Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
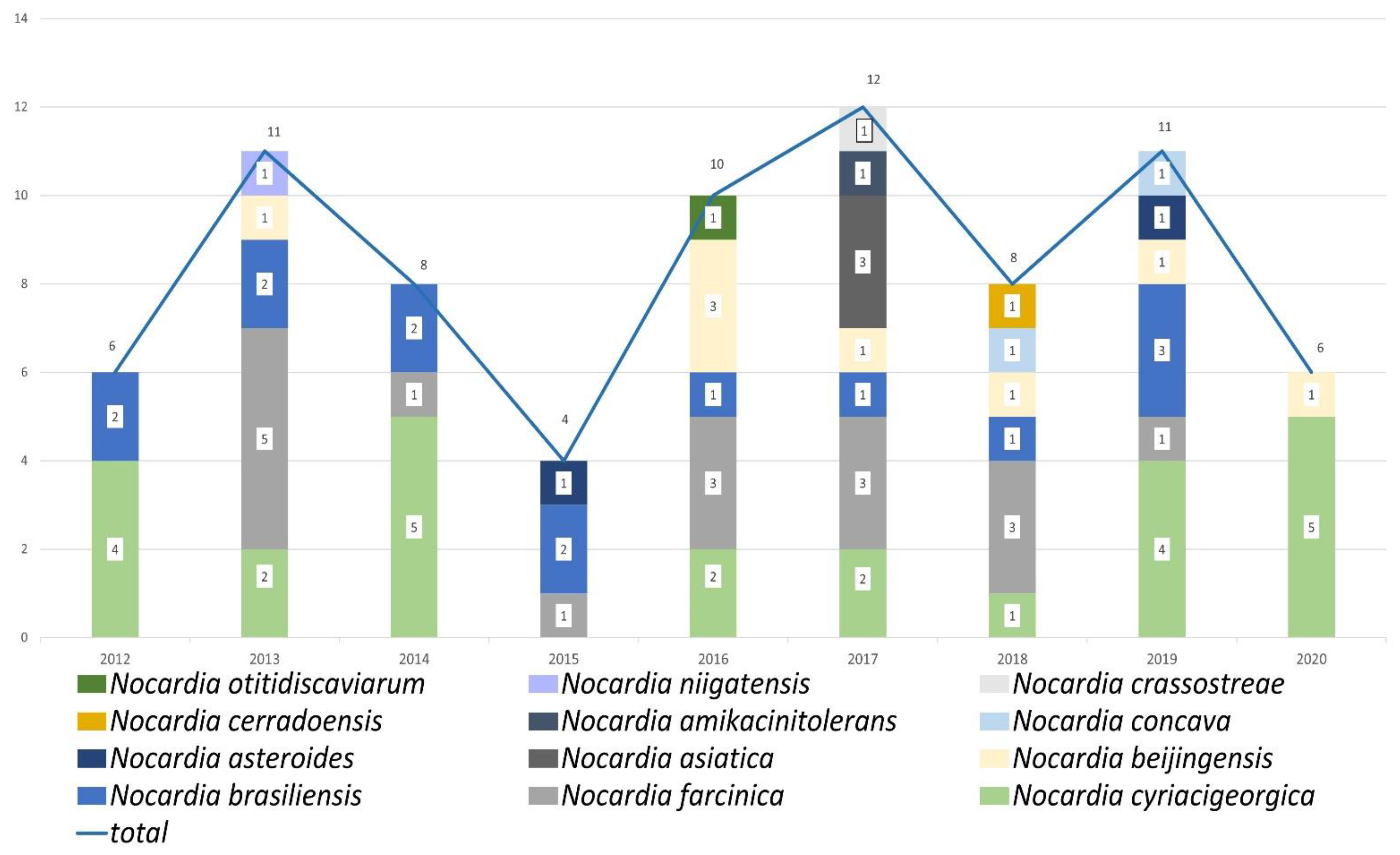
2.2. Distribution of Nocardia Species
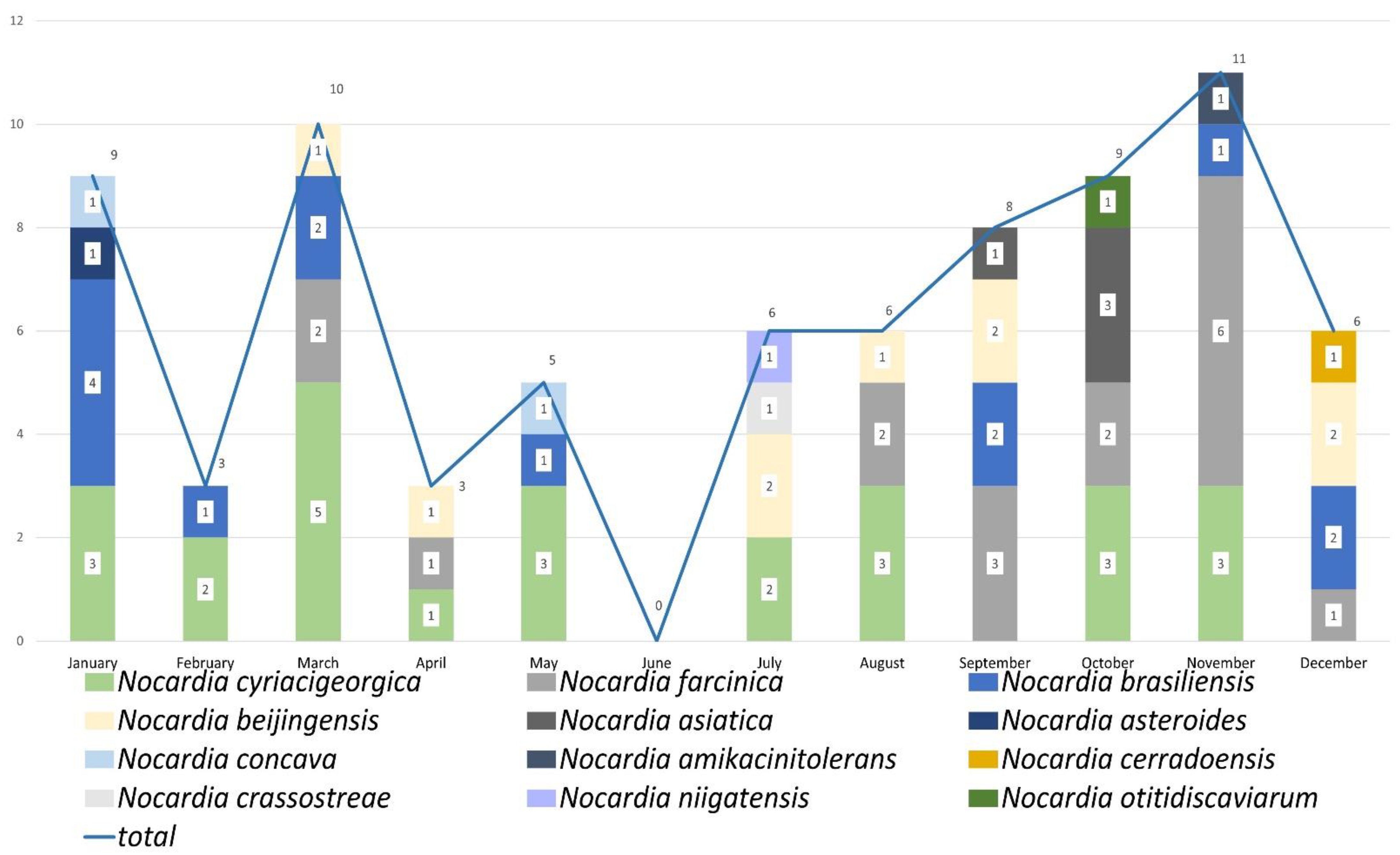
2.3. Nocardia Species Profile by Analysis of Years and Months
2.4. Antibiotic Susceptibility Profiles
2.5. PFGE for N. cyriacigeorgica
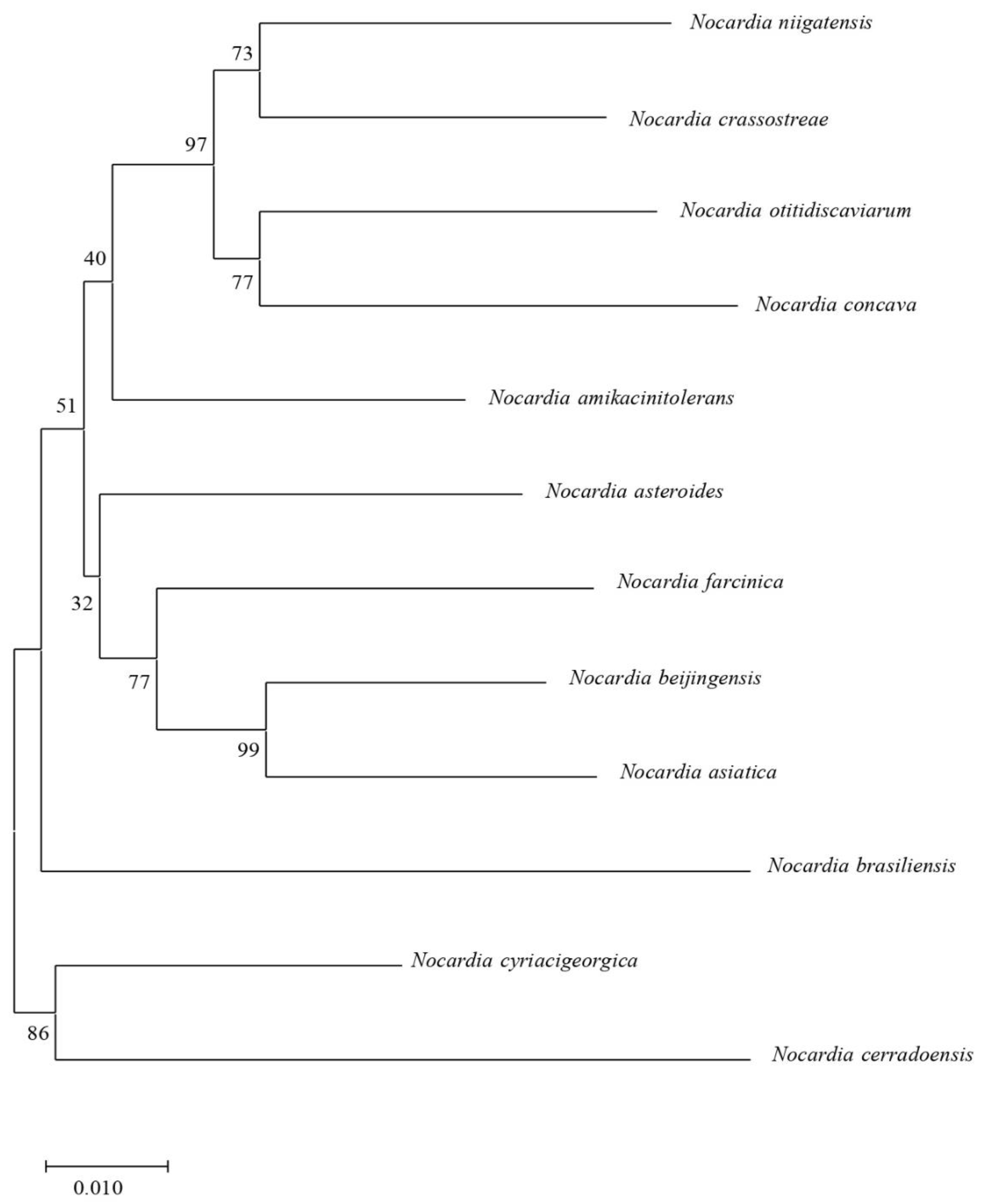
2.6. Phylogenetic Tree by MLSA Scheme
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Housekeeping Gene Selection, DNA Extraction, PCR, and Sequencing
4.3. Construction of Phylogenetic Tree
4.4. Pulsed-Field Gel Electrophoresis (PFGE) Analysis
4.5. Antimicrobial Susceptibility Test
4.6. Antimicrobial Susceptibility Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown-Elliott, B.A.; Brown, J.M.; Conville, P.S.; Wallace, R.J., Jr. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 2006, 19, 259–282. [Google Scholar] [CrossRef] [PubMed]
- Duggal, S.D.; Chugh, T.D. Nocardiosis: A neglected disease. Med. Princ. Pract. 2020, 29, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.; Jenney, A.W.; Spelman, D.W. Nocardia bacteremia: A single-center retrospective review and a systematic review of the literature. Int. J. Infect. Dis. 2020, 92, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, X.; Xu, H.; Sun, L.; Li, C.; Guo, W.; Xiang, L.; Luo, G.; Cui, Y.; Lu, B. Clinical features, identification, antimicrobial resistance patterns of Nocardia species in China: 2009–2017. Diagn. Microbiol. Infect. Dis. 2019, 94, 165–172. [Google Scholar] [CrossRef]
- Lebeaux, D.; Bergeron, E.; Berthet, J.; Djadi-Prat, J.; Mouniée, D.; Boiron, P.; Lortholary, O.; Rodriguez-Nava, V. Antibiotic susceptibility testing and species identification of Nocardia isolates: A retrospective analysis of data from a French expert laboratory, 2010–2015. Clin. Microbiol. Infect. 2019, 25, 489–495. [Google Scholar] [CrossRef]
- Van den Bogaart, L.; Manuel, O. Antibiotic therapy for difficult-to-treat infections in lung transplant recipients: A practical approach. Antibiotics 2022, 11, 612. [Google Scholar] [CrossRef]
- Conville, P.S.; Brown-Elliott, B.A.; Smith, T.; Zelazny, A.M. The complexities of Nocardia taxonomy and identification. J. Clin. Microbiol. 2017, 56, e01419-17. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Doucet, J.; Witkowska, M.; Richardson, S.E. Antimicrobial susceptibility among clinical Nocardia species identified by multilocus sequence analysis. Antimicrob. Agents Chemother. 2015, 59, 269–275. [Google Scholar] [CrossRef]
