis lung microenvironment alters antibiotic activity: Causes and effects. Eur. Respir. Rev. 2021, 30, 210055. [Google Scholar] [CrossRef] [PubMed]
- Jean-Pierre, F.; Vyas, A.; Hampton, T.H.; Henson, M.A.; O’Toole, G.A. One versus many: Polymicrobial communities and the cystic fibrosis airway. MBio. 2021, 12, e00006-21. [Google Scholar] [CrossRef]
- Bluestone, C.D.; Gates, G.A.; Klein, J.O.; Bluestone, C.D.; Gates, G.A.; Klein, J.O.; Lim, D.J.; Mogi, G.; Ogra, P.L.; Paparella, M.M.; et al. Definitions, terminology, and classification of otitis media. Ann. Otol. Rhinol. Laryngol. 2002, 111, 8–18. [Google Scholar] [CrossRef]
- World Health Organization. Chronic Suppurative Otitis Media: Burden of Illness and Management Options. Available online: https://apps.who.int/iris/handle/10665/42941?locale-attribute=en&utm_source=transaction&utm_medium=email (accessed on 23 November 2022).
- Woodfield, G.; Dugdale, A. Evidence Behind the WHO Guidelines: Hospital Care for Children: What is the Most Effective Antibiotic Regime for Chronic Suppurative Otitis Media in Children? J. Trop. Pediatr. 2007, 54, 151–156. [Google Scholar] [CrossRef]
- Leach, A.J.; Morris, P.S.; Mathews, J.D. Compared to placebo, long-term antibiotics resolve otitis media with effusion (OME) and prevent acute otitis media with perforation (AOMwiP) in a high-risk population: A randomized controlled trial. BMC Pediatr. 2008, 8, 23. [Google Scholar] [CrossRef]
- Bair, K.L.; Shafirstein, G.; Campagnari, A.A. In vitro Photodynamic Therapy of Polymicrobial Biofilms Commonly Associated with Otitis Media. Front. Microbiol. 2020, 11, 558482. [Google Scholar] [CrossRef] [PubMed]
- Chonmaitree, T.; Revai, K.; Grady, J.J.; Clos, A.; Patel, J.A.; Nair, S.; Fan, J.; Henrickson, K.J. Viral upper respiratory tract infection and otitis media complication in young children. Clin. Infect. Dis. 2008, 46, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Massa, H.M.; Cripps, A.W.; Lehmann, D. Otitis media: Viruses, bacteria, biofilms and vaccines. Med. J. Aust. 2009, 191, S44–S49. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, M.G.; Kim, S.S.; Cha, S.H.; Yeo, S.G. Change in Detection Rate of Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa and Their Antibiotic Sensitivities in Patients with Chronic Suppurative Otitis Media. J. Int. Adv. Otol. 2015, 11, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.A. Pathogenesis of Polymicrobial Biofilms. Open Mycol. J. 2011, 5, 39–43. [Google Scholar] [CrossRef]
- Bakaletz, L.O. Developing animal models for polymicrobial diseases. Nat. Rev. Microbiol. 2004, 2, 552–568. [Google Scholar] [CrossRef]
- Brown, J.L.; Johnston, W.; Delaney, C.; Short, B.; Butcher, M.C.; Young, T.; Butcher, J.; Riggio, M.; Culshaw, S.; Ramage, G. Polymicrobial oral biofilm models: Simplifying the complex. J. Med. Microbiol. 2019, 68, 1573–1584. [Google Scholar] [CrossRef]
- Manavathu, E.K.; Vager, D.L.; Vazquez, J.A. Development and antimicrobial susceptibility studies of in vitro monomicrobial and polymicrobial biofilm models with Aspergillus fumigatus and Pseudomonas aeruginosa. BMC Microbiol. 2014, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Jordana-Lluch, E.; Garcia, V.; Kingdon, A.D.; Singh, N.; Alexander, C.; Williams, P.; Hardie, K.R. Simple Polymicrobial Biofilm Keratinocyte Colonization Model for Exploring Interactions Between Commensals, Pathogens and Antimicrobials. Front. Microbiol. 2020, 11, 291. [Google Scholar] [CrossRef]
- Sun, Y.; Smith, E.; Wolcott, R.; Dowd, S.E. Propagation of anaerobic bacteria within an aerobic multi-species chronic wound biofilm model. J Wound Care. 2009, 18, 426–431. [Google Scholar] [CrossRef]
- Brown, J.L.; Townsend, E.; Short, R.D.; Williams, C.; Woodall, C.; Nile, C.J.; Ramage, G. Assessing the inflammatory response to in vitro polymicrobial wound biofilms in a skin epidermis model. npj Biofilms Microbiomes 2022, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Fourie, R.; Albertyn, J.; Sebolai, O.; Gcilitshana, O.; Pohl, C.H. Candida albicans SET3 Plays a Role in Early Biofilm Formation, Interaction with Pseudomonas aeruginosa and Virulence in Caenorhabditis elegans. Front. Cell. Infect. Microbiol. 2021, 11, 680732. [Google Scholar] [CrossRef] [PubMed]
