Detection of Clones B2-ST131-C2 and A-ST617 in Escherichia coli Producing Both CTX-M-15 and CTX-M-27 from Tunisian Community Patients
Abstract
1. Introduction
2. Results
2.1. Prevalence of ESBL-Producing E. coli
2.2. Analysis of β-Lactamase Content
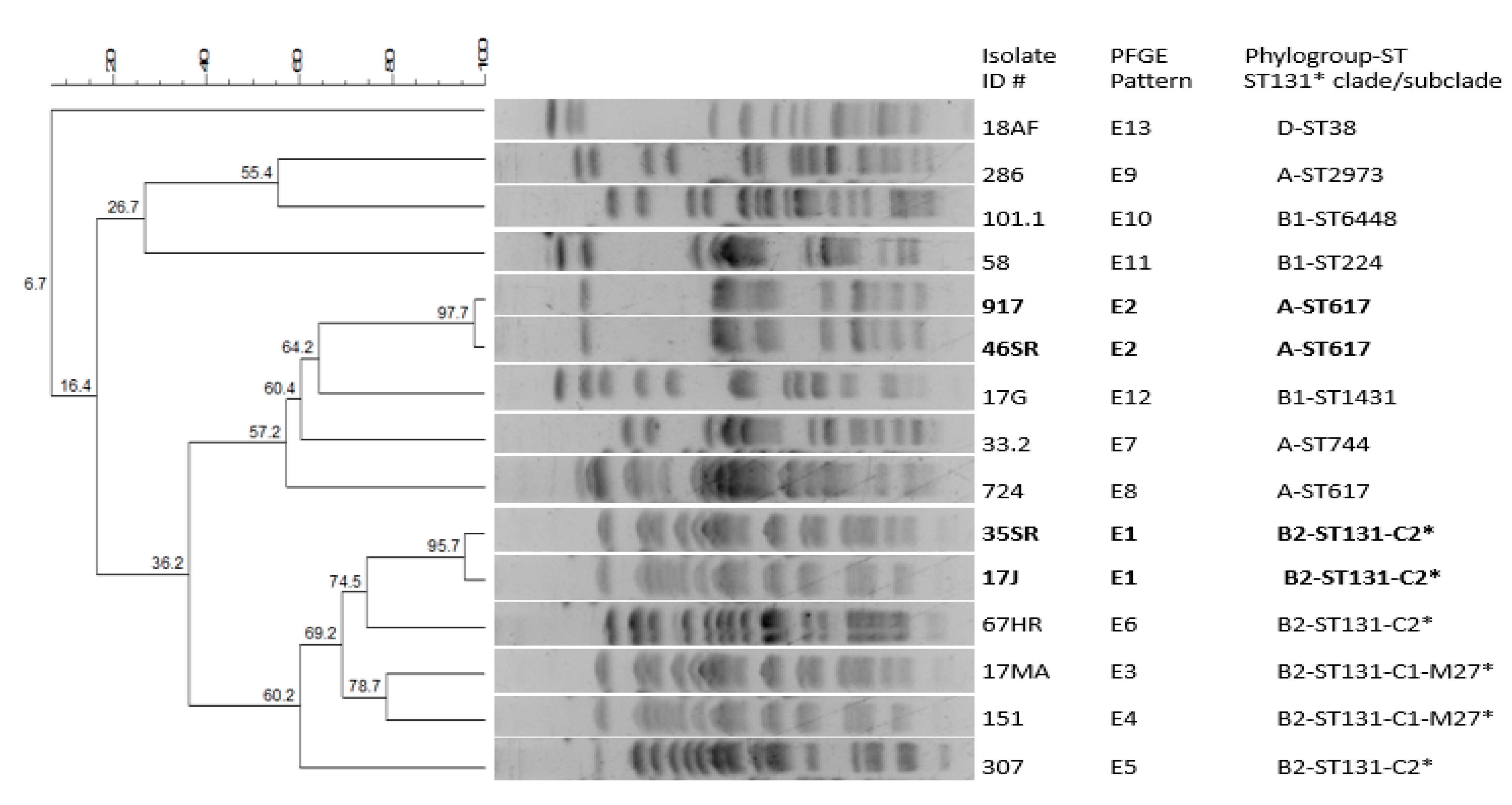
2.3. Molecular Typing Analysis
2.4. Antibiotic Resistance Pattern
2.5. Plasmid Content Analysis
3. Discussion
4. Materials and Methods
4.1. Strains Collection and Identification of E. coli
4.2. Antibiotic Susceptibility Testing
4.3. Molecular Characterization of ß-Lactamase Genes
4.4. Molecular Typing of ESBL-Producing E. coli
4.5. Plasmid Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global Epidemiology of CTX-M β-Lactamases: Temporal and Geographical Shifts in Genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Plasmids and the Spread of Resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Chanoine, M.-H.; Bertrand, X.; Madec, J.-Y. Escherichia Coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Pitout, J.D.D.; Gomi, R.; Matsuda, T.; Noguchi, T.; Yamamoto, M.; Peirano, G.; DeVinney, R.; Bradford, P.A.; Motyl, M.R.; et al. Global Escherichia Coli Sequence Type 131 Clade with BlaCTX-M-27 Gene. Emerg. Infect. Dis. 2016, 22, 1900–1907. [Google Scholar] [CrossRef]
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; De Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia Coli Epidemic Clone ST131. MBio 2016, 7, e02162. [Google Scholar] [CrossRef]
- Merino, I.; Hernández-García, M.; Turrientes, M.-C.; Pérez-Viso, B.; López-Fresneña, N.; Diaz-Agero, C.; Maechler, F.; Fankhauser-Rodriguez, C.; Kola, A.; Schrenzel, J.; et al. Emergence of ESBL-Producing Escherichia Coli ST131-C1-M27 Clade Colonizing Patients in Europe. J. Antimicrob. Chemother. 2018, 73, 2973–2980. [Google Scholar] [CrossRef]
