Canine Saliva as a Possible Source of Antimicrobial Resistance Genes
Abstract
:1. Introduction
2. Materials and Methods
Bioinformatic Analysis
3. Results
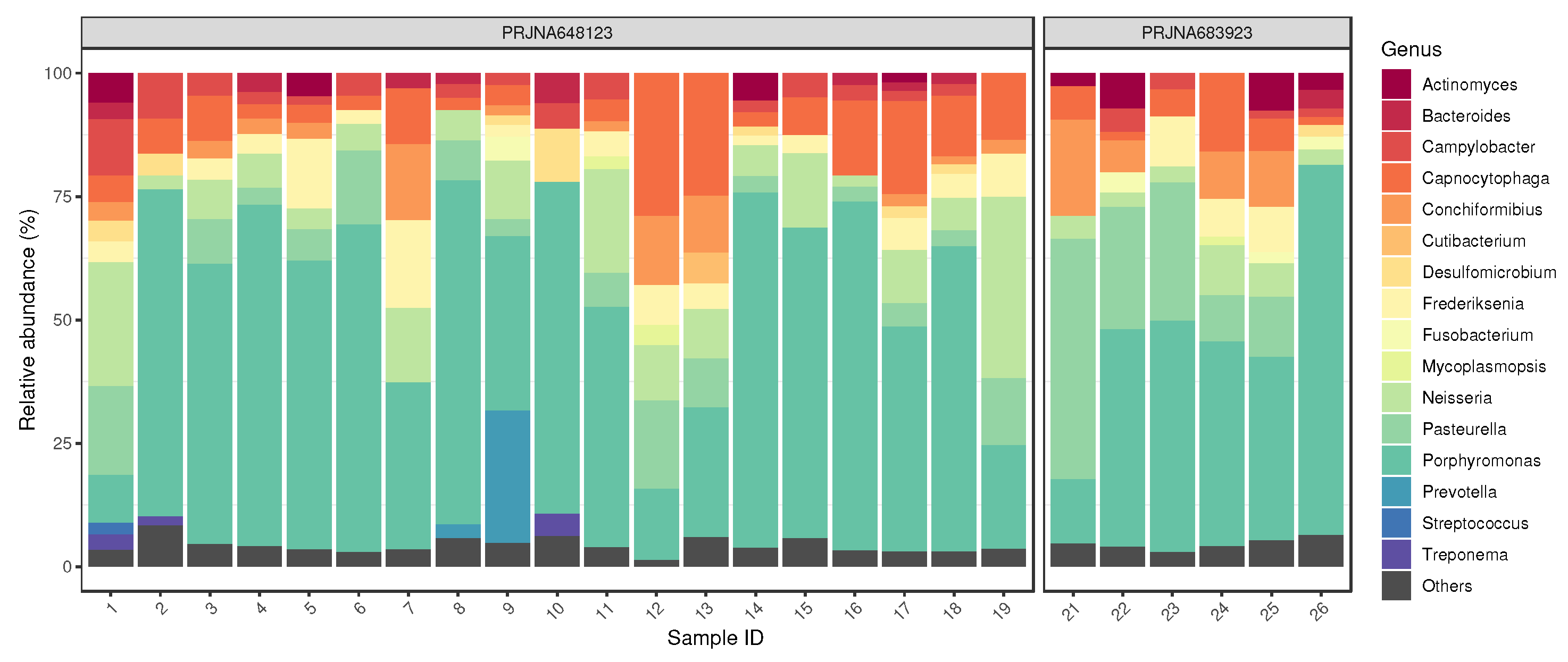
3.1. Bacteriome
3.2. Resistome
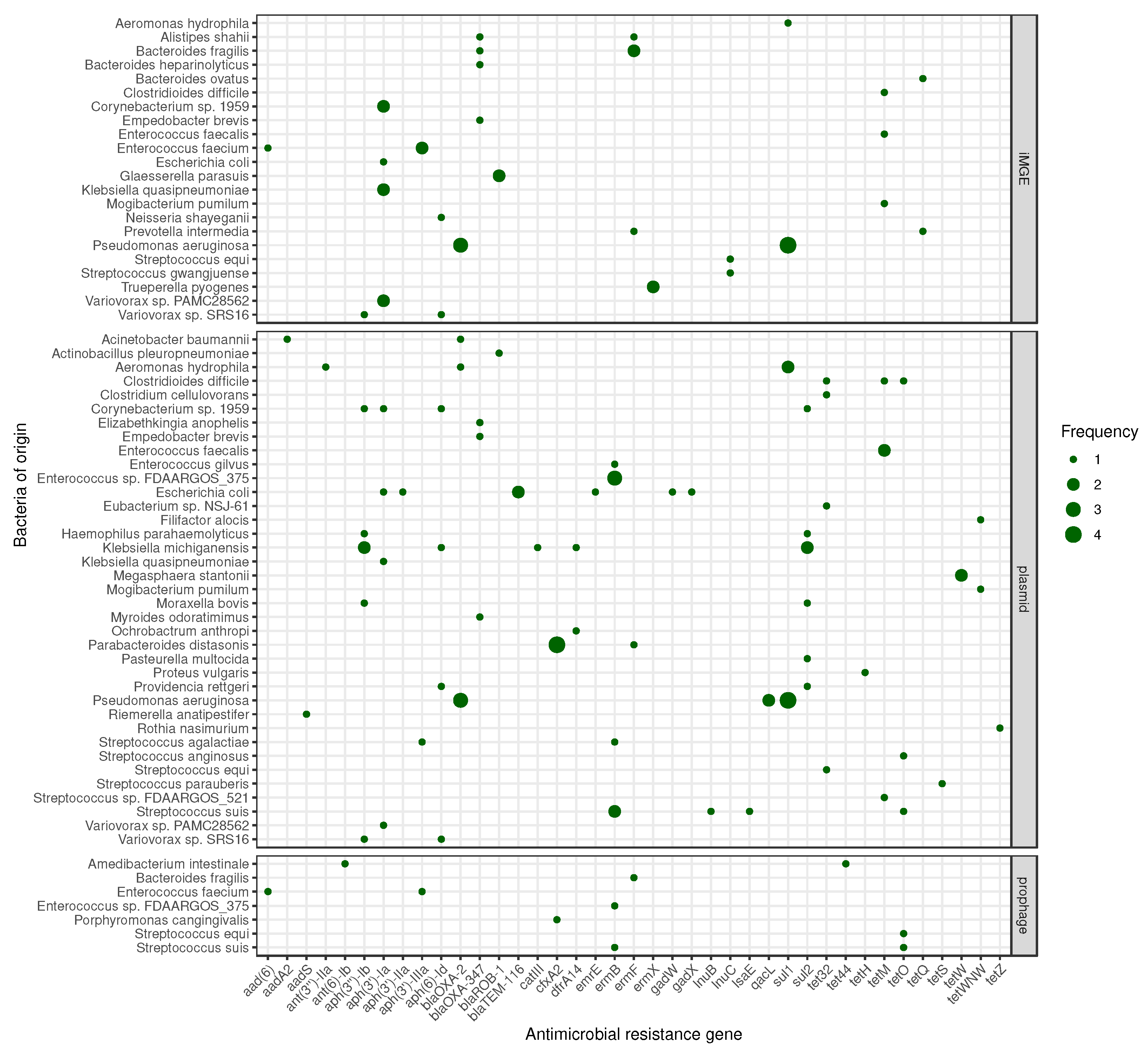
3.3. Mobilome
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). Third joint inter-agency report on integrated analysis of consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals in the EU/EEA. EFSA J. 2021, 19, e06712. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, Á.V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 2020, 10, 7464. [Google Scholar] [CrossRef] [PubMed]
- Rowan, A.N. Companion Animal Statistics in the USA. 2018. Available online: https://animalstudiesrepository.org/cgi/viewcontent.cgi?article=1002&context=demscapop (accessed on 6 August 2022).
- Bedford, E. Number of Dogs in the United States from 2000 to 2017. 2019. Available online: https://www.statista.com/statistics/198100/dogs-in-the-united-states-since-2000 (accessed on 6 March 2022).
- American Pet Products Association. 2021–2022 APPA National Pet Owners Survey. 2022. Available online: https://www.americanpetproducts.org/puBs_survey.asp (accessed on 6 March 2022).