- Mastropaolo, M.D.; Evans, N.P.; Byrnes, M.K.; Stevens, A.M.; Robertson, J.L.; Melville, S.B. Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect Immun. 2005, 73, 6055–6063. [Google Scholar] [CrossRef] [PubMed]
- Daep, C.A.; Novak, E.A.; Lamont, R.J.; Demuth, D.R. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011, 79, 67–74. [Google Scholar] [CrossRef]
- Orth, R.K.H.; O’Brien-Simpson, N.M.; Dashper, S.G.; Reynolds, E.C. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol. Oral. Microbiol. 2011, 26, 229–240. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Hong, W.; Pang, B.; Weimer, K.E.D.; Juneau, R.A.; Turner, J.; Swords, W.E. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in Polymicrobial Otitis media occurs via interspecies quorum signaling. MBio 2010, 1, e00102-10. [Google Scholar] [CrossRef]
- Di Giulio, M.; Di Lodovico, S.; Fontana, A.; Traini, T.; Di Campli, E.; Pilato, S.; D’Ercole, S.; Cellini, L. Graphene Oxide affects Staphylococcus aureus and Pseudomonas aeruginosa dual species biofilm in Lubbock Chronic Wound Biofilm model. Sci. Rep. 2020, 10, 18525. [Google Scholar] [CrossRef]
- Kucera, J.; Sojka, M.; Pavlik, V.; Szuskiewicz, K.; Velebny, V.; Klein, P. Multispecies biofilm in an artificial wound bed—A novel model for in vitro assessment of solid antimicrobial dressings. J. Microbiol. Methods 2014, 103, 18–24. [Google Scholar] [CrossRef]
- Glavis-Bloom, J.; Muhammed, M.; Mylonakis, E. Of model hosts and man: Using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv. Exp. Med. Biol. 2012, 710, 11–17. [Google Scholar]
- Marsh, E.K.; May, R.C. Caenorhabditis elegans, a Model Organism for Investigating Immunity. Appl. Environ. Microbiol. 2012, 78, 2075–2081. [Google Scholar] [CrossRef]
- Holt, J.E.; Houston, A.; Adams, C.; Edwards, S.; Kjellerup, B.V. Role of extracellular polymeric substances in polymicrobial biofilm infections of Staphylococcus epidermidis and Candida albicans modelled in the nematode Caenorhabditis elegans. Pathog. Dis. 2017, 75, ftx052. [Google Scholar] [CrossRef] [PubMed]
- Bragonzi, A.; Farulla, I.; Paroni, M.; Twomey, K.B.; Pirone, L.; Lore, N.I.; Bianconi, I.; Dalmastri, C.; Ryan, R.P.; Bevivino, A. Modelling Co-Infection of the Cystic Fibrosis Lung by Pseudomonas aeruginosa and Burkholderia cenocepacia Reveals Influences on Biofilm Formation and Host Response. PLoS ONE 2012, 7, e52330. [Google Scholar] [CrossRef] [PubMed]
- Kesavalu, L.; Sathishkumar, S.; Bakthavatchalu, V.; Matthews, C.; Dawson, D.; Steffen, M.; Ebersole, J.L. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infect. Immun. 2007, 75, 1704–1712. [Google Scholar] [CrossRef]
- Kimizuka, R.; Kato, T.; Ishihara, K.; Okuda, K. Mixed infections with Porphyromonas gingivalis and Treponema denticola cause excessive inflammatory responses in a mouse pneumonia model compared with monoinfections. Microbes Infect. 2003, 5, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Chae, S.-W.; Go, Y.Y.; Im, G.J.; Song, J.-J. In vitro Multi-Species Biofilms of Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa and Their Host Interaction during In vivo Colonization of an Otitis Media Rat Model. Front. Cell. Infect. Microbiol. 2017, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Doualeh, M.; Payne, M.; Litton, E.; Raby, E.; Currie, A. Molecular Methodologies for Improved Polymicrobial Sepsis Diagnosis. Int. J. Mol. Sci. 2022, 23, 4484. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Evangelista III, J.S.; Schmieder, R.; Bailey, B.; Haynes, M.; Furlan, M.; Maughan, H.; Edwards, R.; Rohwer, F.; Conrad, D. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 2014, 52, 425–437. [Google Scholar] [CrossRef]
- Xu, P.; Gunsolley, J. Application of metagenomics in understanding oral health and disease. Virulence. 2014, 5, 424–432. [Google Scholar] [CrossRef]
- de Vos, M.G.; Zagorski, M.; McNally, A.; Bollenbach, T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. USA 2017, 114, 10666–10671. [Google Scholar] [CrossRef]