- Marín, M.; Ruiz, A.; Iglesias, C.; Quiroga, L.; Cercenado, E.; Martín-Rabadán, P.; Bouza, E.; Rodríguez-Sánchez, B. Identification of Nocardia species from clinical isolates using MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2018, 24, 1342.e5–1342.e8. [Google Scholar] [CrossRef]
- Tan, Y.E.; Chen, S.C.; Halliday, C.L. Antimicrobial susceptibility profiles and species distribution of medically relevant Nocardia species: Results from a large tertiary laboratory in Australia. J. Glob. Antimicrob. Resist. 2020, 20, 110–117. [Google Scholar] [CrossRef]
- Lai, C.C.; Liu, W.L.; Ko, W.C.; Chen, Y.H.; Tan, H.R.; Huang, Y.T.; Hsueh, P.R. Multicenter study in Taiwan of the in vitro activities of nemonoxacin, tigecycline, doripenem, and other antimicrobial agents against clinical isolates of various Nocardia species. Antimicrob. Agents Chemother. 2011, 55, 2084–2091. [Google Scholar] [CrossRef]
- Liu, W.L.; Lai, C.C.; Ko, W.C.; Chen, Y.H.; Tang, H.J.; Huang, Y.L.; Huang, Y.T.; Hsueh, P.R. Clinical and microbiological characteristics of infections caused by various Nocardia species in Taiwan: A multicenter study from 1998 to 2010. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1341–1347. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Cui, Q.; Wu, W.; Li, G.; Chen, D.; Xiang, L.; Qu, J.; Shi, D.; Lu, B. Epidemiology and Antimicrobial Resistance Profiles of the Nocardia Species in China, 2009 to 2021. Microbiol. Spectr. 2022, 10, e0156021. [Google Scholar] [CrossRef]
- Tremblay, J.; Thibert, L.; Alarie, I.; Valiquette, L.; Pépin, J. Nocardiosis in Quebec, Canada, 1988–2008. Clin. Microbiol. Infect. 2011, 17, 690–696. [Google Scholar] [CrossRef]
- Uhde, K.B.; Pathak, S.; McCullum, I.; Jannat-Khah, D.P., Jr.; Shadomy, S.V.; Dykewicz, C.A.; Clark, T.A.; Smith, T.L.; Brown, J.M. Antimicrobial-resistant Nocardia isolates, United States, 1995–2004. Clin. Infect. Dis. 2010, 51, 1445–1448. [Google Scholar] [CrossRef]
- Minero, M.V.; Marín, M.; Cercenado, E.; Rabadán, P.M.; Bouza, E.; Muñoz, P. Nocardiosis at the turn of the century. Medicine 2009, 88, 250–261. [Google Scholar] [CrossRef]