- Ho, J.; Hussain, S.; Sparagano, O. Did the COVID-19 pandemic spark a public interest in pet adoption? Front. Vet. Sci. 2021, 8, 647308. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.L.; Thibault, M.; Hong, J. Characterizing pet acquisition and retention during the COVID-19 pandemic. Front. Vet. Sci. 2021, 8, 781403. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H. AVMA Pet Ownership and Demographics Sourcebook, 2017–2018 ed.; American Veterinary Medical Association: Schaumburg, IL, USA, 2017. [Google Scholar]
- Overgaauw, P.A.; Vinke, C.M.; van Hagen, M.A.; Lipman, L.J. A one health perspective on the human–companion animal relationship with emphasis on zoonotic aspects. Int. J. Environ. Res. Public Health 2020, 17, 3789. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, J.; Sacks, J.J.; White, D.; Kresnow, M.j. Dog bites: Still a problem? Inj. Prev. 2008, 14, 296–301. [Google Scholar] [CrossRef]
- Loder, R.T. The demographics of dog bites in the United States. Heliyon 2019, 5, e01360. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, J.; Hoopes, J.; Epp, T. Scoping decades of dog evidence: A scoping review of dog bite-related sequelae. Can. J. Public Health 2019, 110, 364–375. [Google Scholar] [CrossRef]
- Tulloch, J.S.; Owczarczak-Garstecka, S.C.; Fleming, K.M.; Vivancos, R.; Westgarth, C. English hospital episode data analysis (1998–2018) reveal that the rise in dog bite hospital admissions is driven by adult cases. Sci. Rep. 2021, 11, 1767. [Google Scholar] [CrossRef]
- Sarenbo, S.; Svensson, P.A. Bitten or struck by dog: A rising number of fatalities in Europe, 1995–2016. Forensic Sci. Int. 2021, 318, 110592. [Google Scholar] [CrossRef] [PubMed]
- Overall, K.L.; Love, M. Dog bites to humans—Demography, epidemiology, injury, and risk. J. Am. Vet. Med. Assoc. 2001, 218, 1923–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guardabassi, L.; Schwarz, S.; Lloyd, D.H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.H. Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 2007, 45, S148–S152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, K.; Pedersen, K.; Jensen, H.; Finster, K.; Jensen, V.F.; Heuer, O.E. Occurrence of antimicrobial resistance in bacteria from diagnostic samples from dogs. J. Antimicrob. Chemother. 2007, 60, 775–781. [Google Scholar] [CrossRef] [Green Version]
- Damborg, P.; Sørensen, A.H.; Guardabassi, L. Monitoring of antimicrobial resistance in healthy dogs: First report of canine ampicillin-resistant Enterococcus Faecium Clonal Complex 17. Vet. Microbiol. 2008, 132, 190–196. [Google Scholar] [CrossRef]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; Van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar]
- Li, Y.; Fernández, R.; Durán, I.; Molina-López, R.A.; Darwich, L. Antimicrobial resistance in bacteria isolated from cats and dogs from the Iberian Peninsula. Front. Microbiol. 2021, 11, 621597. [Google Scholar] [CrossRef] [PubMed]
- Goh, S. Clostridium Difficile: Methods and Protocols; Chapter Phage Transduction; Springer: New York, NY, USA, 2016; pp. 177–185. [Google Scholar]
- Ostrander, E.A.; Wang, G.D.; Larson, G.; Vonholdt, B.M.; Davis, B.W.; Jagannathan, V.; Hitte, C.; Wayne, R.K.; Zhang, Y.P. Dog10K: An international sequencing effort to advance studies of canine domestication, phenotypes and health. Natl. Sci. Rev. 2019, 6, 810–824. [Google Scholar] [CrossRef]
- Morrill, K.; Hekman, J.; Li, X.; McClure, J.; Logan, B.; Goodman, L.; Gao, M.; Dong, Y.; Alonso, M.; Carmichael, E.; et al. Ancestry-inclusive dog genomics challenges popular breed stereotypes. Science 2022, 376, eabk0639. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahti, L.; Shetty, S. Microbiome R Package, 2012–2019. Available online: https://www.bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 29 September 2022).
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Johansson, M.H.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Bolduc, B.; Zayed, A.A.; Varsani, A.; Dominguez-Huerta, G.; Delmont, T.O.; Pratama, A.A.; Gazitúa, M.C.; Vik, D.; Sullivan, M.B.; et al. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 2021, 9, 37. [Google Scholar] [CrossRef]
- Thomas, N.; Brook, I. Animal bite-associated infections: Microbiology and treatment. Expert Rev. Anti. Infect. Ther. 2011, 9, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Abrahamian, F.M.; Goldstein, E.J. Microbiology of animal bite wound infections. Clin. Microbiol. Rev. 2011, 24, 231–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- European Medicines Agency. European Surveillance of Veterinary Antimicrobial Consumption. Available online: https://www.ema.europa.eu/en/veterinary-regulatory/overview/antimicrobial-resistance/european-surveillance-veterinary-antimicrobial-consumption-esvac (accessed on 6 March 2022).
