High-Throughput Transcriptomic Profiling Reveals the Inhibitory Effect of Hydroquinine on Virulence Factors in Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Hydroquinine Preparation
2.3. RNA Extraction
2.4. Transcriptomic Analysis and Differential Expression Gene (DEG) Analysis
2.5. Complementary DNA (cDNA) Synthesis and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.6. Motility Assays
2.7. Detection of Quorum Sensing (QS) Signaling-Related Virulence Factors
2.8. Statistical Analysis
3. Results
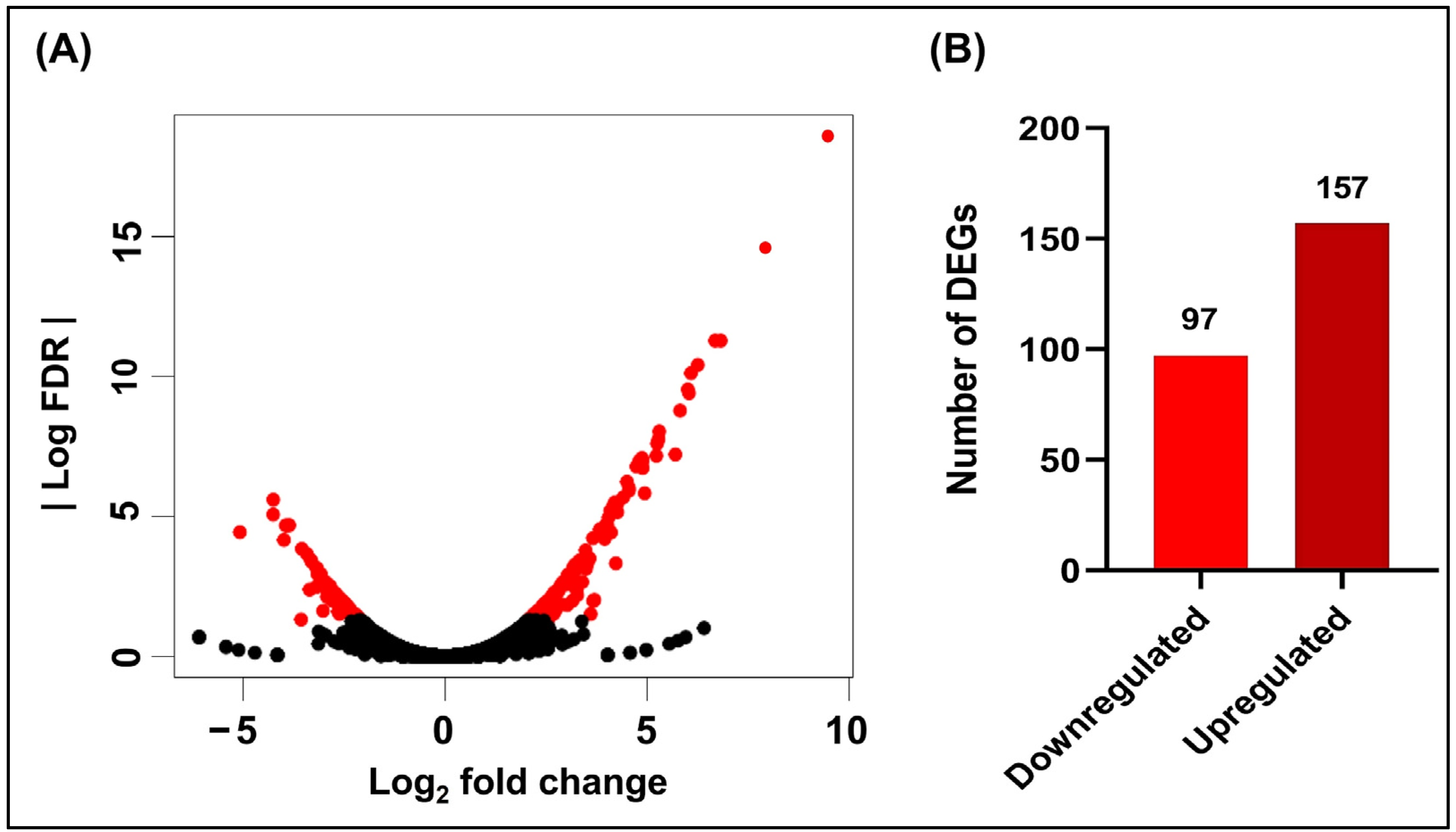