- Chen, Y.C.; Lee, C.H.; Chien, C.C.; Chao, T.L.; Lin, W.C.; Liu, J.W. Pulmonary nocardiosis in southern Taiwan. J. Microbiol. Immunol. Infect. 2013, 46, 441–447. [Google Scholar] [CrossRef]
- Wei, M.; Xu, X.; Yang, J.; Wang, P.; Liu, Y.; Wang, S.; Yang, C.; Gu, L. MLSA phylogeny and antimicrobial susceptibility of clinical Nocardia isolates: A multicenter retrospective study in China. BMC Microbiol. 2021, 21, 342. [Google Scholar] [CrossRef]
- Valdezate, S.; Garrido, N.; Carrasco, G.; Medina-Pascual, M.J.; Villalón, P.; Navarro, A.M.; Saéz-Nieto, J.A. Epidemiology and susceptibility to antimicrobial agents of the main Nocardia species in Spain. J. Antimicrob. Chemother. 2017, 72, 754–761. [Google Scholar]
- McGuinness, S.L.; Whiting, S.E.; Baird, R.; Currie, B.J.; Ralph, A.P.; Anstey, N.M.; Price, R.N.; Davis, J.S.; Tong, S.Y.C. Nocardiosis in the tropical northernterritory of Australia, 1997–2014. Open Forum Infect. Dis. 2016, 3, ofw208. [Google Scholar] [CrossRef]
- Grau, S.; Fondevilla, E.; Echeverría-Esnal, D.; Alcorta, A.; Limon, E.; Gudiol, F.; VINCat Program group. Widespread increase of empirical carbapenem use in acute care hospitals in Catalonia, Spain. Enferm. Infecc. Microbiol. Clin. 2019, 37, 36–40. [Google Scholar] [CrossRef]
- Rhodes, N.J.; Wagner, J.L.; Davis, S.L.; Bosso, J.A.; Goff, D.A.; Rybak, M.J.; Scheetz, M.H.; MAD-ID Research Network. Trends in and predictors of carbapenem consumption across north American hospitals: Results from a multicenter survey by the MAD-ID research network. Antimicrob. Agents Chemother. 2019, 63, e00327-19. [Google Scholar] [CrossRef]
- Yew, W.W.; Wong, P.C.; Kwan, S.Y.; Chan, C.Y.; Li, M.S. Two cases of Nocardia asteroides sternotomy infection treated with ofloxacin and a review of other active antimicrobial agents. J. Infect. 1991, 23, 297–302. [Google Scholar] [CrossRef]
- Exmelin, L.; Malbruny, B.; Vergnaud, M.; Prosvost, F.; Boiron, P.; Morel, C. Molecular study of nosocomial nocardiosis outbreak involving heart transplant recipients. J. Clin. Microbiol. 1996, 34, 1014–1016. [Google Scholar] [CrossRef]
- Blümel, J.; Blümel, E.; Yassin, A.F.; Schmidt-Rotte, H.; Schaal, K.P. Typing of Nocardia farcinica by pulsed-field gel electrophoresis reveals an endemic strain as source of hospital infections. J. Clin. Microbiol. 1998, 36, 118–122. [Google Scholar] [CrossRef]