- McGreevy, P.; Thomson, P.; Dhand, N.K.; Raubenheimer, D.; Masters, S.; Mansfield, C.S.; Baldwin, T.; Soares Magalhaes, R.J.; Rand, J.; Hill, P.; et al. VetCompass Australia: A national big data collection system for veterinary science. Animals 2017, 7, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radford, A.; Noble, P.; Coyne, K.; Gaskell, R.; Jones, P.; Bryan, J.; Setzkorn, C.; Tierney, Á.; Dawson, S. Antibacterial prescribing patterns in small animal veterinary practice identified via SAVSNET: The small animal veterinary surveillance network. Vet. Rec. 2011, 169, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckland, E.L.; O’Neill, D.; Summers, J.; Mateus, A.; Church, D.; Redmond, L.; Brodbelt, D. Characterisation of antimicrobial usage in cats and dogs attending UK primary care companion animal veterinary practices. Vet. Rec. 2016, 179, 489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odoi, A.; Samuels, R.; Carter, C.N.; Smith, J. Antibiotic prescription practices and opinions regarding antimicrobial resistance among veterinarians in Kentucky, USA. PLoS ONE 2021, 16, e0249653. [Google Scholar] [CrossRef]
- Sobierajski, T.; Mazińska, B.; Chajęcka-Wierzchowska, W.; Śmiałek, M.; Hryniewicz, W. Antimicrobial and Antibiotic Resistance from the Perspective of Polish Veterinary Students: An Inter-University Study. Antibiotics 2022, 11, 115. [Google Scholar] [CrossRef]
- Hughes, L.A.; Williams, N.; Clegg, P.; Callaby, R.; Nuttall, T.; Coyne, K.; Pinchbeck, G.; Dawson, S. Cross-sectional survey of antimicrobial prescribing patterns in UK small animal veterinary practice. Prev. Vet. Med. 2012, 104, 309–316. [Google Scholar] [CrossRef]
- Jacob, M.E.; Hoppin, J.A.; Steers, N.; Davis, J.L.; Davidson, G.; Hansen, B.; Lunn, K.F.; Murphy, K.M.; Papich, M.G. Opinions of clinical veterinarians at a US veterinary teaching hospital regarding antimicrobial use and antimicrobial-resistant infections. J. Am. Vet. Med. Assoc. 2015, 247, 938–944. [Google Scholar] [CrossRef]
- Hardefeldt, L.Y.; Selinger, J.; Stevenson, M.A.; Gilkerson, J.R.; Crabb, H.; Billman-Jacobe, H.; Thursky, K.; Bailey, K.E.; Awad, M.; Browning, G.F. Population wide assessment of antimicrobial use in dogs and cats using a novel data source—A cohort study using pet insurance data. Vet. Microbiol. 2018, 225, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Hopman, N.E.; Mughini-Gras, L.; Speksnijder, D.C.; Wagenaar, J.A.; van Geijlswijk, I.M.; Broens, E.M. Attitudes and perceptions of Dutch companion animal veterinarians towards antimicrobial use and antimicrobial resistance. Prev. Vet. Med. 2019, 170, 104717. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.M.; Zhuo, A.; Govendir, M.; Rowbotham, S.J.; Labbate, M.; Degeling, C.; Gilbert, G.L.; Dominey-Howes, D.; Ward, M.P. Factors influencing the behaviour and perceptions of Australian veterinarians towards antibiotic use and antimicrobial resistance. PLoS ONE 2019, 14, e0223534. [Google Scholar] [CrossRef]
- Samuels, R.; Qekwana, D.N.; Oguttu, J.W.; Odoi, A. Antibiotic prescription practices and attitudes towards the use of antimicrobials among veterinarians in the City of Tshwane, South Africa. PeerJ 2021, 9, e10144. [Google Scholar] [CrossRef] [PubMed]
- Kvaale, M.; Grave, K.; Kristoffersen, A.; Norström, M. The prescription rate of antibacterial agents in dogs in Norway–geographical patterns and trends during the period 2004–2008. J. Vet. Pharmacol. Ther. 2013, 36, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Tompson, A.C.; Mateus, A.L.; Brodbelt, D.C.; Chandler, C.I. Understanding antibiotic use in companion animals: A literature review identifying avenues for future efforts. Front. Vet. Sci. 2021, 8, 719547. [Google Scholar] [CrossRef] [PubMed]
- Singleton, D.; Sánchez-Vizcaíno, F.; Dawson, S.; Jones, P.; Noble, P.; Pinchbeck, G.; Williams, N.; Radford, A. Patterns of antimicrobial agent prescription in a sentinel population of canine and feline veterinary practices in the United Kingdom. Vet. J. 2017, 224, 18–24. [Google Scholar] [CrossRef] [Green Version]
- Ellis, R.; Ellis, C. Dog and cat bites. Am. Fam. Physician 2014, 90, 239–243. [Google Scholar]
- Malahias, M.; Jordan, D.; Hughes, O.; Khan, W.S.; Hindocha, S. Bite injuries to the hand: Microbiology, virology and management. Open Orthop. J. 2014, 8, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Aziz, H.; Rhee, P.; Pandit, V.; Tang, A.; Gries, L.; Joseph, B. The current concepts in management of animal (dog, cat, snake, scorpion) and human bite wounds. J. Trauma Acute Care Surg. 2015, 78, 641–648. [Google Scholar] [CrossRef]
- Jakeman, M.; Oxley, J.A.; Owczarczak-Garstecka, S.C.; Westgarth, C. Pet dog bites in children: Management and prevention. BMJ Paediatr. Open 2020, 4, e000726. [Google Scholar] [CrossRef] [PubMed]
