Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/Ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Bacteriological Culture
2.3. Antimicrobial Susceptibility and ESBL/Ampc Phenotype
2.4. DNA Extraction
2.5. Determination of Phylogenetic Groups
2.6. Determination of ESBL- and AmpC-Associated Genes
2.7. Whole Genome Sequencing
2.8. Sequence Analysis
2.9. Statistical Analyses
3. Results
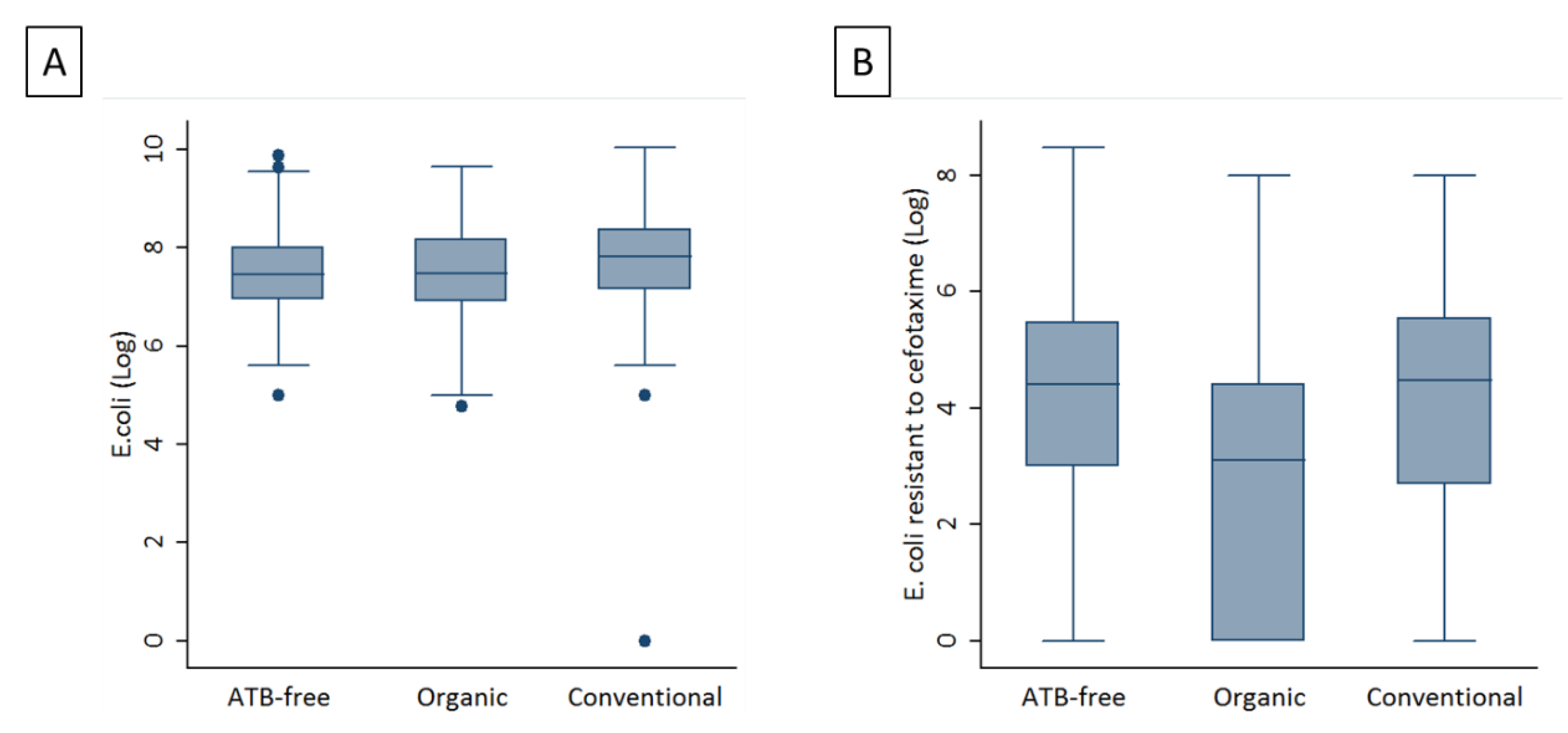
3.1. E. coli loads in the Three Management Systems
3.2. Antimicrobial Susceptibility Testing Results
3.3. Molecular Analyses
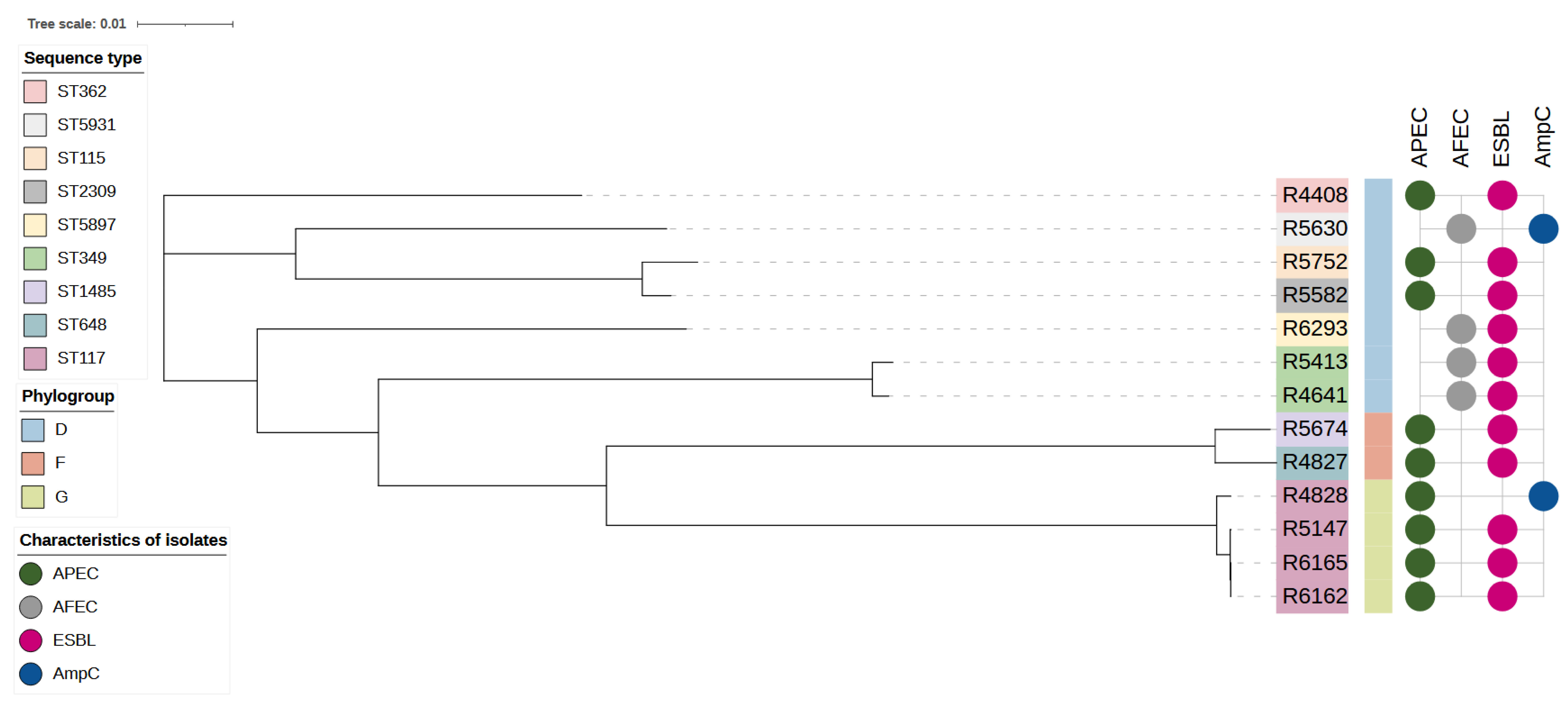
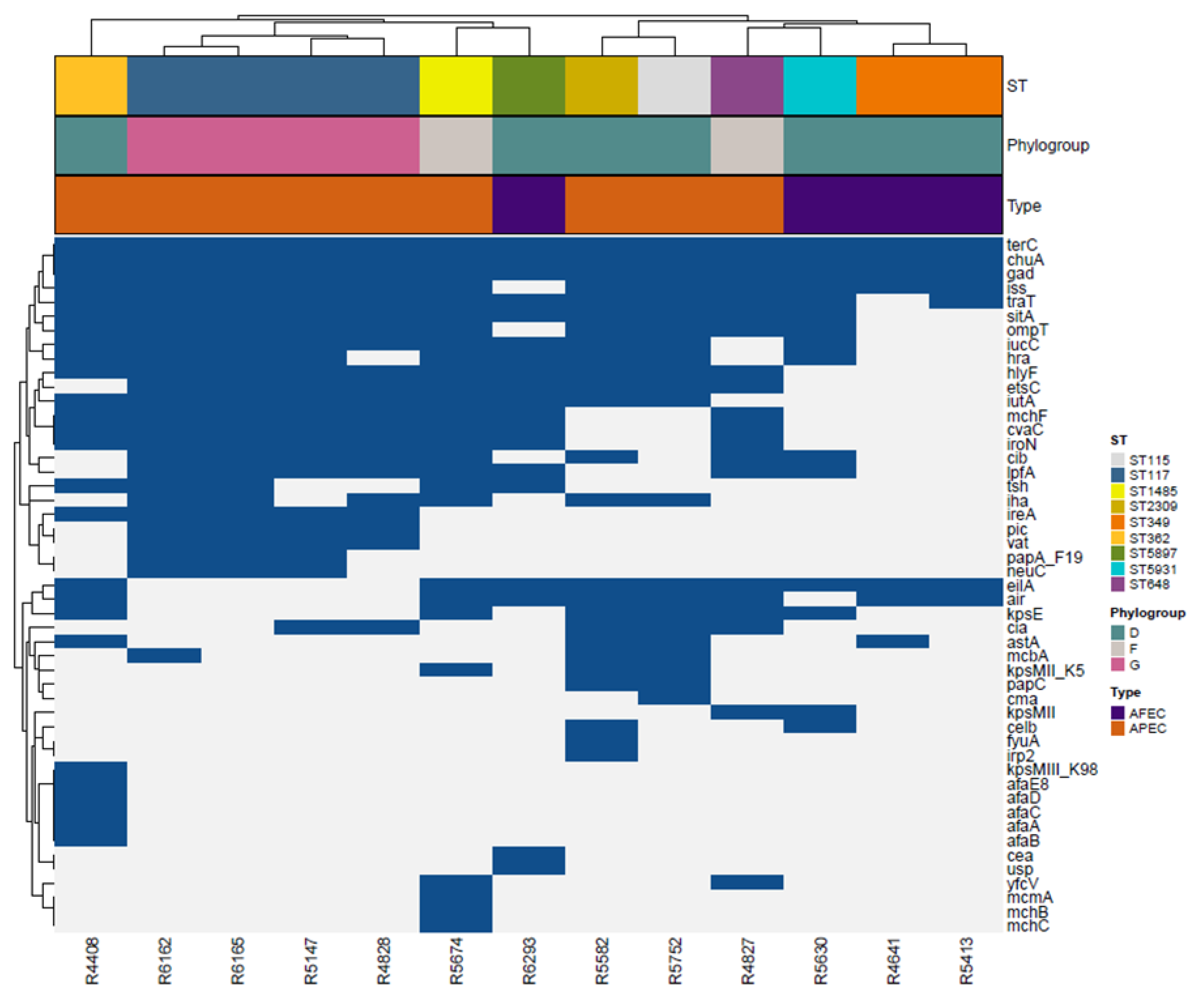
3.4. Whole Genome Sequencing (WGS)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; WHO: Genève, Zwitserland, 2019; ISBN 9789241515528.
- Dame-Korevaar, A.; Fischer, E.A.J.; Stegeman, A.; Mevius, D.; van Essen-Zandbergen, A.; Velkers, F.; van der Goot, J. Dynamics of CMY-2 producing E. coli in a broiler parent flock. Vet. Microbiol. 2017, 203, 211–214. [Google Scholar] [CrossRef]
- EFSA. The European union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, e05598. EFSA J. 2019, 17, e05598. [Google Scholar] [CrossRef]