- Wenger, P.N.; Brown, J.M.; McNeil, M.M.; Jarvis, W.R. Nocardia farcinica sternotomy site infections in patients following open heart surgery. J. Infect. Dis. 1998, 178, 1539–1543. [Google Scholar] [CrossRef]
- Apostolou, A.; Bolcen, S.J.; Dave, V.; Jani, N.; Lasker, B.A.; Tan, C.G.; Montana, B.; Brown, J.M.; Genese, C.A. Nocardia cyriacigeorgica infections attributable to unlicensed cosmetic procedures—An emerging public health problem? Clin. Infect. Dis. 2012, 55, 251–253. [Google Scholar] [CrossRef]
- Wright, L.; Katouli, M.; Kurtböke, D.İ. Isolation and characterization of Nocardiae associated with foaming coastal marine waters. Pathogens 2021, 10, 579. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Kang, Y.; Yazawa, K.; Gonoi, T.; Mikami, Y. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J. Med. Microbiol. 2010, 59, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, H.; Zhang, E.; Sintchenko, V.; Xiao, M.; Sorrell, T.C.; Chen, X.; Chen, S.C. secA1 gene sequence polymorphisms for species identification of Nocardia species and recognition of intraspecies genetic diversity. J. Clin. Microbiol. 2010, 48, 3928–3934. [Google Scholar] [CrossRef] [PubMed]
- McTaggart, L.R.; Richardson, S.E.; Witkowska, M.; Zhang, S.X. Phylogeny and identification of Nocardia species on the basis of multilocus sequence analysis. J. Clin. Microbiol. 2010, 48, 4525–4533. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Gnanam, H.; Rajapandian, S.; Gunasekaran, R.; Roshni Prithiviraj, S.; Ravindran, R.S.; Sen, S.; Prajna, L. Molecular identification of Nocardia species causing endophthalmitis using multilocus sequence analysis (MLSA): A 10-year perspective. J. Med. Microbiol. 2020, 9, 728–738. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes, 1st ed.; Approved standard M62; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Wallace, R.J.; Steele, L.C., Jr.; Sumter, G.; Smith, J.M. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 1988, 32, 1776–1779. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, X.; Du, P.; Li, G.; Li, L.; Li, Z. Susceptibility profiles of Nocardia spp. to antimicrobial and antituberculotic agents detected by a microplate Alamar Blue assay. Sci. Rep. 2017, 7, 43660. [Google Scholar] [CrossRef]

