Bactericidal and Anti-Biofilm Activity of the FtsZ Inhibitor C109 against Acinetobacter baumannii
Abstract
:1. Introduction
2. Results
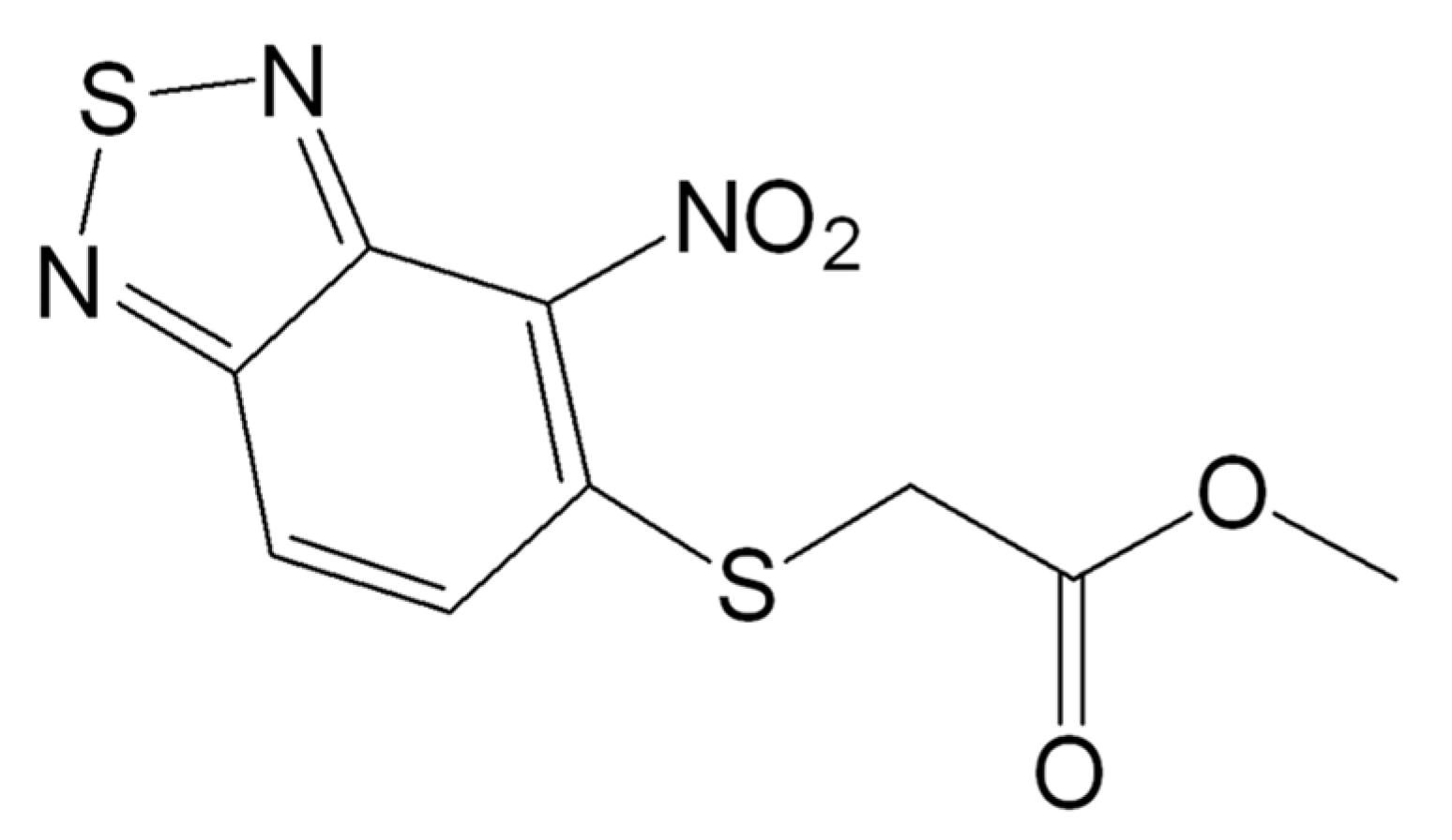
2.1. Activity of C109 against Sensitive and Resistant A. baumannii Strains
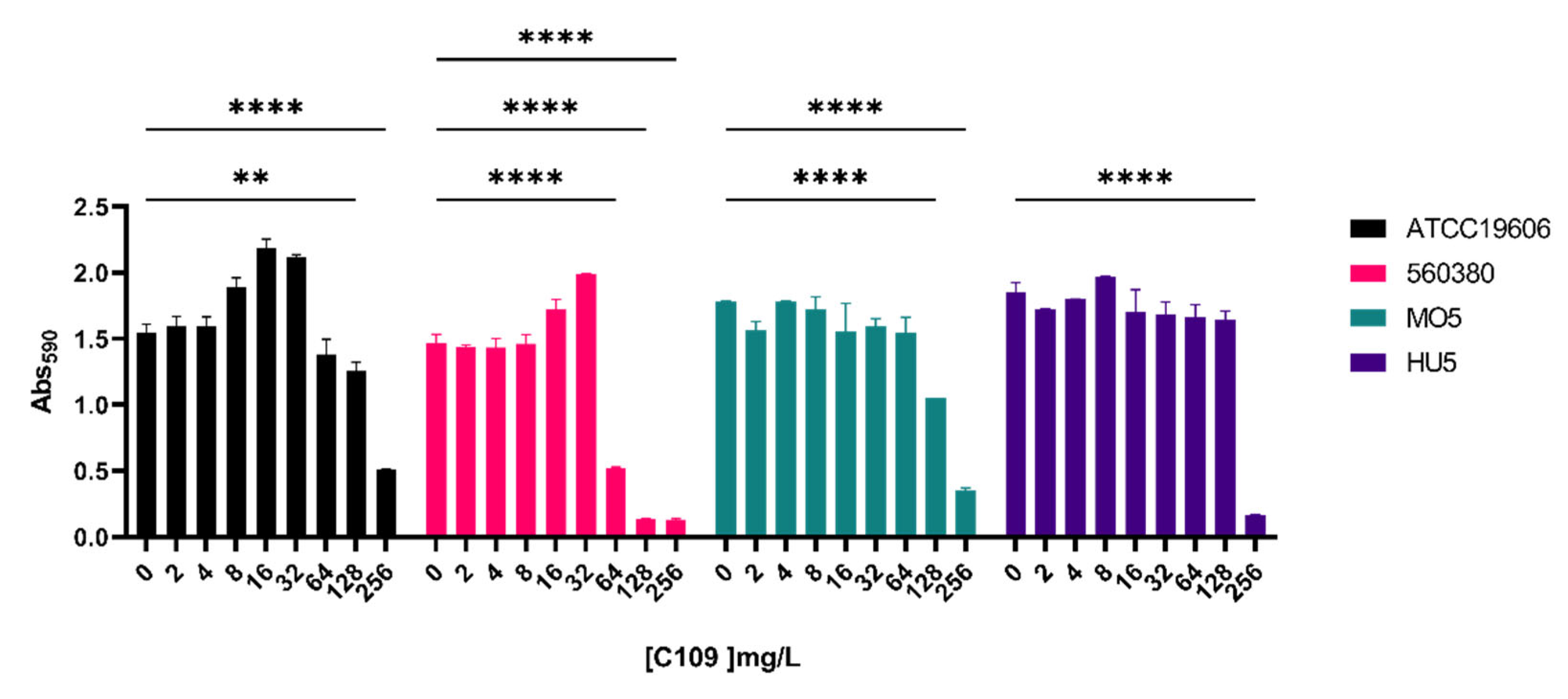
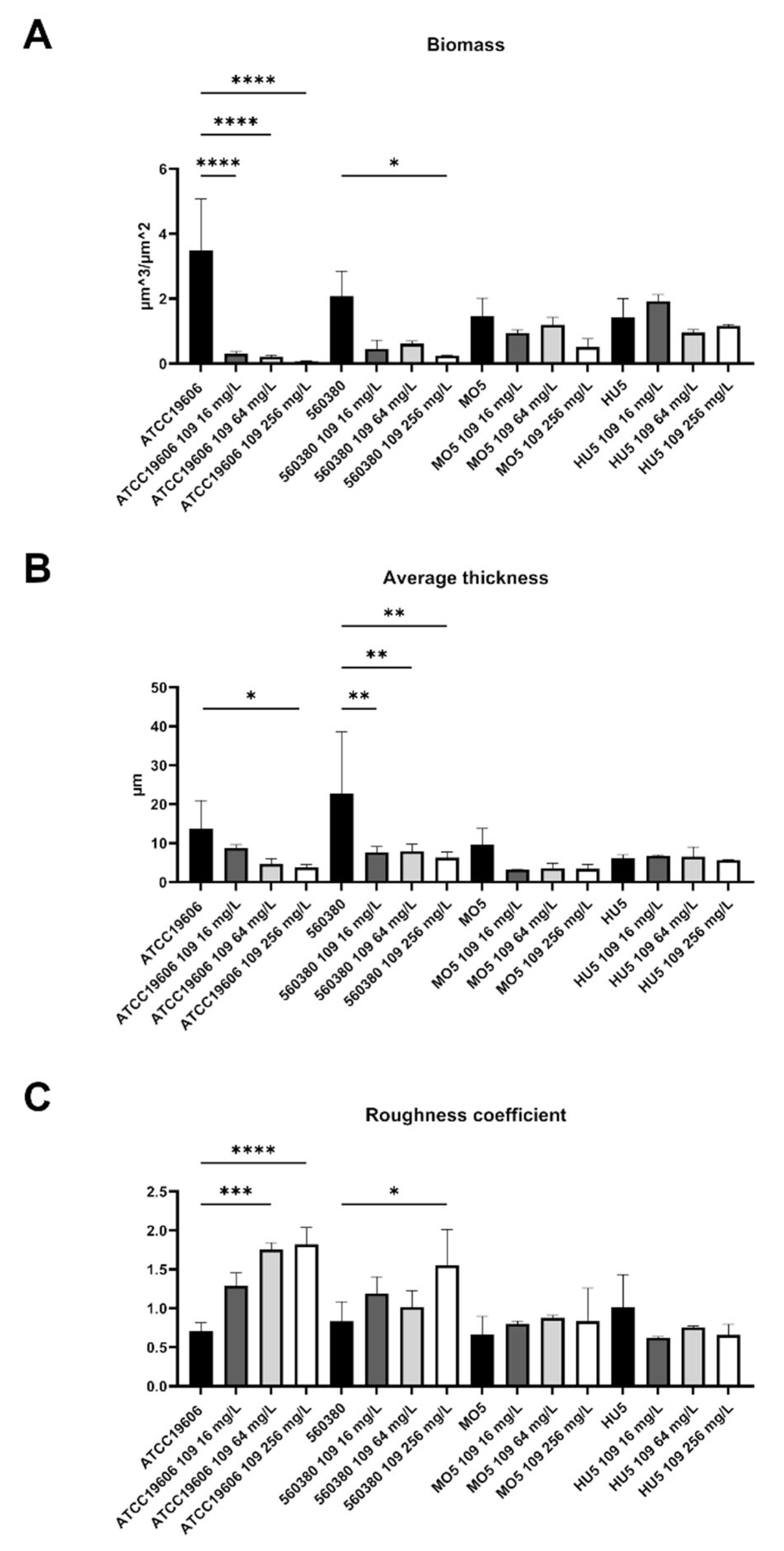
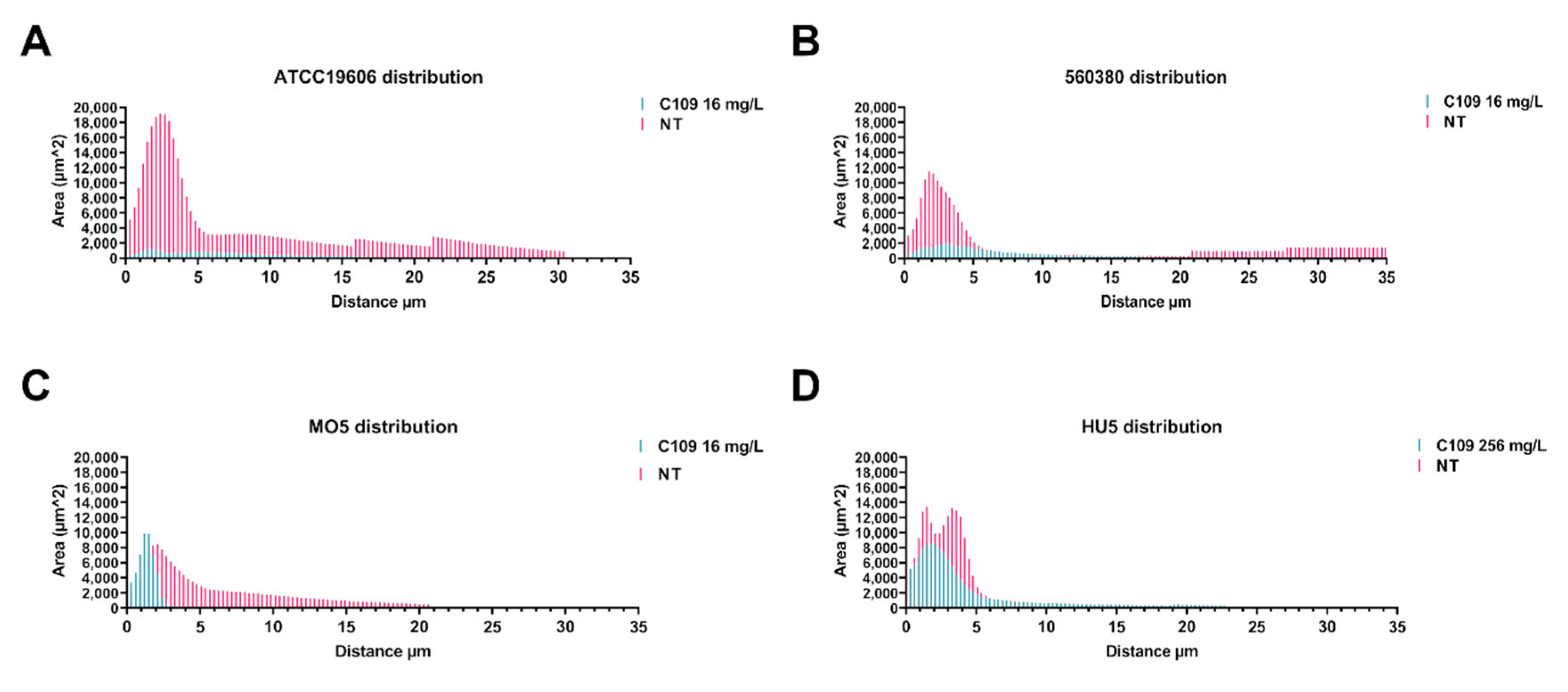
2.2. C109 Biofilm Inhibitory Activity against A. baumannii Strains
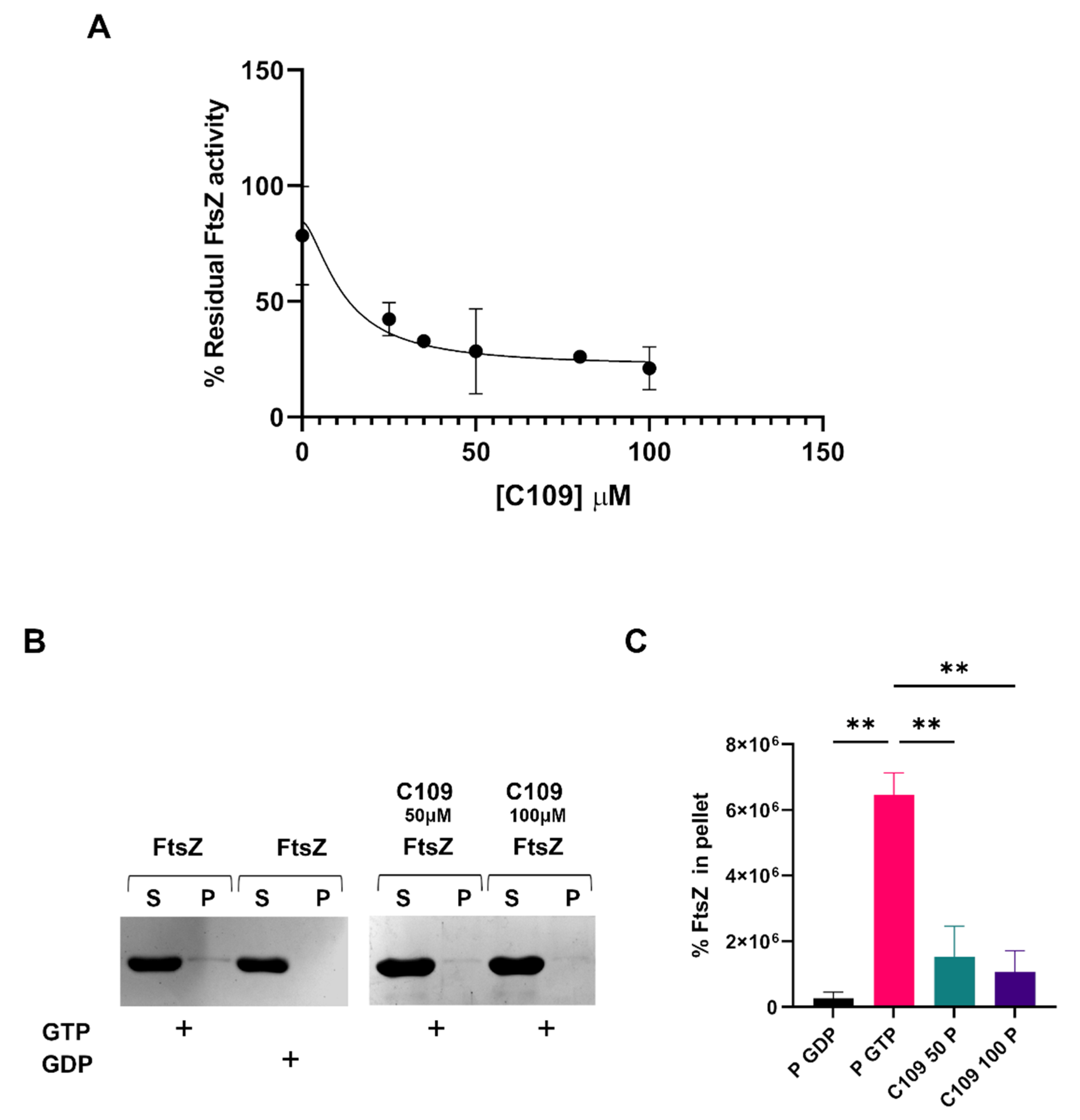
2.3. C109 Activity on Purified A. baumannii FtsZ
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Media
4.2. A. baumannii Clinical Isolates Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Determination
4.3. In Vitro Biofilm Inhibition Test in 96-Well Microtiter Plates
4.4. Biofilm Evaluation by Confocal Laser Scanning Microscopy
4.5. Cloning, Expression and Purification of the A. baumannii FtsZ Protein
4.6. In Vitro FtsZ GTPase Activity
4.7. In Vitro FtsZ Polymerization Assay
4.8. Statistical Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ibrahim, S.; Al-Saryi, N.; Al-Kadmy, I.M.S.; Aziz, S.N. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 2021, 48, 6987–6998. [Google Scholar] [CrossRef]
- Brasiliense, D.; Cayô, R.; Streling, A.P.; Nodari, C.S.; Souza, C.; Leal, C.; Gales, A.C. Outbreak of Acinetobacter colistiniresistens bloodstream infections in a neonatal intensive care unit. J. Glob. Antimicrob. Resist. 2021, 24, 257–259. [Google Scholar] [CrossRef]
- Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Bravata-Alcantará, J.C.; Sosa-Hernández, O.; Delgado-Balbuena, L.; León-García, G.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Castro-Escarpulli, G.; et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries 2021, 15, 58–68. [Google Scholar] [CrossRef]
- Jean, S.S.; Chang, Y.C.; Lin, W.C.; Lee, W.S.; Hsueh, P.R.; Hsu, C.W. Epidemiology, treatment, and prevention of nosocomial bacterial pneumonia. J. Clin. Med. 2020, 9, 275. [Google Scholar] [CrossRef] [Green Version]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021, 10, 373. [Google Scholar] [CrossRef]
- Hassan, R.M.; Salem, S.T.; Hassan, S.I.M.; Hegab, A.S.; Elkholy, Y.S. Molecular characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates from Egyptian patients. PLoS ONE 2021, 16, e0251508. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Stracquadanio, S.; Bonomo, C.; Marino, A.; Bongiorno, D.; Privitera, G.F.; Bivona, D.A.; Mirabile, A.; Bonacci, P.G.; Stefani, S. Acinetobacter baumannii and cefiderocol, between cidality and adaptability. Microbiol. Spectr. 2022, 10, e0234722. [Google Scholar] [CrossRef]
- Katip, W.; Meechoui, M.; Thawornwittayakom, P.; Chinwong, D.; Oberdorfer, P. Efficacy and safety of high loading dose of colistin in Multidrug-Resistant Acinetobacter baumannii: A prospective cohort study. J. Intensive Care Med. 2019, 34, 996–1002. [Google Scholar] [CrossRef]