- Ai, D.; Huang, R.; Wen, J.; Li, C.; Zhu, J.; Xia, L.C. Integrated metagenomic data analysis demonstrates that a loss of diversity in oral microbiota is associated with periodontitis. BMC Genom. 2017, 18, 1041. [Google Scholar] [CrossRef]
- Suryaletha, K.; John, J.; Radhakrishnan, M.P.; George, S.; Thomas, S. Metataxonomic approach to decipher the polymicrobial burden in diabetic foot ulcer and its biofilm mode of infection. Int. Wound J. 2018, 15, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Ogarkov, O.; Khromova, P.; Sinkov, V.; Orlova, E.; Zhdanova, S.; Jiao, W.W.; Shen, A.; Mokrousov, I. Metagenomic analysis of the lung tuberculomas microbiome: Paucibacillary bacterial community. Int. J. Mycobacteriol. 2021, 10, 13. [Google Scholar] [CrossRef]
- Chen, M.F.; Chang, C.H.; Chiang-Ni, C.; Hsieh, P.H.; Shih, H.N.; Ueng, S.W.; Chang, Y. Rapid analysis of bacterial composition in prosthetic joint infection by 16S rRNA metagenomic sequencing. Bone Joint Res. 2019, 8, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Sengupta, D.J.; Rosenthal, C.; Costa, G.; Spangler, J.; Sims, E.H.; Jacobs, M.A.; Miller, S.I.; Hoogestraat, D.R.; Cookson, B.T.; et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS ONE 2013, 8, e65226. [Google Scholar] [CrossRef]
- Stebner, A.; Ensser, A.; Geißdörfer, W.; Bozhkov, Y.; Lang, R. Molecular diagnosis of polymicrobial brain abscesses with 16S-rDNA-based next-generation sequencing. Clin. Microbiol. Infect. 2021, 27, 2776–2782. [Google Scholar] [CrossRef]
- Ruppé, E.; Lazarevic, V.; Girard, M.; Mouton, W.; Ferry, T.; Laurent, F.; Schrenzel, J. Clinical metagenomics of bone and joint infections: A proof of concept study. Sci. Rep. 2017, 7, 7718. [Google Scholar] [CrossRef]
- Xie, F.; Duan, Z.; Zeng, W.; Xie, S.; Xie, M.; Fu, H.; Ye, Q.; Xu, T.; Xie, L. Clinical metagenomics assessments improve diagnosis and outcomes in community-acquired pneumonia. BMC Infect. Dis. 2021, 21, 352. [Google Scholar] [CrossRef]
- Tan, C.C.S.; Acman, M.; van Dorp, L.; Balloux, F. Metagenomic evidence for a polymicrobial signature of sepsis. Microb. Genom. 2021, 7, 000642. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, W.; Ye, K.; Guo, L.; Xia, H.; Guan, Y.; Chai, L.; Shi, W.; Zhai, C.; Wang, J.; et al. Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell. Infect. Microbiol. 2021, 11, 541092. [Google Scholar] [CrossRef]
- Nelson, M.T.; Pope, C.E.; Marsh, R.L.; Wolter, D.J.; Weiss, E.J.; Hager, K.R.; Vo, A.T.; Brittnacher, M.J.; Radey, M.C.; Hayden, H.S.; et al. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep. 2019, 26, 2227–2240. [Google Scholar] [CrossRef]
- Zhao, M.; Tang, K.; Liu, F.; Zhou, W.; Fan, J.; Yan, G.; Qin, S.; Pang, Y. Metagenomic next-generation sequencing improves diagnosis of osteoarticular infections from abscess specimens: A multicenter retrospective study. Front. Microbiol. 2020, 11, 2034. [Google Scholar] [CrossRef] [PubMed]
- Bijtenhoorn, P.; Mayerhofer, H.; Müller-Dieckmann, J.; Utpatel, C.; Schipper, C.; Hornung, C.; Szesny, M.; Grond, S.; Thürmer, A.; Brzuszkiewicz, E.; et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 2011, 6, e26278. [Google Scholar] [CrossRef] [PubMed]
- Schipper, C.; Hornung, C.; Bijtenhoorn, P.; Quitschau, M.; Grond, S.; Streit, W.R. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liang, M.; Wang, L.; Chen, R.; Li, H.; Liu, X. Aii810, a novel cold-adapted N-acylhomoserine lactonase discovered in a metagenome, can strongly attenuate Pseudomonas aeruginosa virulence factors and biofilm formation. Front. Microbiol. 2017, 8, 1950. [Google Scholar] [CrossRef] [PubMed]
- Bijtenhoorn, P.; Schipper, C.; Hornung, C.; Quitschau, M.; Grond, S.; Weiland, N.; Streit, W.R. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. 2011, 155, 86–94. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Hou, X.; Quan, C.; Chen, M. Widespread existence of quorum sensing inhibitors in marine bacteria: Potential drugs to combat pathogens with novel strategies. Mar. Drugs 2019, 17, 275. [Google Scholar] [CrossRef] [PubMed]
- Yaniv, K.; Golberg, K.; Kramarsky-Winter, E.; Marks, R.; Pushkarev, A.; Béjà, O.; Kushmaro, A. Functional marine metagenomic screening for anti-quorum sensing and anti-biofilm activity. Biofouling 2017, 33, 1–3. [Google Scholar]