- Zangari, A.; Cerigioni, E.; Nino, F.; Guidi, R.; Gulia, C.; Piergentili, R.; Ilari, M.; Mazzoni, N.; Cobellis, G. Dog bite injuries in a tertiary care children’s hospital: A seven-year review. Pediatr. Int. 2021, 63, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Monroy, A.; Behar, P.; Nagy, M.; Poje, C.; Pizzuto, M.; Brodsky, L. Head and neck dog bites in children. Otolaryngol. Head Neck Surg. 2009, 140, 354–357. [Google Scholar] [CrossRef]
- Cummings, P. Antibiotics to prevent infection in pateints with dog bite wounds: A meta-analysis of randomized trials. Ann. Emerg. Med. 1994, 23, 535–540. [Google Scholar] [CrossRef]
- Medeiros, I.M.; Saconato, H. Antibiotic prophylaxis for mammalian bites. Cochrane Database Syst. Rev. 2001. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L.; Bisno, A.L.; Chambers, H.F.; Everett, E.D.; Dellinger, P.; Goldstein, E.J.C.; Gorbach, S.L.; Hirschmann, J.V.; Kaplan, E.L.; Montoya, J.G.; et al. Practice Guidelines for the Diagnosis and Management of Skin and Soft-Tissue Infections. Clin. Infect. Dis. 2005, 41, 1373–1406. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Palmer, J. Dog bites. BMJ 2007, 334, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner Jr, R.C.; Day, N.P.; et al. Global antibiotic consumption and usage in humans, 2000–18: A spatial modelling study. Lancet Planet. Health 2021, 5, e893–e904. [Google Scholar] [CrossRef]
- Di Conza, J.A.; Badaracco, A.; Ayala, J.; Rodriguez, C.; Famiglietti, Á.; Gutkind, G.O. β-lactamases produced by amoxicillin-clavulanate-resistant enterobacteria isolated in Buenos Aires, Argentina: A new blaTEM gene. Rev. Argent Microbiol. 2014, 46, 210–217. [Google Scholar] [CrossRef] [Green Version]
- Davies, T.J.; Stoesser, N.; Sheppard, A.E.; Abuoun, M.; Fowler, P.; Swann, J.; Quan, T.P.; Griffiths, D.; Vaughan, A.; Morgan, M.; et al. Reconciling the potentially irreconcilable? Genotypic and phenotypic amoxicillin-clavulanate resistance in Escherichia Coli. Antimicrob. Agents Chemother. 2020, 64, e02026-19. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front. Microbiol. 2019, 10, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knafl, D.; Winhofer, Y.; Lötsch, F.; Weisshaar, S.; Steininger, C.; Burgmann, H.; Thalhammer, F. Tigecycline as last resort in severe refractory Clostridium difficile infection: A case report. J. Hosp. Infect. 2016, 92, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Wong, D.; Malhotra, S. Intravenous fosfomycin as salvage therapy for osteomyelitis caused by multidrug-resistant Pseudomonas aeruginosa. Am. J. Health-Syst. Pharm. 2021, 78, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine; World Health Organization: Geneva, Switzerland, 2019.
- Chen, C.; Cui, C.Y.; Wu, X.T.; Fang, L.X.; He, Q.; He, B.; Long, T.F.; Liao, X.P.; Chen, L.; Liu, Y.H.; et al. Spread of tet (X5) and tet (X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 2021, 253, 108954. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, Y.; Liu, D.; Yang, D.; Liu, Z.; Wang, Y.; Wang, J.; Wang, X.; Xu, X.; Li, X.; et al. Abundance of tigecycline resistance genes and association with antibiotic residues in Chinese livestock farms. J. Hazard. Mater. 2021, 409, 124921. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Lv, Y.; Cui, L.; Li, Y.; Li, T.; Song, H.; Hao, Y.; Shen, J.; Wang, Y.; et al. Novel plasmid-mediated tet (X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 2019, 64, e01326-19. [Google Scholar] [CrossRef]
- Verrier, L. Dog licks man. Lancet 1970, 295, 615. [Google Scholar] [CrossRef]
- Vallejo, J.R.; Santos-Fita, D.; González, J.A. The therapeutic use of the dog in Spain: A review from a historical and cross-cultural perspective of a change in the human-dog relationship. J. Ethnobiol. Ethnomed. 2017, 13, 47. [Google Scholar] [CrossRef] [Green Version]
- Benjamin, N.; Pattullo, S.; Weller, R.; Smith, L.; Ormerod, A. Wound licking and nitric oxide. Lancet 1997, 349, 1776. [Google Scholar] [CrossRef]
- Dhasarathan, P.; Arunkumar, R.; Blessy Jesubell, R.; Sowmya, P. Analysis of compounds and screening for anti-microbial, anti-inflammatory activity from saliva of Canis lupus Fam. Int. J. Ethnomed. Pharm. Res. 2013, 1, 1–6. [Google Scholar]
- Akpomie, O.; Ukoha, P.; Nwafor, O.; Umukoro, G. Saliva of different dog breeds as antimicrobial agents against microorganisms isolated from wound infections. Anim. Sci. J. 2011, 2, 18–22. [Google Scholar]
- Bager, F.; Bortolaia, V.; Ellis-Iversen, J.; Hendriksen, R.; Borck Høg, B.; Jensen, L.; Korsgaard, H.; Pedersen, K.; Dalby, T.; Træholt Franck, K.; et al. DANMAP 2016—Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark; Statens Serum Institut: Copenhagen, Denmark; National Veterinary Institute: Uppsala, Sweden; Technical University of Denmark National Food Institute: Kongens Lyngby, Denmark; Technical University of Denmark: Kongens Lyngby, Denmark, 2017.