| Characteristics | |
|---|---|
| Gender, n (%) | |
| Male | 49 (63.6) |
| Age (years) | |
| Median (range) | 76 (31–97) |
| Mean ± standard deviation | 70.4 ± 15.7 |
| Specimen type, n (%) | |
| Pus | 21 (27.3) |
| Sputum | 14 (18.2) |
| Wound | 11 (14.3) |
| Blood | 7 (9.1) |
| Abscess | 5 (6.5) |
| Bronchial washing | 7 (9.1) |
| Corneal ulcer | 4 (5.2) |
| Pleural effusion | 4 (5.2) |
| Synovial fluid | 2 (2.6) |
| Cerebrospinal fluid | 1 (1.3) |
| Bone tissue | 1 (1.3) |
| Site of involvement, n (%) | |
| Lung | 35 (45.5) |
| Central nervous system | 9 (11.7) |
| Skin and soft tissue | 19 (24.7) |
| Bone and joint | 7 (9.1) |
| Blood stream | 7 (9.1) |
| Disseminated (including blood stream) | 18 (23.3) |
| Nocardia Species | No. of Isolates | Drug Patterns Types | Antimicrobial Susceptibility Pattern | |
|---|---|---|---|---|
| Non-Susceptible (%) | Susceptible (%) | |||
| N. farcinica | 18 | V | IPM (100) | SXT (100) |
| FEP (100) | LZD (100) | |||
| DOX (100) | AN (100) | |||
| TOB (100) | ||||
| CLR (100) | ||||
| N. cyriacigeorgica | 25 | VI | CIP (100) | SXT (100) |
| IPM (100) | LZD (100) | |||
| MXF (100) | AN (100) | |||
| FEP (100) | TOB (100) | |||
| AMC (100) | ||||
| CLR (92) | ||||
| N. brasiliensis | 13 | VIII | CIP (100) | SXT (100) |
| IPM (100) | LZD (100) | |||
| FEP (100) | AN (100) | |||
| CRO (92) | TOB (100) | |||
| DOX (100) | ||||
| CLR (92) | ||||
| N. otitidiscaviarium | 1 | VII | CIP (100) | SXT (100) |
| IPM (100) | LZD (100) | |||
| FEP (100) | AN (100) | |||
| AMC (100) | TOB (100) | |||
| CRO (100) | ||||
| CLR (100) | ||||
| Antimicrobial Agent | Species (No. of Strains Tested) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N. cyriacigeorgica (25) | N. brasiliensis (13) | N. farcinica (18) | N. niigatensis (1) | N. asteroides (2) | N. beijingensis (9) | N. otitidiscaviarum (1) | N. crassostreae (1) | N. concava (2) | N. cerradoensis (1) | N. asiatica (3) | N. amikacinitolerans (1) | |
| Trimethoprim/ Sulfamethoxazole (SXT) | ||||||||||||
| Resistant [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intermediate [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 25 (100) | 13 (100) | 18 (100) | 1 (100) | 2 (100) | 9 (100) | 1 (100) | 1 (100) | 2 (100) | 1 (100) | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 0.25/4.75 | 0.5/9.5 | 1/19 | 0.25/4.75 | 0.25/4.75 | |||||||