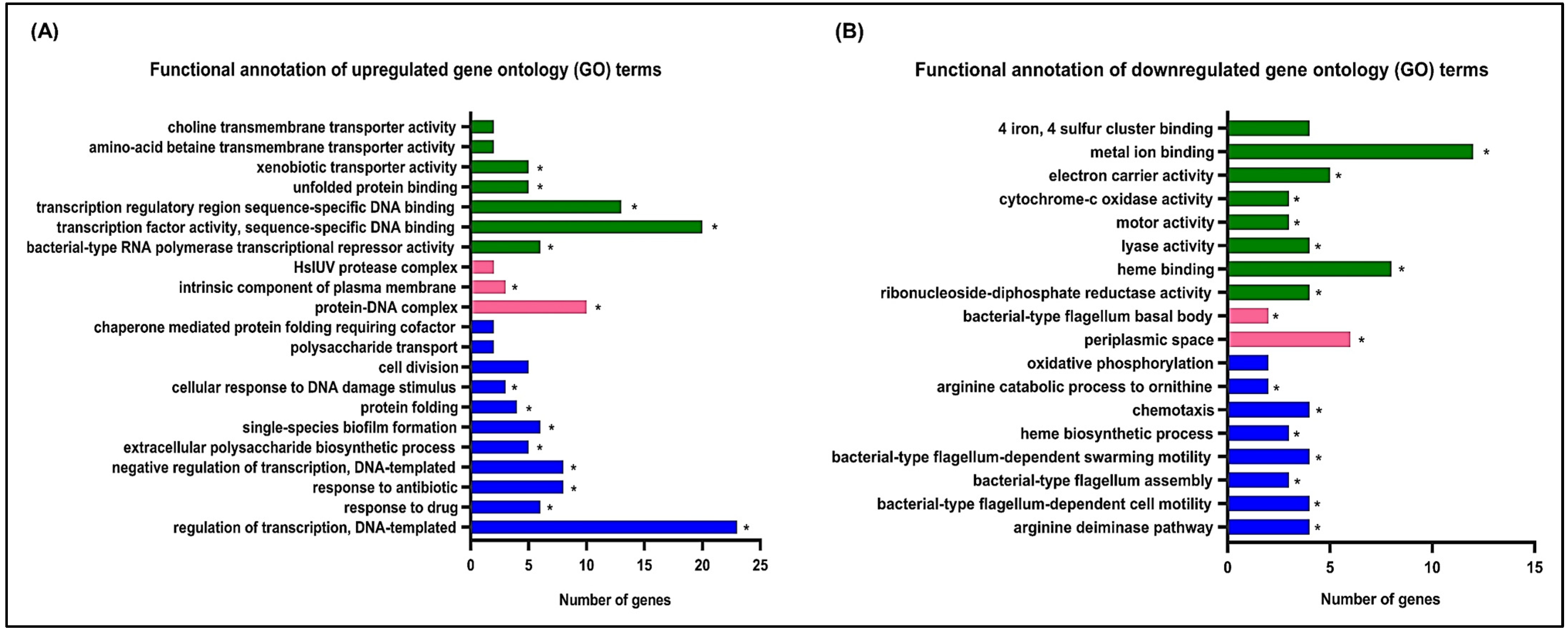
3.1. Effects of Hydroquinine on the Transcriptomic Profile of P. aeruginosa ATCC 27853
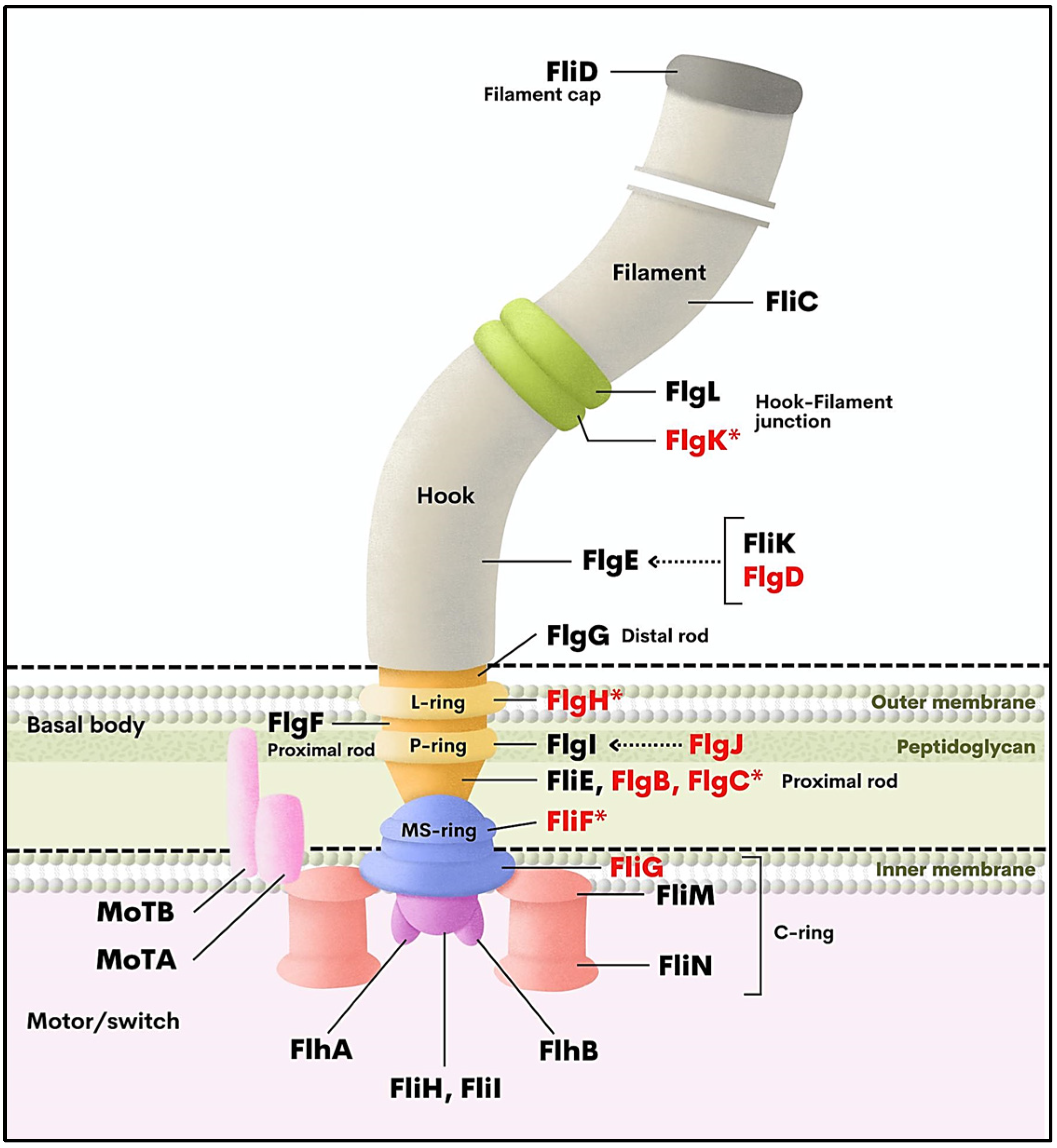
3.2. Validation of Downregulation of Flagella-Related Genes by qRT-PCR
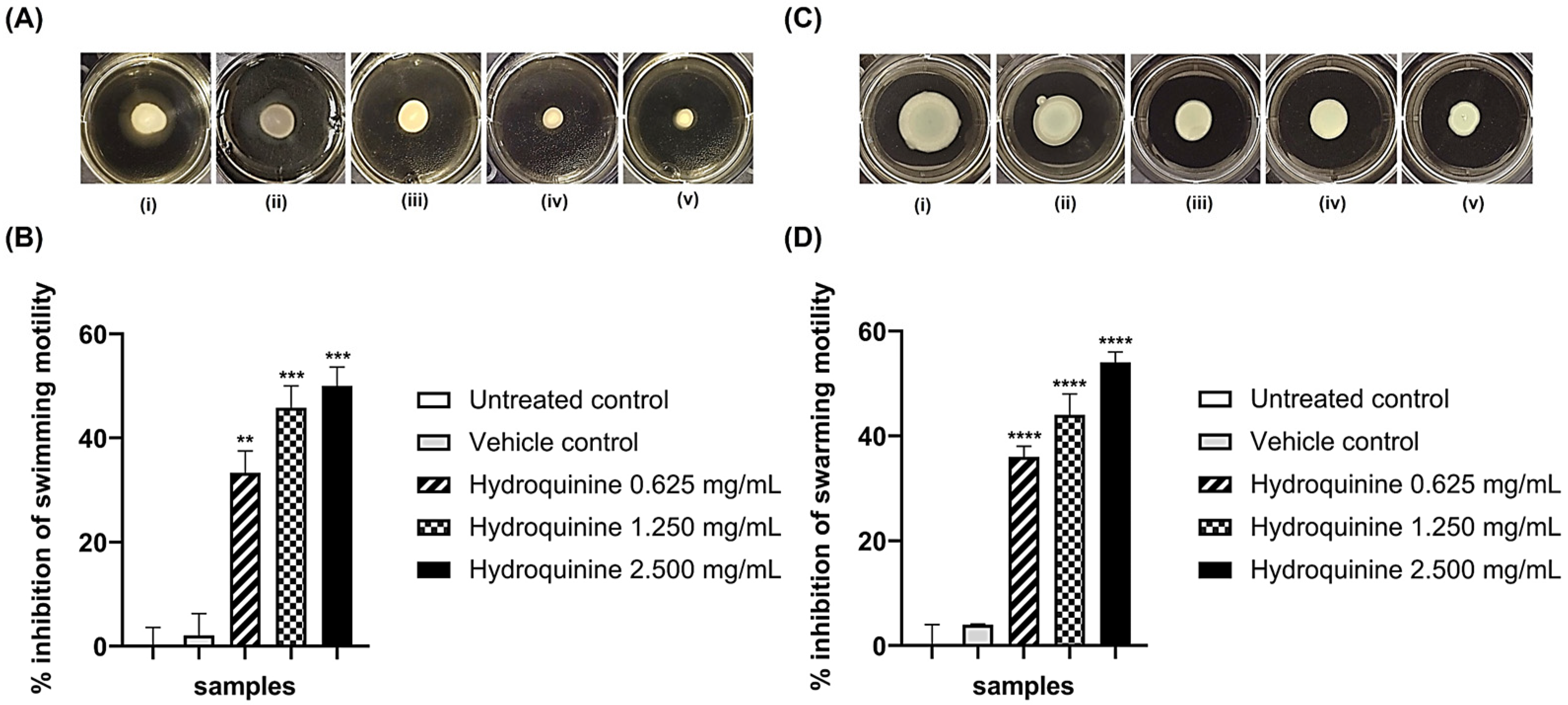
3.3. Hydroquinine Disrupts Motility in Both P. aeruginosa Strains
3.4. Inhibitory Effects of Hydroquinine on the Virulence Factor Productions and QS-Related Genes Were Exhibited in Both P. aeruginosa Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 15 August 2022).