- Hopman, N.E.; Portengen, L.; Heederik, D.J.; Wagenaar, J.A.; Van Geijlswijk, I.M.; Broens, E.M. Time trends, seasonal differences and determinants of systemic antimicrobial use in companion animal clinics (2012–2015). Vet. Microbiol. 2019, 235, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.E. Antibiotic use across the globe. Nat. Hum. Behav. 2018, 2, 373. [Google Scholar] [CrossRef] [PubMed]

| ID | BioProject | Run | Bacterial Read Count |
|---|---|---|---|
| 1 | PRJNA648123 | SRR12330029 | 2,900,387 |
| 2 | SRR12330041 | 16,153,172 | |
| 3 | SRR12330042 | 13,072,781 | |
| 4 | SRR12330043 | 13,774,332 | |
| 5 | SRR12330044 | 6,123,646 | |
| 6 | SRR12330045 | 16,707,766 | |
| 7 | SRR12330098 | 18,826,266 | |
| 8 | SRR12330104 | 27,598,592 | |
| 9 | SRR12330220 | 9,938,948 | |
| 10 | SRR12330260 | 17,642,933 | |
| 11 | SRR12330298 | 17,277,697 | |
| 12 | SRR12330356 | 13,988,719 | |
| 13 | SRR12330364 | 17,378,513 | |
| 14 | SRR12330377 | 12,155,726 | |
| 15 | SRR12330378 | 34,183,357 | |
| 16 | SRR12330382 | 22,353,314 | |
| 17 | SRR12330383 | 22,886,951 | |
| 18 | SRR12330384 | 18,328,656 | |
| 19 | SRR12330385 | 6,631,504 | |
| 20 | PRJNA683923 | SRR13340534 | 0 |
| 21 | SRR13340535 | 6,752,169 | |
| 22 | SRR13340537 | 8,245,374 | |
| 23 | SRR13340538 | 41,212,470 | |
| 24 | SRR13340539 | 13,028,655 | |
| 25 | SRR13340540 | 6,964,460 | |
| 26 | SRR13340541 | 6,279,921 |
| ARG(s) | Frequency | Drug Class | |
|---|---|---|---|
| n | % | ||
| aac(6’)-Im | 2 | 7.7 | aminoglycoside |
| aad(6) | 2 | 7.7 | aminoglycoside |
| aadA2 | 1 | 3.8 | aminoglycoside |
| aadA3 | 1 | 3.8 | aminoglycoside |
| aadA5 | 1 | 3.8 | aminoglycoside |
| aadA15 | 1 | 3.8 | aminoglycoside |
| aadS | 12 | 46.2 | aminoglycoside |
| acrA | 1 | 3.8 | cephalosporin, fluoroquinolone, glycylcycline, penam, phenicol, rifamycin, tetracycline, triclosan |
| ant(2”)-Ia | 1 | 3.8 | aminoglycoside |
| ant(3”)-IIa | 1 | 3.8 | aminoglycoside |
| ant(6)-Ib | 1 | 3.8 | aminoglycoside |
| aph(2”)-IIa | 3 | 11.5 | aminoglycoside |
| aph(3”)-Ib | 10 | 38.5 | aminoglycoside |
| aph(3’)-Ia | 5 | 19.2 | aminoglycoside |
| aph(3’)-IIa | 1 | 3.8 | aminoglycoside |
| aph(3’)-IIIa | 4 | 15.4 | aminoglycoside |
| aph(6)-Id | 10 | 38.5 | aminoglycoside |
| bacA | 1 | 3.8 | peptide |
| blaACT-12 | 1 | 3.8 | carbapenem, cephalosporin, cephamycin, penam |
| blaOXA-2 | 12 | 46.2 | carbapenem, cephalosporin, penam |
| blaOXA-85 | 1 | 3.8 | carbapenem, cephalosporin, penam |
| blaOXA-119 | 1 | 3.8 | carbapenem, cephalosporin, penam |
| blaOXA-347 | 16 | 61.5 | carbapenem, cephalosporin, penam |
| blaROB-1 | 21 | 80.8 | cephalosporin, penam |
| blaROB-9 | 1 | 3.8 | cephalosporin, penam |
| blaROB-10 | 3 | 11.5 | cephalosporin, penam |
| blaTEM-116 | 2 | 7.7 | cephalosporin, monobactam, penam, penem |
| catIII | 2 | 7.7 | phenicol |
| cfxA2 | 20 | 76.9 | cephamycin |
| cmlA9 | 1 | 3.8 | phenicol |
| dfrA14 | 3 | 11.5 | diaminopyrimidine |
| emrE | 1 | 3.8 | macrolide |
| emrK | 1 | 3.8 | tetracycline |
| ereA | 1 | 3.8 | macrolide |
| ermB | 9 | 34.6 | lincosamide, macrolide, streptogramin |
| ermF | 18 | 69.2 | lincosamide, macrolide, streptogramin |
| ermG | 2 | 7.7 | lincosamide, macrolide, streptogramin |