- Ewers, C.; de Jong, A.; Prenger-Berninghoff, E.; El Garch, F.; Leidner, U.; Tiwari, S.K.; Semmler, T. Genomic Diversity and Virulence Potential of ESBL- and AmpC-β-Lactamase-Producing Escherichia coli Strains from Healthy Food Animals Across Europe. Front. Microbiol. 2021, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.; Ricci, A.; Auce, Z.; Beechinor, J.G.; Bergendahl, H.; Da, D.; Hederov, J.; Hekman, P.; Breathnach, R.; Persson, E.L.; et al. EMA and EFSA Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). EFSA J. 2017, 15, e04666. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; Lestón, L.; Mora, A. Microbiological risk assessment of Turkey and chicken meat for consumer: Significant differences regarding multidrug resistance, mcr or presence of hybrid aEPEC/ExPEC pathotypes of E. coli. Food Control 2021, 123, 107713. [Google Scholar] [CrossRef]
- Apostolakos, I.; Feudi, C.; Eichhorn, I.; Palmieri, N.; Fasolato, L.; Schwarz, S.; Piccirillo, A. High-resolution characterisation of ESBL/pAmpC-producing Escherichia coli isolated from the broiler production pyramid. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S. Urinary tract infections attributed to diverse ExPEC strains in food animals: Evidence and data gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, M.P.V.; Saidenberg, A.B.; Moreno, A.M.; Ferreira, A.J.P.; Vieira, M.A.M.; Gomes, T.A.T.; Knöbl, T. Pandemic extra-intestinal pathogenic Escherichia coli (ExPEC) clonal group O6-B2-ST73 as a cause of avian colibacillosis in Brazil. PLoS ONE 2017, 12, e0178970. [Google Scholar] [CrossRef] [Green Version]