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The mortality burden of multidrug-resistant pathogens in India: A retrospective, observational study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Park, B.K.; Kim, S.K.; Han, S.B.; Lee, J.W.; Lee, D.G.; Chung, N.G.; Cho, B.; Jeong, D.C.; Kang, J.H. Clinical characteristics and outcomes of Pseudomonas aeruginosa bacteremia in febrile neutropenic children and adolescents with the impact of antibiotic resistance: A retrospective study. BMC Infect. Dis. 2017, 17, 500. [Google Scholar] [CrossRef] [PubMed]
- Parker, C.M.; Kutsogiannis, J.; Muscedere, J.; Cook, D.; Dodek, P.; Day, A.G.; Heyland, D.K. Ventilator-associated pneumonia caused by multidrug-resistant organisms or Pseudomonas aeruginosa: Prevalence, incidence, risk factors, and outcomes. J. Crit. Care 2008, 23, 18–26. [Google Scholar] [CrossRef]
- Paz-Zarza, V.M.; Mangwani-Mordani, S.; Martínez-Maldonado, A.; Álvarez-Hernández, D.; Solano-Gálvez, S.G.; Vázquez-López, R. Pseudomonas aeruginosa: Pathogenicity and antimicrobial resistance in urinary tract infection. Rev. Chil. Infectol. 2019, 36, 180–189. [Google Scholar] [CrossRef]
- Parchand, S.M.; Agrawal, D.; Chatterjee, S.; Gangwe, A.; Mishra, M.; Agrawal, D. Post-cataract surgery cluster endophthalmitis due to multidrug-resistant Pseudomonas aeruginosa: A retrospective cohort study of six clusters. Indian J. Ophthalmol. 2020, 68, 1424–1431. [Google Scholar] [CrossRef]
- Meschiari, M.; Franconi, I.; Bacca, E.; Bianco, V.; Orlando, G.; Cuomo, G.; Bedini, A.; Mussini, C. Ceftazidime/avibactam and ceftolozane/tazobactam for the treatment of extensively drug-resistant Pseudomonas aeruginosa post-neurosurgical infections: Three cases and a review of the literature. Infection 2021, 49, 549–553. [Google Scholar] [CrossRef]
- Mirzaei, B.; Bazgir, Z.N.; Goli, H.R.; Iranpour, F.; Mohammadi, F.; Babaei, R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res. Notes 2020, 13, 380. [Google Scholar] [CrossRef]
- Peña, C.; Cabot, G.; Gómez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A.; et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef]
- Sader, H.; Castanheira, M.; Duncan, L.; Flamm, R. Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa Isolates from United States medical centers stratified by infection type: Results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program, 2015–2016. Diagn. Microbiol. Infect. Dis. 2018, 92, 69–74. [Google Scholar] [CrossRef]
- Walkty, A.; Lagace-Wiens, P.; Adam, H.; Baxter, M.; Karlowsky, J.; Mulvey, M.R.; McCracken, M.; Zhanel, G.G. Antimicrobial susceptibility of 2906 Pseudomonasaeruginosa clinical isolates obtained from patients in Canadian hospitals over a period of 8 years: Results of the Canadian Ward surveillance study (CANWARD), 2008–2015. Diagn. Microbiol. Infect. Dis. 2017, 87, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Naik, P.; Pandey, S.; Gagan, S.; Biswas, S.; Joseph, J. Virulence factors in multidrug (MDR) and Pan-drug resistant (XDR) Pseudomonas aeruginosa: A cross-sectional study of isolates recovered from ocular infections in a high-incidence setting in southern India. J. Ophthalmic Inflamm. Infect. 2021, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial virulence factors: Secreted for survival. Indian J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant natural products targeting bacterial virulence factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef]