- Sheldon, J.R.; Skaar, E.P. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, e1008995. [Google Scholar] [CrossRef]
- Kumar, S.; Answer, R.; Azzi, A. Virulence potential and treatment options of multidrug-resistant (MDR) Acinetobacter baumannii. Microorganisms 2021, 9, 2104. [Google Scholar] [CrossRef]
- Kim, S.; Lee, D.W.; Jin, J.S.; Kim, J. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Glob. Antimicrob. Resist. 2020, 22, 32–39. [Google Scholar] [CrossRef]
- Styles, K.M.; Thummeepak, R.; Leungtongkam, U.; Smith, S.E.; Christie, G.S.; Millard, A.; Moat, J.; Dowson, C.G.; Wellington, E.M.H.; Sitthisak, S.; et al. Investigating bacteriophages targeting the opportunistic pathogen Acinetobacter baumannii. Antibiotics 2020, 9, 200. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [Green Version]
- Wachino, J.I.; Jin, W.; Kimura, K.; Arakawa, Y. Intercellular transfer of chromosomal antimicrobial resistance genes between Acinetobacter baumannii strains mediated by prophages. Antimicrob. Agents Chemother. 2019, 63, e00334-19. [Google Scholar] [CrossRef] [Green Version]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides: New hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef]
- Chen, S.P.; Chen, E.H.; Yang, S.Y.; Kuo, P.S.; Jan, H.M.; Yang, T.C.; Hsieh, M.Y.; Lee, K.T.; Lin, C.H.; Chen, R.P. A systematic study of the stability, safety, and efficacy of the de novo designed antimicrobial peptide PepD2 and its modified derivatives against Acinetobacter baumannii. Front. Microbiol. 