- Skunca, D.; Tomasevic, I.; Nastasijevic, I.; Tomovic, V.; Djekic, I. Life cycle assessment of the chicken meat chain. J. Clean. Prod. 2018, 184, 440–450. [Google Scholar] [CrossRef]
- Mellata, M.; Dho-Moulin, M.; Dozois, C.M.; Curtiss, R.; Brown, P.K.; Arné, P.; Brée, A.; Desautels, C.; Fairbrother, J.M. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 2003, 71, 536–540. [Google Scholar] [CrossRef]
- Gambi, L.; Rossini, R.; Menandro, M.L.; Franzo, G.; Valentini, F.; Tosi, G.; D’Incau, M.; Fiorentini, L. Virulence Factors and Antimicrobial Resistance Profile of Escherichia coli Isolated from Laying Hens in Italy. Animals 2022, 12, 1812. [Google Scholar] [CrossRef] [PubMed]
- EFSA; ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar] [CrossRef]
- EMA. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017. Trends from 2010 to 2017; Ninth ESVAC Rep.-EMA/294674/2019; European Medicines Agency: Amsterdam, The Netherland, 2019. [Google Scholar]
- Unaitalia Giornata Europea Antibiotici, l’impegno Della Filiera Avicola, −82% Dal 2011. Available online: https://www.unaitalia.com/giornata-europea-antibiotici-limpegno-della-filiera-avicola-82-dal-2011/ (accessed on 22 February 2022).