- Alonso, B.; Fernández-Barat, L.; Di Domenico, E.G.; Marín, M.; Cercenado, E.; Merino, I.; De Pablos, M.; Muñoz, P.; Guembe, M. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect. Dis. 2020, 20, 909. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Mühlen, S.; Dersch, P. Anti-virulence strategies to target bacterial infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [CrossRef]
- Gonelimali, F.D.; Lin, J.; Miao, W.; Xuan, J.; Charles, F.; Chen, M.; Hatab, S.R. Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front. Microbiol. 2018, 9, 1639. [Google Scholar] [CrossRef]
- Gyawali, R.; Ibrahim, S.A. Natural products as antimicrobial agents. Food Control 2014, 46, 412–429. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Cho, M.H.; Kim, J.-A.; Lee, J. 7-fluoroindole as an antivirulence compound against Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2012, 329, 36–44. [Google Scholar] [CrossRef]
- Monte, J.; Abreu, A.C.; Borges, A.; Simões, L.C.; Simões, M. Antimicrobial activity of selected phytochemicals against Escherichia coli and Staphylococcus aureus and their biofilms. Pathogens 2014, 3, 473–498. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, M.H.; Lee, J. 3-indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ. Microbiol. 2011, 13, 62–73. [Google Scholar] [CrossRef]
- Norizan, S.N.; Yin, W.F.; Chan, K.G. Caffeine as a potential quorum sensing inhibitor. Sensors 2013, 13, 5117–5129. [Google Scholar] [CrossRef]
- Park, J.; Kaufmann, G.F.; Bowen, J.P.; Arbiser, J.L.; Janda, K.D. Solenopsin A, a venom alkaloid from the fire ant solenopsis invicta, inhibits quorum-sensing signaling in Pseudomonas aeruginosa. J. Infect. Dis. 2008, 198, 1198–1201. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I.; Khan, M.S.; Al-Shabib, N.A. Trigonella foenum-graceum (Seed) extract interferes with quorum sensing regulated traits and biofilm formation in the strains of Pseudomonas aeruginosa and Aeromonas hydrophila. Evid.-Based Complement. Altern. Med. 2015, 2015, 879540. [Google Scholar] [CrossRef]
- Kraikongjit, S.; Jongjitvimol, T.; Mianjinda, N.; Sirithep, N.; Kaewbor, T.; Jumroon, N.; Jongjitwimol, J. Antibacterial effect of plant resin collected from Tetrigona apicalis (Smith, 1857) in Thung Salaeng Luang National Park, Phitsanulok. Walailak J. Sci. Tech. 2017, 15, 599–607. [Google Scholar] [CrossRef]
- Jongjitvimol, T.; Kraikongjit, S.; Paensuwan, P.; Jongjitwimol, J. In vitro biological profiles and chemical contents of ethanolic nest entrance extracts of thai stingless bees Tetrigona apicalis. Online J. Biol. Sci. 2020, 20, 157–165. [Google Scholar] [CrossRef]
- Murauer, A.; Ganzera, M. Quantitative determination of major alkaloids in Cinchona bark by Supercritical Fluid Chromatography. J. Chromatogr. A 2018, 1554, 117–122. [Google Scholar] [CrossRef]
- Rattanachak, N.; Weawsiangsang, S.; Jongjitvimol, T.; Baldock, R.A.; Jongjitwimol, J. Hydroquinine possesses antibacterial activity, and at half the MIC, induces the overexpression of RND-type efflux pumps using Multiplex Digital PCR in Pseudomonas aeruginosa. Trop. Med. Int. Dis. 2022, 7, 156. [Google Scholar] [CrossRef]
- Mitchell, G.; Lafrance, M.; Boulanger, S.; Séguin, D.L.; Guay, I.; Gattuso, M.; Marsault, E.; Bouarab, K.; Malouin, F. Tomatidine acts in synergy with aminoglycoside antibiotics against multiresistant Staphylococcus aureus and prevents virulence gene expression. J. Antimicrob. Chemother. 2012, 67, 559–568. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature counts: An efficient general purpose program for assigning sequence reads to genomic features. J. Bioinform. 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Wang, Y.; Luo, Z.; Chen, L.; Tan, R.; Wang, Y.; Wu, Y. Meloxicam inhibits biofilm formation and enhances antimicrobial agents efficacy by Pseudomonas aeruginosa. MicrobiologyOpen 2018, 7, e00545. [Google Scholar] [CrossRef]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2006, 22, 1B.1.1–1B.1.18. [Google Scholar] [CrossRef] [PubMed]