- Gupta, S.; Sharma, A.K.; Jaiswal, S.K.; Sharma, V.K. Prediction of biofilm inhibiting peptides: An in silico approach. Front. Microbiol. 2016, 7, 949. [Google Scholar] [CrossRef]
- Atanaki, F.; Behrouzi, S.; Ariaeenejad, S.; Boroomand, A.; Kavousi, K. BIPEP: Sequence-based prediction of biofilm inhibitory peptides using a combination of NMR and physicochemical descriptors. ACS Omega 2020, 5, 7290–7297. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel Strategies for the Prevention and Treatment of Biofilm Related Infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural Anti-biofilm Agents: Strategies to Control Biofilm-Forming Pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; O’Toole, G.A. “It takes a village”: Mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J. Bacteriol. 2020, 202, e00530-19. [Google Scholar] [CrossRef] [PubMed]
- Weigel, L.M.; Donlan, R.M.; Shin, D.H.; Jensen, B.; Clark, N.C.; McDougal, L.K.; Zhu, W.; Musser, K.A.; Thompson, J.; Kohlerschmidt, D.; et al. High-Level Vancomycin-Resistant Staphylococcus aureus Isolates Associated with a Polymicrobial Biofilm. Antimicrob. Agents Chemother. 2007, 51, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Ready, D.; Pratten, J.; Roberts, A.P.; Bedi, R.; Mullany, P.; Wilson, M. Potential Role of Veillonella spp. as a Reservoir of Transferable Tetracycline Resistance in the Oral Cavity. Antimicrob. Agents Chemother. 2006, 50, 2866–2868. [Google Scholar] [CrossRef]
- Nabb, D.L.; Song, S.; Kluthe, K.E.; Daubert, T.A.; Luedtke, B.E.; Nuxoll, A.S. Polymicrobial Interactions Induce Multidrug Tolerance in Staphylococcus aureus Through Energy Depletion. Front. Microbiol. 2019, 10, 2803. [Google Scholar] [CrossRef]
- Sorg, R.A.; Lin, L.; van Doorn, G.S.; Sorg, M.; Olson, J.; Nizet, V.; Veening, J.-W. Collective Resistance in Microbial Communities by Intracellular Antibiotic Deactivation. PLoS Biol. 2016, 14, e2000631. [Google Scholar] [CrossRef]
- Kim, S.W.; Park, S.B.; Im, S.P.; Lee, J.S.; Jung, J.W.; Gong, T.W.; Lazarte, J.M.S.; Kim, J.; Seo, J.-S.; Kim, J.-H.; et al. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 2018, 8, 5402. [Google Scholar] [CrossRef]
- Perez, A.C.; Pang, B.; King, L.B.; Tan, L.; Murrah, K.A.; Reimche, J.L.; Wren, J.T.; Richardson, S.H.; Ghandi, U.; Swords, W.E. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog. Dis. 2014, 70, 280–288. [Google Scholar] [CrossRef]
- Dincer, S.; Uslu, F.M.; Delik, A. Antibiotic Resistance in Biofilm. In Bacterial Biofilms; IntechOpen: London, UK, 2020; Volume 13. [Google Scholar]
- Ma, S.; Moser, D.; Han, F.; Leonhard, M.; Schneider-Stickler, B.; Tan, T. Preparation and antibiofilm studies of curcumin loaded chitosan nanoparticles against polymicrobial biofilms of Candida albicans and Staphylococcus aureus. Carbohydr. Polym. 2020, 241, 116254. [Google Scholar] [CrossRef]
- Lara, H.H.; Lopez-Ribot, J.L. Inhibition of mixed biofilms of Candida albicans and methicillin-resistant Staphylococcus aureus by positively charged silver nanoparticles and functionalized silicone elastomers. Pathogens 2020, 9, 784. [Google Scholar] [CrossRef]
- Galdiero, E.; Ricciardelli, A.; D’Angelo, C.; de Alteriis, E.; Maione, A.; Albarano, L.; Casillo, A.; Corsaro, M.M.; Tutino, M.L.; Parrilli, E. Pentadecanoic acid against Candida albicans-Klebsiella pneumoniae biofilm: Towards the development of an anti-biofilm coating to prevent polymicrobial infections. Res. Microbiol. 2021, 172, 103880. [Google Scholar] [CrossRef] [PubMed]
- Scaffaro, R.; Lopresti, F.; D’Arrigo, M.; Marino, A.; Nostro, A. Efficacy of poly(lactic acid)/carvacrol electrospun membranes against Staphylococcus aureus and Candida albicans in single and mixed cultures. Appl. Microbiol. Biotechnol. 2018, 102, 4171–4181. [Google Scholar] [CrossRef] [PubMed]