- Pesciaroli, M.; Magistrali, C.F.; Filippini, G.; Epifanio, E.M.; Lovito, C.; Marchi, L.; Maresca, C.; Massacci, F.R.; Orsini, S.; Scoccia, E.; et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 2020, 314, 108391. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Kong, L.; Gao, H.; Cheng, X.; Wang, X. A Review of Current Bacterial Resistance to Antibiotics in Food Animals. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef]
- Musa, L.; Patrizia Casagrande, P.; Raffaella, B.; Laura, M.; Sara, B.; David, R.; Maria Luisa, M.; Maria Pia, F.; Marenzoni, M.L.; Franciosini, M.P. Antimicrobial Susceptibility of Escherichia coli Chickens at Slaughter. Animals 2020, 10, 1215. [Google Scholar] [CrossRef]
- McNally, A.; Oren, Y.; Kelly, D.; Pascoe, B.; Dunn, S.; Sreecharan, T.; Vehkala, M.; Välimäki, N.; Prentice, M.B.; Ashour, A.; et al. Combined Analysis of Variation in Core, Accessory and Regulatory Genome Regions Provides a Super-Resolution View into the Evolution of Bacterial Populations. PLoS Genet. 2016, 12, e1006280. [Google Scholar] [CrossRef] [Green Version]
- Schaufler, K.; Semmler, T.W.L.; Trott, D.J.; Pitout, J.; Peirano, G.B.J.; Dolejska, M.; Literak, I.; Fuchs, S.A.N.; Grobbel, M.; Torres, C.; et al. Genomic and Functional Analysis of Emerging Virulent and Multidrug-Resistant Escherichia coli Lineage Sequence Type 648. Antimicrob. Agents Chemother. 2019, 63, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Duse, A.; Waller, K.P.; Emanuelson, U.; Unnerstad, H.E.; Persson, Y.; Bengtsson, B. Risk factors for quinolone-resistant Escherichia coli in feces from preweaned dairy calves and postpartum dairy cows. J. Dairy Sci. 2015, 98, 6387–6398. [Google Scholar] [CrossRef]
- Hasman, H.; Agersø, Y.; Cavaco, L.M.; Hendriksen, R.S.; Bortolaia, V.; Pedersen, S.K. Laboratory Protocol Validation of selective MacConkey agar plates supplemented with 1 mg/L cefotaxime for monitoring of ESBL-and AmpC-producing E. coli in meat and caecal samples. 2017. Available online: https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/282_protocol-for-validation-of-macconkey-and-ctx-agar-plates-nov2015.pdf (accessed on 22 February 2022).
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST Disk Diffusion Method for Antimicrobial Susceptibility Testing. Available online: http://www.eucast.org (accessed on 22 February 2022).
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2018. Available online: https://www.eucast.org/ (accessed on 22 February 2022).
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- The European Committee on Antimicrobial Susceptibility Testing—EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistancesof Clinical and/or Epidemiological Importance. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 22 February 2022).
- EUCAST. Recommendations for MIC Determination of Colistin (polymyxin E) As Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group; EUCAST European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2016. [Google Scholar]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosavian, S.M.; Rezvani-Rad, A. Determining frequency of genes of CTX-M and CTX-M-15 of producing Enterobacteriaceae of isolated extended-spectrum beta-lactamases from clinical samples of patients referred to training hospitals of Medical Sciences University, Khorramabad, Iran. Int. J. Pharm. Investig. 2017, 7, 60–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [Green Version]
- Johnson, T.J.; Miller, E.A.; Flores-Figueroa, C.; Munoz-Aguayo, J.; Cardona, C.; Fransen, K.; Lighty, M.; Gonder, E.; Nezworski, J.; Haag, A.; et al. Refining the definition of the avian pathogenic Escherichia coli (APEC) pathotype through inclusion of high-risk clonal groups. Poult. Sci. 2022, 101, 102009. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Nagard, H.; Le; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genomics 2018, 4, e000192. [Google Scholar] [CrossRef]
- TeamCore R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018.