2021, 12, 678330. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Li, L.; Zhou, Y.; Kraus, C.N. Efficacy of ARV-1502, a proline-rich antimicrobial peptide, in a murine model of bacteremia caused by Multi-Drug Resistant (MDR) Acinetobacter baumannii. Molecules. 2019, 24, 2820. [Google Scholar] [CrossRef] [Green Version]
- de Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef] [Green Version]
- Spencer, J.J.; Pitts, R.E.; Pearson, R.A.; King, L.B. The effects of antimicrobial peptides WAM-1 and LL-37 on multidrug-resistant Acinetobacter baumannii. Pathog. Dis. 2018, 76, fty007. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Grygorcewicz, B.; Roszak, M.; Golec, P.; Śleboda-Taront, D.; Łubowska, N.; Górska, M.; Jursa-Kulesza, J.; Rakoczy, R.; Wojciuk, B.; Dołęgowska, B. Antibiotics act with vB_AbaP_AGC01 phage against Acinetobacter baumannii in human heat-inactivated plasma blood and Galleria mellonella models. Int. J. Mol. Sci. 2020, 21, 4390. [Google Scholar] [CrossRef] [PubMed]
- Zharkova, M.S.; Orlov, D.S.; Golubeva, O.Y.; Chakchir, O.B.; Eliseev, I.E.; Grinchuk, T.M.; Shamova, O.V. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics-A novel way to combat antibiotic resistance? Front. Cell Infect Microbiol. 