- De Alteriis, E.; Lombardi, L.; Falanga, A.; Napolano, M.; Galdiero, S.; Siciliano, A.; Carotenuto, R.; Guida, M.; Galdiero, E. Polymicrobial antibiofilm activity of the membranotropic peptide gH625 and its analogue. Microb. Pathog. 2018, 125, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ai, L.; Zhang, Y.; Cheng, J.; Yu, H.; Li, C.; Zhang, D.; Pan, Y.; Lin, L. The Effects of Antimicrobial Peptide Nal-P-113 on Inhibiting Periodontal Pathogens and Improving Periodontal Status. BioMed Res. Int. 2018, 2018, 1805793. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Therapeutic Potential of Antimicrobial Peptides in Polymicrobial Biofilm-Associated Infections. Int. J. Mol. Sci. 2021, 22, 482. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic action of phage and antibiotics: Parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics 2019, 8, E103. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Wang, L.; Perka, C.; Trampuz, A.; Moreno, M.G. Using Bacteriophages as a Trojan Horse to the Killing of Dual-Species Biofilm Formed by Pseudomonas aeruginosa and Methicillin Resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 695. [Google Scholar] [CrossRef]
- Pekmezovic, M.; Aleksic, I.; Barac, A.; Arsic-Arsenijevic, V.; Vasiljevic, B.; Nikodinovic-Runic, J.; Senerovic, L. Prevention of polymicrobial biofilms composed of Pseudomonas aeruginosa and pathogenic fungi by essential oils from selected Citrus species. Pathog. Dis. 2021, 74, 102. [Google Scholar] [CrossRef]
- Dashper, S.; O’Brien-Simpson, N.; Liu, S.W.; Paolini, R.; Mitchell, H.; Walsh, K.; D’Cruze, T.; Hoffmann, B.; Catmull, D.; Zhu, Y.; et al. Oxantel Disrupts Polymicrobial Biofilm Development of Periodontal Pathogens. Antimicrob Agents Chemother. 2014, 58, 378–385. [Google Scholar] [CrossRef][Green Version]
- Luke-Marshall, N.R.; Hansen, L.A.; Shafirstein, G.; Campagnari, A.A. Antimicrobial Photodynamic Therapy with Chlorin e6 Is Bactericidal against Biofilms of the Primary Human Otopathogens. mSphere 2020, 5, e00492-20. [Google Scholar] [CrossRef]
- Diogo, P.; Fernandes, C.; Caramelo, F.; Mota, M.; Miranda, I.M.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Uliana, M.P.; de Oliveira, K.T.; Santos, J.M.; et al. Antimicrobial Photodynamic Therapy against Endodontic Enterococcus faecalis and Candida albicans Mono and Mixed Biofilms in the Presence of Photosensitizers: A Comparative Study with Classical Endodontic Irrigants. Front. Microbiol. 2017, 8, 498. [Google Scholar] [CrossRef] [PubMed]
- Hager, C.L.; Isham, N.; Schrom, K.P.; Chandra, J.; McCormick, T.; Miyagi, M.; Ghannoum, M.A. Effects of a Novel Probiotic Combination on Pathogenic Bacterial-Fungal Polymicrobial Biofilms. mBio 2019, 10, e00338-19. [Google Scholar] [CrossRef] [PubMed]
- Allaker, R.P. Nanoparticles and the Control of Oral Biofilms. In Nanobiomaterials in Clinical Dentistry, 1st ed.; Subramani, K., Ahmed, W., Hartsfield, J.K., Jr., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 2013, pp. 203–227. [Google Scholar]
- Monteiro, D.R.; Gorup, L.F.; Takamiya, A.S.; Ruvollo-Filho, A.C.; de Camargo, E.R.; Barbosa, D.B. The growing importance of materials that prevent microbial adhesion: Antimicrobial effect of medical devices containing silver. Int. J. Antimicrob. Agents. 2009, 34, 103–110. [Google Scholar] [CrossRef] [PubMed]
- de Jong, W.H.; Borm, P.J. Drug delivery and nanoparticles: Applications and hazards. Int. J. Nanomed. 2008, 3, 133–149. [Google Scholar]
- Han, C.; Romero, N.; Fischer, S.; Dookran, J.; Berger, A.; Doiron, A.L. Recent developments in the use of nanoparticles for treatment of biofilms. Nanotechnol. Rev. 2017, 6, 383–404. [Google Scholar] [CrossRef]
- Yasinta, M.S.; Ginawati, H.L.; Arum, N.A.; Hikmah, H.N.; Sumarsih, S.; Fahmi, M.Z.; Baktir, A. Dual Function of Silver Nanoparticles as Matrix Extracell Removal and Antimicrobial Agent in Polymycrobial Biofilms. Indones. J. Chem. 2021, 21, 286. [Google Scholar] [CrossRef]
- Bhatia, E.; Banerjee, R. Hybrid silver-gold nanoparticles suppress drug resistant polymicrobial biofilm formation and intracellular infection. J. Mater. Chem. B 2020, 8, 4890–4898. [Google Scholar] [CrossRef]
- Chong, W.X.; Lai, Y.X.; Choudhury, M.; Amalraj, F.D. Efficacy of incorporating silver nanoparticles into maxillofacial silicone against Staphylococcus aureus, Candida albicans, and polymicrobial biofilms. J. Prosthet. Dent. 2021, 128, 1114–1120. [Google Scholar] [CrossRef]