- Bai, X.; Zhang, J.; Ambikan, A.; Jernberg, C.; Ehricht, R.; Scheutz, F.; Xiong, Y.; Matussek, A. Molecular Characterization and Comparative Genomics of Clinical Hybrid Shiga Toxin-Producing and Enterotoxigenic Escherichia coli (STEC/ETEC) Strains in Sweden. Sci. Rep. 2019, 9, 5619. [Google Scholar] [CrossRef] [Green Version]
- Laube, H.; Friese, A.; von Salviati, C.; Guerra, B.; Käsbohrer, A.; Kreienbrock, L.; Roesler, U. Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 2013, 79, 4815–4820. [Google Scholar] [CrossRef] [Green Version]
- Robé, C.; Blasse, A.; Merle, R.; Friese, A.; Roesler, U.; Guenther, S. Low Dose Colonization of Broiler Chickens With ESBL-/AmpC- Producing Escherichia coli in a Seeder-Bird Model Independent of Antimicrobial Selection Pressure. Front. Microbiol. 2019, 10, 2124. [Google Scholar] [CrossRef]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef]
- Kemmett, K.; Humphrey, T.; Rushton, S.; Close, A.; Wigley, P.; Williams, N.J. A Longitudinal Study Simultaneously Exploring the Carriage of APEC Virulence Associated Genes and the Molecular Epidemiology of Faecal and Systemic E. Coli in Commercial Broiler Chickens. PLoS ONE 2013, 8, e67749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Tippelskirch, P.; Gölz, G.; Projahn, M.; Daehre, K.; Friese, A.; Roesler, U.; Alter, T.; Orquera, S. Prevalence and quantitative analysis of ESBL and AmpC beta-lactamase producing Enterobacteriaceae in broiler chicken during slaughter in Germany. Int. J. Food Microbiol. 2018, 281, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef] [PubMed]
- Gundran, R.S.; Cardenio, P.A.; Villanueva, M.A.; Sison, F.B.; Benigno, C.C.; Kreausukon, K.; Pichpol, D.; Punyapornwithaya, V. Prevalence and distribution of bla(CTX-M), bla(SHV), bla(TEM) genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 2019, 15, 227. [Google Scholar] [CrossRef] [Green Version]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type β-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Massé, J.; Lardé, H.; Fairbrother, J.M.; Roy, J.P.; Francoz, D.; Dufour, S.; Archambault, M. Prevalence of Antimicrobial Resistance and Characteristics of Escherichia coli Isolates From Fecal and Manure Pit Samples on Dairy Farms in the Province of Québec, Canada. Front. Vet. Sci. 2021, 8, 654125. [Google Scholar] [CrossRef] [PubMed]
- Hiki, M.; Usui, M.; Akiyama, T.; Kawanishi, M.; Tsuyuki, M.; Imamura, S.; Sekiguchi, H.; Kojima, A.; Asai, T. Phylogenetic grouping, epidemiological typing, analysis of virulence genes, and antimicrobial susceptibility of Escherichia coli isolated from healthy broilers in Japan. Ir. Vet. J. 2014, 67, 14. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.R.; Johnston, B.D.; Gordon, D.M. Rapid and Specific Detection of the Escherichia coli Sequence Type 648 Complex within Phylogroup F. J. Clin. Microbiol. 2017, 55, 1116–1121. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef]