2019, 9, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulussen, F.M.; Schouten, G.K.; Moertl, C.; Verheul, J.; Hoekstra, I.; Koningstein, G.M.; Hutchins, G.H.; Alkir, A.; Luirink, R.A.; Geerke, D.P.; et al. Covalent proteomimetic inhibitor of the bacterial FtsQB divisome complex. J. Am. Chem. Soc. 2022, 144, 15303–15313. [Google Scholar] [CrossRef] [PubMed]
- Di Somma, A.; Avitabile, C.; Cirillo, A.; Moretta, A.; Merlino, A.; Paduano, L.; Duilio, A.; Romanelli, A. The antimicrobial peptide Temporin L impairs E. coli cell division by interacting with FtsZ and the divisome complex. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129606. [Google Scholar] [CrossRef] [PubMed]
- Trespidi, G.; Scoffone, V.C.; Barbieri, G.; Riccardi, G.; De Rossi, E.; Buroni, S. Molecular characterization of the Burkholderia cenocepacia dcw operon and FtsZ interactors as new targets for novel antimicrobial design. Antibiotics 2020, 9, 841. [Google Scholar] [CrossRef]
- Kumar, M.; Mathur, T.; Barman, T.K.; Chaira, T.; Kumar, R.; Joshi, V.; Pandya, M.; Sharma, L.; Fujii, K.; Bandgar, M.; et al. Novel FtsZ inhibitor with potent activity against Staphylococcus aureus. J. Antimicrob. Chemother. 2021, 76, 2867–2874. [Google Scholar] [CrossRef]
- Monterroso, B.; Zorrilla, S.; Sobrinos-Sanguino, M.; Robles-Ramos, M.A.; López-Álvarez, M.; Margolin, W.; Keating, C.D.; Rivas, G. Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 2019, 20, e45946. [Google Scholar] [CrossRef]