| ermX | 1 | 3.8 | lincosamide, macrolide, streptogramin |
| fosA2 | 1 | 3.8 | fosfomycin |
| gadW | 1 | 3.8 | fluoroquinolone, macrolide, penam |
| gadX | 1 | 3.8 | fluoroquinolone, macrolide, penam |
| lnuB | 2 | 7.7 | lincosamide |
| lnuC | 2 | 7.7 | lincosamide |
| lsaE | 2 | 7.7 | lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| mdtN | 1 | 3.8 | acridine dye, disinfecting agents and intercalating dyes, nucleoside |
| mef(En2) | 12 | 46.2 | macrolide |
| mel | 5 | 19.2 | lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| oqxA | 1 | 3.8 | diaminopyrimidine, fluoroquinolone, glycylcycline, nitrofuran, tetracycline |
| pgpB | 23 | 88.5 | peptide |
| qacL | 2 | 7.7 | disinfecting agents and intercalating dyes |
| SAT-4 | 1 | 3.8 | nucleoside |
| sul1 | 7 | 26.9 | sulfonamide |
| sul2 | 8 | 30.8 | sulfonamide |
| tet32 | 19 | 73.1 | tetracycline |
| tet44 | 1 | 3.8 | tetracycline |
| tetH | 3 | 11.5 | tetracycline |
| tetM | 5 | 19.2 | tetracycline |
| tetO | 18 | 69.2 | tetracycline |
| tetQ | 20 | 76.9 | tetracycline |
| tetS | 1 | 3.8 | tetracycline |
| tetW | 5 | 19.2 | tetracycline |
| tetWNW | 5 | 19.2 | tetracycline |
| tetX | 10 | 38.5 | glycylcycline, tetracycline |
| tetX1 | 1 | 3.8 | tetracycline |
| tetX4 | 1 | 3.8 | glycylcycline, tetracycline |
| tetX5 | 10 | 38.5 | tetracycline |
| tetY | 1 | 3.8 | tetracycline |
| tetZ | 1 | 3.8 | tetracycline |
| ugd | 1 | 3.8 | peptide |
| ARG(s) | Bacteria of Origin |
|---|---|
| aac(6’)-Im | Clostridioides difficile |
| aad(6) | Enterococcus faecium, Staphylococcus aureus |
| aadA2 | Acinetobacter baumannii |
| aadA3 | Neisseria animaloris |
| aadS | Bacteroides fragilis, Capnocytophaga sp. H2931, Capnocytophaga sp. H4358, Capnocytophaga stomatis, Chryseobacterium indologenes, Riemerella anatipestifer |
| acrA | E. coli |
| ant(3”)-IIa | Aeromonas hydrophila |
| ant(6)-Ib | Amedibacterium intestinale |
| aph(2”)-IIa | C. difficile |
| aph(3”)-Ib | Corynebacterium sp. 1959, Haemophilus parahaemolyticus, Klebsiella michiganensis, Moraxella bovis, Variovorax sp. SRS16 |
| aph(3’)-Ia | Corynebacterium sp. 1959, E. coli, Klebsiella quasipneumoniae, Variovorax sp. PAMC28562 |
| aph(3’)-IIa | E. coli |
| aph(3’)-IIIa | E. faecium, S. aureus, Streptococcus agalactiae |
| aph(6)-Id | Corynebacterium sp. 1959, K. michiganensis, Neisseria shayeganii, Providencia rettgeri, Variovorax sp. SRS16 |
| blaOXA-2 | A. baumannii, A. hydrophila, P. aeruginosa |
| blaOXA-85 | Fusobacterium ulcerans |
| blaOXA-119 | Geobacter sulfurreducens |
| blaOXA-347 | Alistipes shahii, B. fragilis, Bacteroides heparinolyticus, Capnocytophaga sp. H2931, Capnocytophaga sp. H4358, C. stomatis, Chryseobacterium sp. POL2, Elizabethkingia anophelis, Empedobacter brevis, Myroides odoratimimus, R. anatipestifer |
| blaROB-1 | Actinobacillus pleuropneumoniae, Conchiformibius steedae, Glaesserella parasuis, Haemophilus haemolyticus |
| blaROB-9 | G. parasuis |
| blaROB-10 | Bibersteinia trehalosi |
| blaTEM-116 | E. coli |
| catIII | K. michiganensis |
| cfxA2 | Capnocytophaga cynodegmi, Parabacteroides distasonis, Porphyromonas cangingivalis, Porphyromonas crevioricanis, Porphyromonas gingivalis, Tannerella forsythia |
| dfrA14 | K. michiganensis, Ochrobactrum anthropi |
| emrE | E. coli |