- Kallonen, T.; Brodrick, H.J.; Harris, S.R.; Corander, J.; Brown, N.M.; Martin, V.; Peacock, S.J.; Parkhill, J. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017, 27, 1437–1449. [Google Scholar] [CrossRef]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian Pathogenic Escherichia coli (APEC): An Overview of Virulence and Pathogenesis Factors, Zoonotic Potential, and Control Strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, C.R.; Prussing, C.; Boerlin, P.; Daignault, D.; Dutil, L.; Reid-Smith, R.J.; Zhanel, G.G.; Manges, A.R. Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg. Infect. Dis. 2012, 18, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Ronco, T.; Stegger, M.; Olsen, R.H.; Sekse, C.; Nordstoga, A.B.; Pohjanvirta, T.; Lilje, B.; Lyhs, U.; Andersen, P.S.; Pedersen, K. Spread of avian pathogenic Escherichia coli ST117 O78:H4 in Nordic broiler production. BMC Genom. 2017, 18, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drugresistant Escherichia coli isolated from veal calves. PLoS ONE 2022, 17, e0265445. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.A.; Rempel, H.; Carrillo, C.D.; Ziebell, K.; Allen, K.; Manges, A.R.; Topp, E.; Diarra, M.S. Virulence Genotype and Phenotype of Multiple Antimicrobial-Resistant Escherichia coli Isolates from Broilers Assessed from a “One-Health” Perspective. J. Food Prot. 2022, 85, 336–354. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Ghosh, H.; Doijad, S.; Yao, Y.; Bunk, B.; Chakraborty, T. Genome Analysis of the Carbapenem- and Colistin-Resistant Escherichia coli Isolate NRZ14408 Reveals Horizontal Gene Transfer Pathways towards Panresistance and Enhanced Virulence. Antimicrob. Agents Chemother. 2017, 61, e02359-16. [Google Scholar] [CrossRef]
| Antibiotic Molecules * | Antibiotic-Free | Organic | Conventional | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R | S | Tot | R | S | Tot | R | S | Tot | |
| AMP | 55 (100) | 0 (0.00) | 55 (100) | 46 (97.87) | 1 (2.13) | 47 (100) | 54 (100) | 0 (0.00) | 54 (100) |
| AMC | 25 (45.45) | 30 (54.54) | 55 (100) | 31 (65.96) | 16 (34.04) | 47 (100) | 35 (64.81) | 19 (35.19) | 54 (100) |
| CTX | 26 (47.27) | 29 (52.73) | 55 (100) | 28 (59.57) | 19 (40.43) | 47 (100) | 39 (72.22) | 15 (27.78) | 54 (100) |
| KZ | 54 (98.18) | 1 (1.82) | 55 (100) | 44 (93.62) | 3 (6.38) | 47 (100) | 54 (100) | 0 (0.00) | 54 (100) |
| C | 17 (30.91) | 38 (69.09) | 55 (100) | 5 (10.64) | 42 (89.36) | 47 (100) | 18 (33.33) | 36 (66.67) | 54 (100) |
| CIP | 10 (18.18) | 45 (81.82) | 55 (100) | 21 (44.68) | 26 (55.32) | 47 (100) | 19 (35.19) | 35 (64.81) | 54 (100) |
| CN | 3 (5.45) | 52 (94.55) | 55 (100) | 4 (8.51) | 43 (91.49) | 47 (100) | 3 (5.56) | 51 (94.44) | 54 (100) |
| K | 4 (7.27) | 51 (92.73) | 55 (100) | 1 (2.13) | 46 (97.87) | 47 (100) | 9 (16.67) | 45 (83.33) | 54 (100) |
| NA | 28 (50.91) | 27 (49.09) | 55 (100) | 29 (61.70) | 18 (38.30) | 47 (100) | 26 (48.15) | 28 (51.85) | 54 (100) |
| S | 21 (38.18) | 34 (61.82) | 55 (100) | 4 (8.51) | 43 (91.49) | 47 (100) | 12 (22.22) | 42 (77.78) | 54 (100) |
| S3 | 38 (69.09) | 17 (30.91) | 55 (100) | 27 (57.45) | 20 (42.55) | 47 (100) | 47 (87.04) | 7 (12.96) | 54 (100) |
| TE | 35 (63.64) | 20 (36.36) | 55 (100) | 29 (61.70) | 18 (38.30) | 47 (100) | 32 (59.26) | 22 (40.74) | 54 (100) |
| SXT | 21 (38.18) | 34 (61.82) | 55 (100) | 14 (29.79) | 33 (70.21) | 47 (100) | 32 (59.26) | 22 (40.74) | 54 (100) |
| Antibiotic Molecules * | Production System | OR | 95% CI | p-Value |
|---|---|---|---|---|
| C | Conventional | 1 | - | - |
| Antibiotic-free | 0.89 | 0.40, 2.00 | 0.786 | |
| Organic | 0.24 | 0.08, 0.71 | 0.007 | |
| CIP | Conventional | 1 | - | - |
| Antibiotic-free | 0.41 | 0.17, 0.99 | 0.045 | |
| Organic | 1.49 | 0.67, 3.32 | 0.33 | |