- Scoffone, V.C.; Ryabova, O.; Makarov, V.; Iadarola, P.; Fumagalli, M.; Fondi, M.; Fani, R.; De Rossi, E.; Riccardi, G.; Buroni, S. Efflux-mediated resistance to a benzothiadiazol derivative effective against Burkholderia cenocepacia. Front. Microbiol. 2015, 6, 815. [Google Scholar] [CrossRef] [Green Version]
- Hogan, A.M.; Scoffone, V.C.; Makarov, V.; Gislason, A.S.; Tesfu, H.; Stietz, M.S.; Brassinga, A.K.C.; Domaratzki, M.; Li, X.; Azzalin, A.; et al. Competitive fitness of essential gene knockdowns reveals a broad-spectrum antibacterial inhibitor of the cell division protein FtsZ. Antimicrob. Agents Chemother. 2018, 62, e01231-18. [Google Scholar] [CrossRef]
- Chiarelli, L.R.; Scoffone, V.C.; Trespidi, G.; Barbieri, G.; Riabova, O.; Monakhova, N.; Porta, A.; Manina, G.; Riccardi, G.; Makarov, V.; et al. Chemical, Metabolic, and Cellular Characterization of a FtsZ Inhibitor Effective Against Burkholderia cenocepacia. Front. Microbiol. 2020, 11, 562. [Google Scholar] [CrossRef]
- Costabile, G.; Provenzano, R.; Azzalin, A.; Scoffone, V.C.; Chiarelli, L.R.; Rondelli, V.; Grillo, I.; Zinn, T.; Lepioshkin, A.; Savina, S.; et al. PEGylated mucus-penetrating nanocrystals for lung delivery of a new FtsZ inhibitor against Burkholderia cenocepacia infection. Nanomed. J. 2020, 23, 102–113. [Google Scholar] [CrossRef]
- Trespidi, G.; Scoffone, V.C.; Barbieri, G.; Marchesini, F.; Abualsha’ar, A.; Coenye, T.; Ungaro, F.; Makarov, V.; Migliavacca, R.; De Rossi, E.; et al. Antistaphylococcal activity of the FtsZ inhibitor C109. Pathogens 2021, 10, 886. [Google Scholar] [CrossRef]