- Wongkaewkhiaw, S.; Taweechaisupapong, S.; Thanaviratananich, S.; Bolscher, J.G.M.; Nazmi, K.; Anutrakunchai, C.; Chareonsudjai, S.; Kanthawong, S. D-LL-31 enhances biofilm-eradicating effect of currently used antibiotics for chronic rhinosinusitis and its immunomodulatory activity on human lung epithelial cells. PLoS ONE 2020, 15, e0243315. [Google Scholar] [CrossRef]
- Nolan, L.M.; Whitchurch, C.B.; Barquist, L.; Katrib, M.; Boinett, C.J.; Mayho, M.; Goulding, D.; Charles, I.G.; Filloux, A.; Parkhill, J.; et al. A global genomic approach uncovers novel components for twitching motility-mediated biofilm expansion in Pseudomonas aeruginosa. bioRxiv 2018, 4, 372805. [Google Scholar] [CrossRef]
- Haiko, J.; Westerlund-Wikström, B. The role of the bacterial flagellum in adhesion and virulence. Biology 2013, 2, 1242–1267. [Google Scholar] [CrossRef]
- Schavemaker, P.E.; Lynch, M. Flagellar energy costs across the tree of life. eLife 2022, 11, e77266. [Google Scholar] [CrossRef] [PubMed]
- Mariano, G.; Faba-Rodriguez, R.; Bui, S.; Zhao, W.; Ross, J.; Tzokov, S.B.; Bergeron, J.R.C. Oligomerization of the FliF domains suggests a coordinated assembly of the bacterial flagellum MS Ring. Front. Microbiol. 2022, 12, 781960. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef]
- Herlihey, F.A.; Clarke, A.J. Controlling autolysis during flagella insertion in gram-negative bacteria. Adv. Exp. Med. Biol. 2017, 925, 41–56. [Google Scholar] [CrossRef]
- Konishi, M.; Kanbe, M.; McMurry, J.L.; Aizawa, S. Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J. Bacteriol. 2009, 191, 6186–6191. [Google Scholar] [CrossRef] [PubMed]
- Bouteiller, M.; Dupont, C.; Bourigault, Y.; Latour, X.; Barbey, C.; Konto-Ghiorghi, Y.; Merieau, A. Pseudomonas Flagella: Generalities and specificities. Int. J. Mol. Sci. 2021, 22, 3337. [Google Scholar] [CrossRef]
- Valentin, J.D.P.; Straub, H.; Pietsch, F.; Lemare, M.; Ahrens, C.H.; Schreiber, F.; Webb, J.S.; van der Mei, H.C.; Ren, Q. Role of the flagellar hook in the structural development and antibiotic tolerance of Pseudomonas aeruginosa biofilms. ISME J. 2022, 16, 1176–1186. [Google Scholar] [CrossRef]
- Dusane, D.H.; Hosseinidoust, Z.; Asadishad, B.; Tufenkji, N. Alkaloids modulate motility, biofilm formation and antibiotic susceptibility of uropathogenic Escherichia coli. PLoS ONE 2014, 9, e112093. [Google Scholar] [CrossRef]
- O’Donnell, J.N.; Bidell, M.R.; Lodise, T.P. Approach to the treatment of patients with serious multidrug-resistant Pseudomonas aeruginosa infections. Pharmacotherapy 2020, 40, 952–969. [Google Scholar] [CrossRef]
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.J.; Forbes, A.; Perkins, A.V.; Davey, A.K.; Chess-Williams, R.; Kiefel, M.J.; Arora, D.; et al. Cellular effects of pyocyanin, a secreted virulence factor of Pseudomonas aeruginosa. Toxins 2016, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; O’Neill, J.C.; Miller, D.M.; Mattmann, M.E.; Blackwell, H.E. Modulation of bacterial quorum sensing with synthetic ligands: Systematic evaluation of N-Acylated homoserine lactones in multiple species and new insights into their mechanisms of action. J. Am. Chem. Soc. 2007, 129, 13613–13625. [Google Scholar] [CrossRef]
- Favre-Bonté, S.; Köhler, T.; Van Delden, C. Biofilm formation by Pseudomonas aeruginosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 2003, 52, 598–604. [Google Scholar] [CrossRef]