| emrK | E. coli |
| ereA | Geobacter daltonii |
| ermB | Enterococcus gilvus, Enterococcus sp. FDAARGOS_375, S. agalactiae, Streptococcus suis |
| ermF | A. shahii, B. fragilis, C. stomatis, C. indologenes, P. distasonis, P. cangingivalis, Prevotella intermedia, R. anatipestifer |
| ermG | C. difficile |
| ermX | Trueperella pyogenes |
| gadW | E. coli |
| gadX | E. coli |
| lnuB | S. suis |
| lnuC | Streptococcus equi, Streptococcus gwangjuense |
| lsaE | S. suis |
| mdtN | E. coli |
| mef(En2) | B. fragilis, P. cangingivalis, P. gingivalis, P. intermedia |
| mel | Streptococcus pluranimalium |
| pgpB | P. gingivalis |
| qacL | P. aeruginosa |
| SAT-4 | S. aureus |
| sul1 | A. hydrophila, P. aeruginosa |
| sul2 | Corynebacterium sp. 1959, H. parahaemolyticus, K. michiganensis, M. bovis, Pasteurella multocida, P. rettgeri |
| tet32 | Blautia hansenii, Bulleidia sp. zg-1006, C. difficile, Clostridium cellulovorans, Eubacterium maltosivorans, Eubacterium sp. NSJ-61, Faecalibacterium prausnitzii, Lachnoanaerobaculum umeaense, Peptoclostridium acidaminophilum, Roseburia intestinalis, Streptococcus anginosus, Streptococcus constellatus, S. equi |
| tet44 | A. intestinale |
| tetH | Proteus vulgaris, Pseudomonas putida |
| tetM | C. difficile, Enterococcus faecalis, Mogibacterium pumilum, Streptococcus sp. FDAARGOS_521 |
| tetO | C. difficile, Enterococcus hirae, Murdochiella vaginalis, Streptococcus acidominimus, S. anginosus, S. constellatus, S. equi, S. suis |
| tetQ | Alistipes indistinctus, Bacteroides dorei, B. heparinolyticus, Bacteroides ovatus, Bacteroides sp. HF-5287, Phocaeicola coprophilus, P. crevioricanis, Prevotella fusca, P. intermedia |
| tetS | Streptococcus parauberis |
| tetW | Enterocloster bolteae, F. prausnitzii, Megasphaera stantonii, S. suis |
| tetWNW | Filifactor alocis, M. pumilum |
| tetX | B. fragilis, P. distasonis, P. intermedia, R. anatipestifer |
| tetX4 | R. anatipestifer |
| tetX5 | B. fragilis, C. stomatis, R. anatipestifer |
| tetZ | Rothia nasimurium |
| ugd | E. coli |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tóth, A.G.; Tóth, I.; Rózsa, B.; Dubecz, A.; Patai, Á.V.; Németh, T.; Kaplan, S.; Kovács, E.G.; Makrai, L.; Solymosi, N. Canine Saliva as a Possible Source of Antimicrobial Resistance Genes. Antibiotics 2022, 11, 1490. https://doi.org/10.3390/antibiotics11111490
Tóth AG, Tóth I, Rózsa B, Dubecz A, Patai ÁV, Németh T, Kaplan S, Kovács EG, Makrai L, Solymosi N. Canine Saliva as a Possible Source of Antimicrobial Resistance Genes. Antibiotics. 2022; 11(11):1490. https://doi.org/10.3390/antibiotics11111490
Chicago/Turabian StyleTóth, Adrienn Gréta, Imre Tóth, Bernadett Rózsa, Attila Dubecz, Árpád V. Patai, Tibor Németh, Selçuk Kaplan, Eszter Gabriella Kovács, László Makrai, and Norbert Solymosi. 2022. "Canine Saliva as a Possible Source of Antimicrobial Resistance Genes" Antibiotics 11, no. 11: 1490. https://doi.org/10.3390/antibiotics11111490
APA StyleTóth, A. G., Tóth, I., Rózsa, B., Dubecz, A., Patai, Á. V., Németh, T., Kaplan, S., Kovács, E. G., Makrai, L., & Solymosi, N. (2022). Canine Saliva as a Possible Source of Antimicrobial Resistance Genes. Antibiotics, 11(11), 1490. https://doi.org/10.3390/antibiotics11111490