- Sallem, N.; Hammami, A.; Mnif, B. Trends in Human Intestinal Carriage of ESBL- and Carbapenemase-Producing Enterobacterales among Food Handlers in Tunisia: Emergence of C1-M27-ST131 Subclades, BlaOXA-48 and BlaNDM. J. Antimicrob. Chemother. 2022, 77, 2142–2152. [Google Scholar] [CrossRef]
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae from Animal Origin and Wastewater in Tunisia: First Detection of O25b-B23-CTX-M-27-ST131 Escherichia Coli and CTX-M-15/OXA-204-Producing Citrobacter Freundii from Wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Hammami, S.; Saidani, M.; Ferjeni, S.; Aissa, I.; Slim, A.; Boutiba-Ben Boubaker, I. Characterization of Extended Spectrum β-Lactamase-Producing Escherichia Coli in Community-Acquired Urinary Tract Infections in Tunisia. Microb. Drug Resist. 2013, 19, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Guermazi-Toumi, S.; Boujlel, S.; Assoudi, M.; Issaoui, R.; Tlili, S.; Hlaiem, M.E. Susceptibility Profiles of Bacteria Causing Urinary Tract Infections in Southern Tunisia. J. Glob. Antimicrob. Resist. 2018, 12, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Pitout, J.D.D.; Peirano, G.; DeVinney, R.; Noguchi, T.; Yamamoto, M.; Gomi, R.; Matsuda, T.; Nakano, S.; Nagao, M.; et al. Rapid Identification of Different Escherichia Coli Sequence Type 131 Clades. Antimicrob. Agents Chemother. 2017, 61, e00179-17. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon Sequence Typing of IncF Plasmids Carrying Virulence and Resistance Determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Hayakawa, K.; Gattu, S.; Marchaim, D.; Bhargava, A.; Palla, M.; Alshabani, K.; Gudur, U.M.; Pulluru, H.; Bathina, P.; Sundaragiri, P.R.; et al. Epidemiology and Risk Factors for Isolation of Escherichia Coli Producing CTX-M-Type Extended-Spectrum β-Lactamase in a Large U.S. Medical Center. Antimicrob. Agents Chemother. 2013, 57, 4010–4018. [Google Scholar] [CrossRef]
- Laboratory of AntibioResistance in Tunisia (LART). Available online: https://www.infectiologie.org.tn/resistance.php (accessed on 16 July 2022).