| MIC90 [µg/mL] | 0.5/9.5 | 0.5/9.5 | 2/38 | 1/19 | 0.25/4.75 | |||||||
| Linezolid (LZD) | ||||||||||||
| Resistant [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intermediate [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 25 (100) | 13 (100) | 18 (100) | 1 (100) | 2 (100) | 9 (100) | 1 (100) | 1 (100) | 2 (100) | 1 (100) | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 2 | 2 | 2 | 1 | 1 | |||||||
| MIC90 [µg/mL] | 2 | 4 | 4 | 2 | 1 | |||||||
| Ciprofloxacin (CIP) | ||||||||||||
| Resistant [n (%)] | 25 (100) | 13 (100) | 4 (22.2) | 0 | 2 (100) | 5 (55.6) | 1 (100) | 1 (100) | 1 (50) | 0 | 3 (100) | 1 (100) |
| Intermediate [n (%)] | 0 | 0 | 3 (16.7) | 1 (100) | 0 | 1 (11.1) | 0 | 0 | 1 (50) | 0 | 0 | 0 |
| Susceptible [n (%)] | 0 | 0 | 11 (61.1) | 0 | 0 | 3 (33.3) | 0 | 0 | 0 | 1 (100) | 0 | 0 |
| MIC50 [µg/mL] | >4 | >4 | 1 | 4 | >4 | |||||||
| MIC90 [µg/mL] | >4 | >4 | >4 | >4 | >4 | |||||||
| Imipenem (IPM) | ||||||||||||
| Resistant [n (%)] | 21 (84.0) | 10 (76.9) | 14 (77.8) | 1 (100) | 0 | 5 (55.5) | 1 (100) | 1 (100) | 2 (100) | 0 | 0 | 1 (100) |
| Intermediate [n (%)] | 4 (16.0) | 3 (23.1) | 4 (22.2) | 0 | 2 (100) | 1 (11.2) | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 0 | 0 | 0 | 0 | 0 | 3 (33.3) | 0 | 0 | 0 | 1 (100) | 3 (100) | 0 |
| MIC50 [µg/mL] | 16 | 32 | 16 | 16 | 2 | |||||||
| MIC90 [µg/mL] | 16 | >64 | 32 | 64 | 4 | |||||||
| Moxifloxacin (MXF) | ||||||||||||
| Resistant [n (%)] | 24 (96.0) | 0 | 4 (22.2) | 0 | 1 (50) | 2 (22.3) | 0 | 0 | 0 | 0 | 0 | 1 (100) |
| Intermediate [n (%)] | 1 (4.0) | 11 (84.6) | 0 | 0 | 1 (50) | 3 (33.3) | 1 (100) | 0 | 0 | 1 (100) | 0 | 0 |
| Susceptible [n (%)] | 0 | 2 (15.4) | 14 (77.8) | 1 (100) | 0 | 4 (44.4) | 0 | 1 (100) | 2 (100) | 0 | 3 (100) | 0 |
| MIC50 [µg/mL] | 4 | 2 | 0.25 | 2 | 8 | |||||||
| MIC90 [µg/mL] | 8 | 2 | 4 | >8 | >8 | |||||||
| Cefepime (FEP) | ||||||||||||
| Resistant [n (%)] | 16 (64.0) | 12 (92.3) | 16 (88.8) | 1 (100) | 2 (100) | 4 (44.4) | 1 (100) | 1 (100) | 2 (100) | 0 | 0 | 1 (100) |
| Intermediate [n (%)] | 9 (36.0) | 1 (7.8) | 2 (11.2) | 0 | 0 | 2 (22.3) | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 0 | 0 | 0 | 0 | 0 | 3 (33.3) | 0 | 0 | 0 | 1 (100) | 3 (100) | 0 |