- Vandecandelaere, I.; Van Acker, H.; Coenye, T. A microplate-based system as in vitro model of biofilm growth and quantification. Methods Mol. Biol. 2016, 1333, 53–66. [Google Scholar]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [Green Version]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Tomlinson, B.R.; Denham, G.A.; Torres, N.J.; Brzozowski, R.S.; Allen, J.L.; Jackson, J.K.; Eswara, P.J.; Shaw, L.N. Assessing the role of cold-shock protein C: A novel regulator of Acinetobacter baumannii biofilm formation and virulence. Infect. Immun. 2022, 90, e0037622. [Google Scholar] [CrossRef]
- Chai, W.C.; Whittall, J.J.; Polyak, S.W.; Foo, K.; Li, X.; Dutschke, C.J.; Ogunniyi, A.D.; Ma, S.; Sykes, M.J.; Semple, S.J.; et al. Cinnamaldehyde derivatives act as antimicrobial agents against Acinetobacter baumannii through the inhibition of cell division. Front. Microbiol. 2022, 29, 967949. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. EUCAST Reading Guide for Broth Microdilution. Version 2.0. 2022. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2022_manuals/Reading_guide_BMD_v_4.0_2022.pdf (accessed on 1 January 2022).
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef] [Green Version]
- Eales, M.G.; Ferrari, E.; Goddard, A.D.; Lancaster, L.; Sanderson, P.; Miller, C. Mechanistic and phenotypic studies of bicarinalin, BP100 and colistin action on Acinetobacter baumannii. Res. Microbiol. 2018, 169, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scoffone, V.C.; Irudal, S.; AbuAlshaar, A.; Piazza, A.; Trespidi, G.; Barbieri, G.; Makarov, V.; Migliavacca, R.; De Rossi, E.; Buroni, S. Bactericidal and Anti-Biofilm Activity of the FtsZ Inhibitor C109 against Acinetobacter baumannii. Antibiotics 2022, 11, 1571. https://doi.org/10.3390/antibiotics11111571
Scoffone VC, Irudal S, AbuAlshaar A, Piazza A, Trespidi G, Barbieri G, Makarov V, Migliavacca R, De Rossi E, Buroni S. Bactericidal and Anti-Biofilm Activity of the FtsZ Inhibitor C109 against Acinetobacter baumannii. Antibiotics. 2022; 11(11):1571. https://doi.org/10.3390/antibiotics11111571
Chicago/Turabian StyleScoffone, Viola Camilla, Samuele Irudal, Aseel AbuAlshaar, Aurora Piazza, Gabriele Trespidi, Giulia Barbieri, Vadim Makarov, Roberta Migliavacca, Edda De Rossi, and Silvia Buroni. 2022. "Bactericidal and Anti-Biofilm Activity of the FtsZ Inhibitor C109 against Acinetobacter baumannii" Antibiotics 11, no. 11: 1571. https://doi.org/10.3390/antibiotics11111571
APA StyleScoffone, V. C., Irudal, S., AbuAlshaar, A., Piazza, A., Trespidi, G., Barbieri, G., Makarov, V., Migliavacca, R., De Rossi, E., & Buroni, S. (2022). Bactericidal and Anti-Biofilm Activity of the FtsZ Inhibitor C109 against Acinetobacter baumannii. Antibiotics, 11(11), 1571. https://doi.org/10.3390/antibiotics11111571