- Warrier, A.; Mazumder, N.; Prabhu, S.; Satyamoorthy, K.; Murali, T.S. Photodynamic therapy to control microbial biofilms. Photodiagnosis Photodyn. Ther. 2021, 33, 102090. [Google Scholar] [CrossRef]
- Hamblin, M.R. Antimicrobial photodynamic inactivation: A bright new technique to kill resistant microbes. Curr. Opin. Microbiol. 2016, 33, 67–73. [Google Scholar] [CrossRef]
- Wainwright, M.; Maisch, T.; Nonell, S.; Plaetzer, K.; Almeida, A.; Tegos, G.P.; Hamblin, M.R. Photoantimicrobials—Are we afraid of the light? Lancet Infect. Dis. 2017, 17, e49–e55. [Google Scholar] [CrossRef] [PubMed]
- Morley, S.; Griffiths, J.; Philips, G.; Moseley, H.; O’Grady, C.; Mellish, K.; Lankester, C.L.; Faris, B.; Young, R.J.; Brown, S.B.; et al. Phase IIa randomized, placebo-controlled study of antimicrobial photodynamic therapy in bacterially colonized, chronic leg ulcers and diabetic foot ulcers: A new approach to antimicrobial therapy. Br. J. Dermatol. 2013, 168, 617–624. [Google Scholar] [PubMed]
- Alvarenga, L.H.; Gomes, A.C.; Carribeiro, P.; Godoy-Miranda, B.; Noschese, G.; Ribeiro, M.S.; Kato, I.T.; Bussadori, S.K.; Pavani, C.; Gearldo, Y.G.E.; et al. Parameters for antimicrobial photodynamic therapy on periodontal pocket—Randomized clinical trial. Photodiagn. Photodyn. Ther. 2019, 27, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.B.; Costa, D.H.; Miyakawa, W.; Delgado, M.G.T.; Garcez, A.S.; Yoshimura, T.M.; Ribeiro, M.S.; Nunez, S.C. Photodynamic Activity on Biofilm in Endotracheal Tubes of Patients Admitted to an Intensive Care Unit. Photochem. Photobiol. 2020, 96, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Fimple, J.L.; Fontana, C.R.; Foschi, F.; Ruggeiro, K.; Song, X.; Pagonis, T.C.; Tanner, A.C.R.; Kent, R.; Doukas, A.G.; Stashenko, P.P. Photodynamic Treatment of Endodontic Polymicrobial Infection In Vitro. J. Endod. 2008, 34, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liang, Q.; Su, G.; Zhang, Y.; Wang, Z.; Baudouin, C.; Labbe, A. In vitro effect of toluidine blue antimicrobial photodynamic chemotherapy on Staphylococcus epidermidis and Staphylococcus aureus isolated from ocular surface infection. Transl. Vis. Sci. Technol. 2019, 8, 45. [Google Scholar] [CrossRef]
- Akhtar, F.; Khan, A.U.; Qazi, B.; Kulanthaivel, S.; Mishra, P.; Akhtar, K.; Ali, A. A nano phototheranostic approach of toluidine blue conjugated gold silver core shells mediated photodynamic therapy to treat diabetic foot ulcer. Sci. Rep. 2021, 11, 24464. [Google Scholar] [CrossRef]
- Usacheva, M.; Layek, B.; Rahman Nirzhor, S.S.; Prabha, S. Nanoparticle-Mediated Photodynamic Therapy for Mixed Biofilms. J. Nanomater. 2016, 2016, 4752894. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Scott, M.G. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA 2000, 97, 8856–8861. [Google Scholar] [CrossRef]
- Park, S.C.; Park, Y.; Hahm, K.S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef]
- di Luca, M.; Maccari, G.; Nifosí, R. Treatment of microbial biofilms in the post-antibiotic era: Prophylactic and therapeutic use of antimicrobial peptides and their design by bioinformatics tools. Pathog. Dis. 2014, 70, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Duplantier, A.J.; van Hoek, M.L. The human cathelicidin antimicrobial peptide LL-37 as a potential treatment for polymicrobial infected wounds. Front. Immunol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.W.; Rehm, B.H.A.; Hancock, R.E.W. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.S.; Jenssen, H. Human cathelicidin LL-37 prevents bacterial biofilm formation. Future Med. Chem. 2012, 4, 1587–1599. [Google Scholar] [CrossRef]
- Lee, J.K.; Chang, S.W.; Perinpanayagam, H.; Lim, S.-M.; Park, Y.-J.; Han, S.H.; Baek, S.-H.; Zhu, Q.; Bae, K.-S.; Kum, K.-Y. Antibacterial efficacy of a human β-defensin-3 peptide on multispecies biofilms. J. Endod. 2013, 39, 1625–1629. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Brackman, G.; Coenye, T. Quorum Sensing Inhibitors as Anti-Biofilm Agents. Curr. Pharm. Des. 2014, 21, 5–11. [Google Scholar] [CrossRef]
- Edris, A.E. Pharmaceutical and therapeutic potentials of essential oils and their individual volatile constituents: A review. Phyther. Res. 2007, 21, 308–323. [Google Scholar] [CrossRef]