- Wu, Z.; Zheng, R.; Zhang, J.; Wu, S. Transcriptional profiling of Pseudomonas aeruginosa PAO1 in response to anti-biofilm and anti-infection agent exopolysaccharide EPS273. J. Appl. Microbiol. 2021, 130, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.K.; Yeung, A.T.; Hancock, R.E. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J. Biotechnol. 2014, 191, 121–130. [Google Scholar] [CrossRef]




| Primer Name | Oligonucleotide Sequences (5′ to 3′) | Annealing Temperature (°C) | References |
|---|---|---|---|
| flgH F | CGAGCAGAACCTCTACGACG | 57.5 | This study |
| flgH R | TCGGGTTGTTGGTGGTCATG | 57.5 | This study |
| flgK F | CCAGCAAGCTGAATTCCAGC | 56.0 | This study |
| flgK R | GGTCGTCTCGATATCGCTGG | 56.0 | This study |
| fliF F | AGATGTACAACCCGGACCAG | 57.5 | This study |
| fliF R | TCGGATCGATGATGGTCTGG | 57.5 | This study |
| flgC F | TTCTCCACCATGTTCCAGCAG | 57.5 | This study |
| flgC R | TCCTCGACCACGTTCACATTG | 57.5 | This study |
| rhlI F | CAGGAATTCGACCAGTTCGACC | 58.5 | This study |
| rhlI R | CGAAGACGTCCTTGAGCAGG | 58.5 | This study |
| rhlR F | GTAGCGAGATGCAGCCGATC | 57.0 | This study |
| rhlR R | CCTTGGGATAGGTGCCATGG | 57.0 | This study |
| 16s rRNA F | CATGGCTCAGATTGAACGCTG | 58.0 | [31] |
| 16s rRNA R | GCTAATCCGACCTAGGCTCATC | 58.0 | [31] |
| Gene Name | Gene Product | Log2 FC 1 | FDR 2 | p Value |
|---|---|---|---|---|
| flgC | Flagellar basal-body rod protein FlgC | −2.93 | 3.90 × 10−3 | 6.66 × 10−5 |
| flgB | Flagellar basal-body rod protein FlgB | −2.38 | 2.43 × 10−2 | 8.00 × 10−4 |
| flgJ | Flagellar protein FlgJ | −2.33 | 2.41 × 10−2 | 8.00 × 10−4 |
| flgD | Flagellar basal-body rod modification protein FlgD | −2.33 | 2.76 × 10−2 | 1.00 × 10−3 |
| fliF | Flagellar M-ring outer membrane protein precursor FliF | −2.26 | 2.51 × 10−2 | 1.00 × 10−3 |
| fliG | Flagellar motor-switch protein 1 FliG | −2.23 | 2.29 × 10−2 | 1.20 × 10−3 |
| flgK | Flagellar hook-associated protein 1 FlgK | −2.23 | 3.22 × 10−2 | 1.50 × 10−3 |
| flgH | Flagellar L-ring protein precursor FlgH | −2.18 | 4.13 × 10−2 | 1.80 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rattanachak, N.; Weawsiangsang, S.; Daowtak, K.; Thongsri, Y.; Ross, S.; Ross, G.; Nilsri, N.; Baldock, R.A.; Pongcharoen, S.; Jongjitvimol, T.; et al. High-Throughput Transcriptomic Profiling Reveals the Inhibitory Effect of Hydroquinine on Virulence Factors in Pseudomonas aeruginosa. Antibiotics 2022, 11, 1436. https://doi.org/10.3390/antibiotics11101436
Rattanachak N, Weawsiangsang S, Daowtak K, Thongsri Y, Ross S, Ross G, Nilsri N, Baldock RA, Pongcharoen S, Jongjitvimol T, et al. High-Throughput Transcriptomic Profiling Reveals the Inhibitory Effect of Hydroquinine on Virulence Factors in Pseudomonas aeruginosa. Antibiotics. 2022; 11(10):1436. https://doi.org/10.3390/antibiotics11101436
Chicago/Turabian StyleRattanachak, Nontaporn, Sattaporn Weawsiangsang, Krai Daowtak, Yordhathai Thongsri, Sukunya Ross, Gareth Ross, Nungruthai Nilsri, Robert A. Baldock, Sutatip Pongcharoen, Touchkanin Jongjitvimol, and et al. 2022. "High-Throughput Transcriptomic Profiling Reveals the Inhibitory Effect of Hydroquinine on Virulence Factors in Pseudomonas aeruginosa" Antibiotics 11, no. 10: 1436. https://doi.org/10.3390/antibiotics11101436
APA StyleRattanachak, N., Weawsiangsang, S., Daowtak, K., Thongsri, Y., Ross, S., Ross, G., Nilsri, N., Baldock, R. A., Pongcharoen, S., Jongjitvimol, T., & Jongjitwimol, J. (2022). High-Throughput Transcriptomic Profiling Reveals the Inhibitory Effect of Hydroquinine on Virulence Factors in Pseudomonas aeruginosa. Antibiotics, 11(10), 1436. https://doi.org/10.3390/antibiotics11101436