| MIC50 [µg/mL] | 32 | >32 | >32 | 16 | 8 | |||||||
| MIC90 [µg/mL] | >32 | >32 | >32 | 32 | 8 | |||||||
| Cefoxitin (FOX) | ||||||||||||
| MIC range | 64–128 | 16–128 | 64–128 | >128 | 16–64 | 8–32 | >128 | >128 | >128 | 64 | 4–16 | >64 |
| Amoxicillin/clavulanic acid 2:1 ratio (AMC) | ||||||||||||
| Resistant [n (%)] | 23 (92.0) | 1 (7.7) | 3 (16.7) | 1 (100) | 2 (100) | 4 (44.5) | 1 (100) | 1 (100) | 2 (100) | 0 | 3 (100) | 0 |
| Intermediate [n (%)] | 2 (8.0) | 2 (15.4) | 12 (66.6) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 0 | 10 (76.9) | 3 (16.7) | 0 | 0 | 5 (55.5) | 0 | 0 | 0 | 1 (100) | 0 | 1 (100) |
| MIC50 [µg/mL] | 32/16 | 8/4 | 16/8 | 8/4 | >64/32 | |||||||
| MIC90 [µg/mL] | 64/32 | 16/8 | 32/16 | >64/32 | >64/32 | |||||||
| Amikacin (AN) | ||||||||||||
| Resistant [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intermediate [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 25 (100) | 13 (100) | 18 (100) | 1 (100) | 2 (100) | 9 (100) | 1 (100) | 1 (100) | 2 (100) | 1 (100) | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 1 | 1 | 1 | 1 | 1 | |||||||
| MIC90 [µg/mL] | 1 | 1 | 1 | 1 | 1 | |||||||
| Ceftriaxone (CRO) | ||||||||||||
| Resistant [n (%)] | 2 (8.0) | 10 (76.9) | 15 (83.3) | 1 (100) | 0 | 1 (11.1) | 1 (100) | 1 (100) | 2 (100) | 0 | 0 | 1 (100) |
| Intermediate [n (%)] | 8 (32.0) | 2 (15.4) | 1 (5.6) | 0 | 0 | 3 (33.3) | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 15 (60.0) | 1 (7.7) | 2 (11.1) | 0 | 2 (100) | 5 (55.6) | 0 | 0 | 0 | 1 (100) | 3 (100) | 0 |
| MIC50 [µg/mL] | 8 | >64 | >64 | 4 | 4 | |||||||
| MIC90 [µg/mL] | 32 | >64 | >64 | 32 | 4 | |||||||
| Doxycycline (DOX) | ||||||||||||
| Resistant [n (%)] | 0 | 1 (7.7) | 1 (5.6) | 1 (100) | 0 | 0 | 0 | 0 | 2 (100) | 0 | 0 | 0 |
| Intermediate [n (%)] | 17 (68.0) | 12 (92.3) | 17 (94.4) | 0 | 2 (100) | 6 (66.7) | 1 (100) | 1 (100) | 0 | 1 (100) | 0 | 0 |
| Susceptible [n (%)] | 8 (32.0) | 0 | 0 | 0 | 0 | 3 (33.3) | 0 | 0 | 0 | 0 | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 2 | 4 | 4 | 2 | 0.12 | |||||||
| MIC90 [µg/mL] | 4 | 4 | 4 | 4 | 0.12 | |||||||
| Minocycline (MIN) | ||||||||||||
| Resistant [n (%)] | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 (100) | 0 | 0 | 0 |