- Murray, E.J.; Dubern, J.-F.; Chan, W.C.; Chhabra, S.R.; Williams, P. A Pseudomonas aeruginosa PQS quorum-sensing system inhibitor with anti-staphylococcal activity sensitizes polymicrobial biofilms to tobramycin. Cell Chem. Biol. 2022, 29, 1187–1199. [Google Scholar] [CrossRef]
- Desbois, A.P.; Smith, V.J. Antibacterial free fatty acids: Activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. [Google Scholar] [CrossRef]
- Kim, Y.G.; Lee, J.H.; Park, S.; Kim, S.; Lee, J. Inhibition of polymicrobial biofilm formation by saw palmetto oil, lauric acid and myristic acid. Microb. Biotechnol. 2022, 15, 590–602. [Google Scholar] [CrossRef]
- Kumar, P.; Lee, J.H.; Beyenal, H.; Lee, J. Fatty Acids as Antibiofilm and Antivirulence Agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases—Novel tools for study of bacterial biofilms. J. Appl. Microbiol. 1998, 85, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Hardy, L.; Cerca, N.; Jespers, V.; Vaneechoutte, M.; Crucitti, T. Bacterial biofilms in the vagina. Res. Microbiol. 2017, 168, 865–874. [Google Scholar] [CrossRef]
- Ryan, E.M.; Gorman, S.P.; Donnelly, R.F.; Gilmore, B.F. Recent advances in bacteriophage therapy: How delivery routes, formulation, concentration and timing influence the success of phage therapy. J. Pharm. Pharmacol. 2011, 63, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Donlan, R.M. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob. Agents Chemother. 2015, 59, 1127–1137. [Google Scholar] [CrossRef]
- Chhibber, S.; Bansal, S.; Kaur, S. Disrupting the mixed-species biofilm of Klebsiella pneumoniae B5055 and Pseudomonas aeruginosa PAO using bacteriophages alone or in combination with xylitol. Microbiology 2015, 161, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhuyse, B.; Galant, C.; Brichard, B.; Docquier, P.-S.; Djebara, S.; Pirnay, J.-P.; der Linden, D.V.; Merabishvili, M.; Chatzis, O. A case of in situ phage therapy against Staphylococcus aureus in a bone allograft polymicrobial biofilm infection: Outcomes and phage-antibiotic interactions. Viruses 2021, 13, 1898. [Google Scholar] [CrossRef]
- Lin, X.; Chen, X.; Chen, Y.; Jiang, W.; Chen, H. The effect of five probiotic lactobacilli strains on the growth and biofilm formation of Streptococcus mutans. Oral Dis. 2015, 21, e128–e134. [Google Scholar] [CrossRef]
- Matsubara, V.H.; Wang, Y.; Bandara, H.M.H.N.; Mayer, M.P.A.; Samaranayake, L.P. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl. Microbiol. Biotechnol. 2016, 100, 6415–6426. [Google Scholar] [CrossRef]
- Tan, Y.; Leonhard, M.; Moser, D.; Schneider-Stickler, B. Inhibition activity of Lactobacilli supernatant against fungal-bacterial multispecies biofilms on silicone. Microb. Pathog. 2017, 113, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Bidossi, A.; De Grandi, R.; Toscano, M.; Bottagisio, M.; De Vecchi, E.; Gelardi, M.; Drago, L. Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect. Dis. 2018, 18, 653. [Google Scholar] [CrossRef] [PubMed]



| Infections | Microorganisms | In Vitro/In Vivo/Model Systems | References |
|---|---|---|---|
| Skin infections by Staphylococcus aureus and Pseudomonas aeruginosa | Commensal, Staphylococcus epidermidis and Micrococcus luteus and pathogenic Staphylococcus aureus and Pseudomonas aeruginosa | Immortalized keratinocytes (HaCat cells) | [102] |
| Chronic wound infections | MRSA, vancomycin-resistant Enterococcus faecalis (VRE) and P. aeruginosa | Lubbock Chronic Wound Biofilm (LCWB) model | [103] |
| Wound infections | Complex polymicrobial biofilms containing Candida albicans, P. aeruginosa, S. aureus, Staphylococcus hominis, Corynebacterium simulans, Streptococcus agalactiae, Finegoldia magna, Prevotella buccalis, Porphyromonas asaccharolytica, Anaerococcus vaginalis, and Peptoniphilus gorbachii | Skin epidermis model | [104] |
| Observed in the lungs of cystic fibrosis patients | C. albicans and P. aeruginosa | Caenorhabditis elegans (Nematode) | [105] |
| Chronic wound infections | S. aureus, P. aeruginosa, E. faecalis and Finegoldia magna | Mice | [57] |