| S3 | Conventional | 1 | - | - |
| Antibiotic-free | 0.33 | 0.13, 0.89 | 0.024 | |
| Organic | 0.20 | 0.08, 0.54 | <0.001 | |
| SXT | Conventional | 1 | - | - |
| Antibiotic-free | 0.42 | 0.20, 0.92 | 0.028 | |
| Organic | 3.21 | 0.82, 12.50 | 0.082 |
| Genotype | ESBL | AmpC | ESBL/AmpC | NEITHER |
|---|---|---|---|---|
| CTXM-1 | 56 (42.7) | 0 (0) | 0 (0) | 0 (0) |
| CTX-M-1/TEM | 38 (29) | 1 (5.6) | 0 (0) | 2 (50) |
| CTX-M-1/SHV | 2 (1.6) | 0 (0) | 0 (0) | 0 (0) |
| CTX-M-1/TEM/SHV | 2 (1.6) | 0 (0) | 0 (0) | 0 (0) |
| TEM | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| TEM/SHV | 7 (5.3) | 0 (0) | 0 (0) | 0 (0) |
| SHV | 26 (19.8) | 0 (0) | 0 (0) | 1 (25) |
| CIT/FOX/TEM | 0 (0) | 11 (61.1) | 0 (0) | 0 (0) |
| CIT/FOX | 0 (0) | 3 (16.6) | 1 (33.3) | 0 (0) |
| CIT/TEM | 0 (0) | 2 (11.1) | 1 (33.3) | 0 (0) |
| CIT/FOX/TEM/SHV | 0 (0) | 1 (5.6) | 0 (0) | 0 (0) |
| CTX-M-1/CIT | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) |
| NONE | 0 (0) | 0 (0) | 0 (0) | 1 (25) |
| Total | 131 (100) | 18 (100) | 3 (100) | 4 (100) |
| Id | Productive System | Esbl | Ampc | Phlo-Group | St | Serotype | Type | Mutation | Plasmid | Resistance Genes | Virulence Factors |
|---|---|---|---|---|---|---|---|---|---|---|---|
| R4827 | AF | POS | NEG | F | ST648 | O83:H42 | APEC | IncFIB(AP001918), IncFII, IncI1-I(Gamma), p0111 | tet(A), dfrA17, mdf(A), blaCTX-M-1, sul2, aadA5 | air, chuA, cia, cib, cvaC, eilA, etsC, gad, hlyF, iroN, iss, kpsE, kpsMII, lpfA, mchF, ompT, sitA, terC, traT, yfcV | |
| R4828 | AF | NEG | POS | G | ST117 | O8:H4 | APEC | IncB/O/K/Z, IncFIB(AP001918), IncFII, p0111 | aadA1, aac(3)-VIa, blaCMY-2, mdf(A), sul1, tet(A) | chuA, cia, cib, cvaC, etsC, gad, hlyF, iha, ireA, iroN, iss, iucC, iutA, lpfA, mchF, ompT, pic, sitA, terC, traT, vat | |
| R5147 | AF | POS | NEG | G | ST117 | H4 | APEC | gyrA p.S83L | IncFIB(AP001918), IncFII, IncI1-I(Gamma) | sul3, sul2, aadA2b, aadA1, mdf(A), tet(A), cmlA1, blaSHV-12 | chuA, cia, cib, cvaC, etsC, etsC, gad, hlyF, hra, ireA, iroN, iss, iucC, iutA, lpfA, mchF, neuC, ompT, papA_F19, pic, sitA, terC, traT, vat |
| R6162 | AF | POS | NEG | G | ST117 | H4 | APEC | gyrA p.S83L | IncFIB(AP001918), IncFIC(FII), IncFII, IncI1-I(Gamma) | tet(A), cmlA1, blaSHV-12, mdf(A), sul3, sul2, aadA2b, aadA1, aph(3″)-Ib, aph(6)-Id | chuA, cib, cvaC, etsC, gad, hlyF, hra, iha, ireA, iroN, iss, iucC, iutA, lpfA, mcbA, mchF, neuC, ompT, papA_F19, pic, sitA, terC, traT, tsh, vat |
| R6165 | AF | POS | NEG | G | ST117 | H4 | APEC | gyrA p.S83L | IncFIB(AP001918), IncFIC(FII), IncI1-I(Gamma) | mdf(A), tet(A), blaSHV-12, aph(3″)-Ib, aadA2b, aadA1, aph(6)-Id, sul3, sul2, cmlA1 | chuA, cib, cvaC, etsC, gad, hlyF, hra, iha, ireA, iroN, iss, iucC, iutA, lpfA, mchF, neuC, ompT, papA_F19, pic, sitA, terC, traT, tsh, vat |
| R4408 | C | POS | NEG | D | ST362 | O15:H1 | APEC | gyrA p.S83L | IncFIB(AP001918), IncFII, IncI1-I(Gamma) | fosX, mph(B), mdf(A), aadA1, aph(6)-Id, aph(3″)-Ib, blaCTX-M-1, tet(A), sul1, catA1, dfrA1 | afaA, afaB, afaC, afaD, afaE8, air, astA, chuA, cvaC, eilA, gad, hlyF, hra, ireA, iroN, iss, iucC, iutA, kpsE, kpsMIII_K98, mchF, ompT, sitA, terC, traT, tsh |
| R4641 | C | POS | NEG | D | ST349 | O166:H15 | AFEC | gyrA p.S83L | IncI1-I(Gamma), p0111 | blaCTX-M-1, mdf(A) | air, astA, chuA, eilA, gad, iss, terC |