| Intermediate [n (%)] | 17 (68.0) | 11 (84.6) | 17 (94.4) | 1 (100) | 2 (100) | 3 (33.3) | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 8 (32.0) | 2 (15.4) | 1 (5.6) | 0 | 0 | 6 (66.7) | 0 | 0 | 0 | 1 (100) | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 2 | 4 | 4 | 1 | 1 | |||||||
| MIC90 [µg/mL] | 4 | 4 | 4 | 2 | 1 | |||||||
| Tigecycline (TGC) | ||||||||||||
| MIC range | 0.25–2 | 0.25–0.5 | 0.5–4 | 1 | 0.5–1 | 0.12–0.5 | 1 | 2 | 2 (100) | 0.12 | 0.25 | 2 |
| Tobramycin (TOB) | ||||||||||||
| Resistant [n (%)] | 0 | 0 | 16 (88.9) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intermediate [n (%)] | 0 | 0 | 2 (11.1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 25 (100) | 13 (100) | 0 | 1 (100) | 2 (100) | 9 (100) | 1 (100) | 1 (100) | 2 (100) | 1 (100) | 3 (100) | 1 (100) |
| MIC50 [µg/mL] | 1 | 1 | 16 | 1 | 1 | |||||||
| MIC90 [µg/mL] | 1 | 1 | >16 | 1 | 1 | |||||||
| Clarithromycin (CLR) | ||||||||||||
| Resistant [n (%)] | 22 (88.0) | 9 (69.2) | 18 (100) | 1 (100) | 2 (100) | 2 (22.2) | 1 (100) | 1 (100) | 0 | 0 | 1 (33.3) | 1 (100) |
| Intermediate [n (%)] | 1 (4.0) | 3 (23.0) | 0 | 0 | 0 | 2 (22.2) | 0 | 0 | 0 | 0 | 0 | 0 |
| Susceptible [n (%)] | 2 (8.0) | 1 (7.8) | 0 | 0 | 0 | 5 (55.6) | 0 | 0 | 2 (100) | 1 (100) | 2 (66.7) | 0 |
| MIC50 [µg/mL] | >16 | 8 | >16 | 1 | 1 | |||||||
| MIC90 [µg/mL] | >16 | >16 | >16 | 16 | 16 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, S.-F.; Chen, F.-J.; Lan, I.-C.; Chien, C.-C.; Lee, C.-H. Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility. Antibiotics 2022, 11, 1438. https://doi.org/10.3390/antibiotics11101438
Kuo S-F, Chen F-J, Lan I-C, Chien C-C, Lee C-H. Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility. Antibiotics. 2022; 11(10):1438. https://doi.org/10.3390/antibiotics11101438
Chicago/Turabian StyleKuo, Shu-Fang, Fang-Ju Chen, I-Chia Lan, Chun-Chih Chien, and Chen-Hsiang Lee. 2022. "Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility" Antibiotics 11, no. 10: 1438. https://doi.org/10.3390/antibiotics11101438
APA StyleKuo, S.-F., Chen, F.-J., Lan, I.-C., Chien, C.-C., & Lee, C.-H. (2022). Epidemiology of Nocardia Species at a Tertiary Hospital in Southern Taiwan, 2012 to 2020: MLSA Phylogeny and Antimicrobial Susceptibility. Antibiotics, 11(10), 1438. https://doi.org/10.3390/antibiotics11101438