| Diabetes-associated manifestations such as lower-limb amputations due to wound infections | Escherichia coli, Bacteroides fragilis, and Clostridium perfringens | Human type 2 diabetes model of mice | [106] |
| Periodontal disease | Porphyromonas gingivalis and Streptococcus gordonii | Murine model of periodontitis | [107] |
| Chronic periodontitis | P. gingivalis and Treponema denticola | Murine model of periodontitis | [108] |
| Otitis media | Haemophilus influenzae and Moraxella catarrhalis | Chinchilla infection model of otitis media | [109] |
| Acute otitis media | Moraxella catarrhalis, Streptococcus pneumoniae and non-typeable H. influenza | In vitro nasopharyngeal colonization model | [32] |
| Strategies | Polymicrobial Biofilm | Mechanism | References |
|---|---|---|---|
| Curcumin loaded with chitosan nanoparticle | Candida albicans and Staphylococcus aureus | Biofilm disruption | [153] |
| AgNP functionalised silicone elastomer | C. albicans and MRSA | Biofilm inhibition | [154] |
| Pentadecanoic acid coated on polydimethylsiloxane (PDMS) surface | C. albicans-Klebsiella pneumoniae | Polymicrobial biofilm prevention | [155] |
| Electrospun membranes of poly (lactic acid) and carvacrol | C. albicans and S. aureus | Decrease in the CFUs, biomass, and metabolic activity of 24- and 48-h biofilms in both single and mixed biofilms | [156] |
| Gh625-GCGKKK Peptide | Candida tropicalis–Serratia marcescens and C. tropicalis–S. aureus | Reduced biofilm architecture, interfering cell adhesion, prevention of long-term formation of polymicrobial biofilm on silicone surface | [157] |
| Synthetic cationic AMP, Nal-P-113 | Streptococcus gordonii, Fusobacterium nucleatum, Porphyromonas gingivalis, | Bactericidal activity in both planktonic and polymicrobial biofilm states | [158] |
| Cholic acid peptide conjugates (CAPs) | C. albicans and S. aureus | Reduces interkingdom polymicrobial biofilm formation and also active towards persister cells as well as stationery cells | [159] |
| Lytic phage, EPA1 with antibiotics such as (gentamicin, kanamycin, tetracycline, chloramphenicol, erythromycin, ciprofloxacin, and meropenem) | P. aeruginosa and S. aureus | Mono and dual species biofilm Inhibition | [160] |
| Phages, PYO and Sb-1 with Ciprofloxacin | P. aeruginosa and S. aureus | Biofilm matrix inhibition | [161] |
| Pompia and grapefruit essential oils | P. aeruginosa, Aspergillus fumigatus or Scedosporium apiospermum | Mono and Polymicrobial biofilm inhibition | [162] |
| Oxantel | Treponema denticola, Porphyromonas gingivalis and Tannerella forsythia. | Polymicrobial biofilm disruption | [163] |
| Antimicrobial Photodynamic therapy- Chlorin e6 | H. influenzae, S. pneumoniae and M. catarrhalis | Biofilm disruption | [164] |
| Zn(II)chlorin e6 methyl ester (Zn(II)e6Me) | Enterococcus faecalis and C. albicans | Loss of biofilm biomass | [165] |
| Probiotics, Saccharomyces boulardii, Lactobacillus acidophilus, Bifidobacterium breve, and Lactobacillus rhamnosus | C. albicans or C. tropicalis combined with E. coli and S. marcescens | Inhibition of candidal pathogenic determinants, prevent adhesion and biofilm formation | [166] |
| Glycoside hydrolases, α-amylase and cellulase | P. aeruginosa and S. aureus | Breakdown of complex sugars and disruption of mono and coculture biofilm | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1731. https://doi.org/10.3390/antibiotics11121731
Anju VT, Busi S, Imchen M, Kumavath R, Mohan MS, Salim SA, Subhaswaraj P, Dyavaiah M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics. 2022; 11(12):1731. https://doi.org/10.3390/antibiotics11121731
Chicago/Turabian StyleAnju, V T, Siddhardha Busi, Madangchanok Imchen, Ranjith Kumavath, Mahima S. Mohan, Simi Asma Salim, Pattnaik Subhaswaraj, and Madhu Dyavaiah. 2022. "Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies" Antibiotics 11, no. 12: 1731. https://doi.org/10.3390/antibiotics11121731
APA StyleAnju, V. T., Busi, S., Imchen, M., Kumavath, R., Mohan, M. S., Salim, S. A., Subhaswaraj, P., & Dyavaiah, M. (2022). Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics, 11(12), 1731. https://doi.org/10.3390/antibiotics11121731