| R5413 | C | POS | NEG | D | ST349 | H15 | AFEC | gyrA p.S83L | IncFII(29), IncI1-I(Gamma), p0111 | sul2, aph(3″)-Ib, aph(6)-Id, dfrA14, blaCTX-M-1, mdf(A) | air, chuA, eilA, gad, iss, terC, traT |
| R5582 | C | POS | NEG | D | ST2309 | O15:H6 | APEC | Col156, IncFIB(AP001918), IncFII, IncI1-I(Gamma), IncN, IncY | aadA2b, aadA1, sul3, mdf(A), tet(A), cmlA1, blaTEM-1B, blaSHV-12, qnrS1 | air, astA, celb, chuA, ciacib, eilA, etsC, fyuA, gad, hlyF, hra, iha, irp2, iss, iucC, iutA, kpsE, kpsMII_K5, mcbA, ompT, papC, sitA, terC, traT | |
| R5752 | C | POS | NEG | D | ST115 | O50:H6 | APEC | Col(pHAD28), IncB/O/K/Z, IncFIB(AP001918), IncFII, IncI1-I(Gamma), p0111 | aadA5, blaCTX-M-1, mdf(A), sul2, qnrB19, dfrA17 | air, astA, chuA, cia, cma, eilA, etsC, gad, hlyF, hra, iha, iss, iucC, iutA, kpsE, kpsMII_K5, mcbA, ompT, papC, sitA, terC, traT | |
| R6293 | C | POS | NEG | D | ST5897 | H31 | AFEC | IncFIA(HI1), IncFIB(AP001918), IncFIC(FII), IncFII(pHN7A8), IncI1-I(Gamma), IncX1 | tet(A), catA1, cmlA1, mdf(A), aadA1, aph(3′)-Ia, aadA2b, sul3, blaTEM-106, blaTEM-126, blaCTX-M-1, blaTEM-220, blaTEM-1B, blaTEM-135, | air, cea, chuA, cvaC, eilA, etsC, gad, hlyF, hra, iroN, iucC, iutA, lpfA, mchF, sitA, terC, traT, tsh, usp | |
| R5630 | O | NEG | POS | D | ST5931 | O1:H1 | AFEC | gyrA p.S83L | Col156, Col8282, IncB/O/K/Z, IncI2(Delta) | sul1, blaCMY-2, blaTEM-1C, mdf(A), aac(3)-VIa, aadA1, dfrA1, tet(A) | celb, chuA, cib, eilA, gad, hra, iha, iss, iucC, kpsE, kpsMII, lpfA, ompT, sitA, terC, traT |
| R5674 | O | POS | NEG | F | ST1485 | O83:H42 | APEC | gyrA p.S83L, gyrA p.D87Y, parC p.S80I | IncFIA, IncFIB(AP001918), IncFIC(FII), IncI1-I(Gamma) | blaCTX-M-1, blaTEM-1B, tet(A), mdf(A), sul2, aph(6)-Id, aph(3″)-Ib, dfrA14 | air, chuA, cib, cvaC, eilA, etsC, gad, hlyF, hra, iha, iroN, iss, iucC, iutA, kpsE, kpsMII_K5, lpfA, mchB, mchC, mchF, mcmA, ompT, sitA, terC, traT, tsh, yfcV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tofani, S.; Albini, E.; Blasi, F.; Cucco, L.; Lovito, C.; Maresca, C.; Pesciaroli, M.; Orsini, S.; Scoccia, E.; Pezzotti, G.; et al. Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/Ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms. Antibiotics 2022, 11, 1484. https://doi.org/10.3390/antibiotics11111484
Tofani S, Albini E, Blasi F, Cucco L, Lovito C, Maresca C, Pesciaroli M, Orsini S, Scoccia E, Pezzotti G, et al. Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/Ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms. Antibiotics. 2022; 11(11):1484. https://doi.org/10.3390/antibiotics11111484
Chicago/Turabian StyleTofani, Silvia, Elisa Albini, Francesca Blasi, Lucilla Cucco, Carmela Lovito, Carmen Maresca, Michele Pesciaroli, Serenella Orsini, Eleonora Scoccia, Giovanni Pezzotti, and et al. 2022. "Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/Ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms" Antibiotics 11, no. 11: 1484. https://doi.org/10.3390/antibiotics11111484
APA StyleTofani, S., Albini, E., Blasi, F., Cucco, L., Lovito, C., Maresca, C., Pesciaroli, M., Orsini, S., Scoccia, E., Pezzotti, G., Magistrali, C. F., & Massacci, F. R. (2022). Assessing the Load, Virulence and Antibiotic-Resistant Traits of ESBL/Ampc E. coli from Broilers Raised on Conventional, Antibiotic-Free, and Organic Farms. Antibiotics, 11(11), 1484. https://doi.org/10.3390/antibiotics11111484