- Szymankiewicz, M.; Stefaniuk, E.; Baraniak, A.; Nowikiewicz, T. Clinical and Molecular Findings of Infections Caused by Extended-Spectrum β-Lactamase-Producing Enterobacterales in Patients with Solid Tumors: A Single-Center Study. Microb. Drug Resist. 2021, 27, 1470–1481. [Google Scholar] [CrossRef]
- Decano, A.G.; Downing, T. An Escherichia Coli ST131 Pangenome Atlas Reveals Population Structure and Evolution across 4,071 Isolates. Sci. Rep. 2019, 9, 17394. [Google Scholar] [CrossRef]
- Takawira, F.T.; Pitout, J.D.; Thilliez, G.; Mashe, T.; Gutierrez, A.V.; Kingsley, R.A.; Peirano, G.; Matheu, J.; Midzi, S.M.; Mwamakamba, L.W.; et al. Molecular Epidemiology of Extended-Spectrum Beta-Lactamase-Producing Extra-Intestinal Pathogenic Escherichia Coli Strains over a 2-Year Period (2017–2019) from Zimbabwe. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 7, 343–355. [Google Scholar] [CrossRef]
- Zurfluh, K.; Stevens, M.J.A.; Stephan, R.; Nüesch-Inderbinen, M. Complete and Assembled Genome Sequence of an NDM-5- and CTX-M-15-Producing Escherichia Coli Sequence Type 617 Isolated from Wastewater in Switzerland. J. Glob. Antimicrob. Resist. 2018, 15, 105–106. [Google Scholar] [CrossRef]
- Dziri, R.; Klibi, N.; Alonso, C.A.; Jouini, A.; Ben Said, L.; Chairat, S.; Bellaaj, R.; Boudabous, A.; Ben Slama, K.; Torres, C. Detection of CTX-M-15-Producing Escherichia Coli Isolates of Lineages ST131-B2 and ST167-A in Environmental Samples of a Tunisian Hospital. Microb. Drug Resist. 2016, 22, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Maamar, E.; Ferjani, S.; Jendoubi, A.; Hammami, S.; Hamzaoui, Z.; Mayonnove-Coulange, L.; Saidani, M.; Kammoun, A.; Rehaiem, A.; Ghedira, S.; et al. High Prevalence of Gut Microbiota Colonization with Broad-Spectrum Cephalosporin Resistant Enterobacteriaceae in a Tunisian Intensive Care Unit. Front. Microbiol. 2016, 7, 1859. [Google Scholar] [CrossRef]
- Chenouf, N.S.; Carvalho, I.; Messaï, C.R.; Ruiz-Ripa, L.; Mama, O.M.; Titouche, Y.; Zitouni, A.; Hakem, A.; Torres, C. Extended Spectrum β-Lactamase-Producing Escherichia Coli and Klebsiella Pneumoniae from Broiler Liver in the Center of Algeria, with Detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia Coli. Microb. Drug Resist. 2021, 27, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence Type ST131 and ST10 Complex (ST617) Predominant among CTX-M-15-Producing Escherichia Coli Isolates from Nigeria. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia Coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-Spectrum β-Lactamase-Producing Escherichia Coli in Human-Derived and Foodchain-Derived Samples from England, Wales, and Scotland: An Epidemiological Surveillance and Typing Study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Cameron, A.; Mangat, R.; Mostafa, H.H.; Taffner, S.; Wang, J.; Dumyati, G.; Stanton, R.A.; Daniels, J.B.; Campbell, D.; Lutgring, J.D.; et al. Detection of CTX-M-27 β-Lactamase Genes on Two Distinct Plasmid Types in ST38 Escherichia Coli from Three U.S. States. Antimicrob. Agents Chemother. 2021, 65, e0082521. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Benlabidi, S.; Hassen, A.; Torres, C.; Hammami, S. Genetic Characterization of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae from a Biological Industrial Wastewater Treatment Plant in Tunisia with Detection of the Colistin-Resistance Mcr-1 Gene. FEMS Microbiol. Ecol. 2021, 97, fiaa231. [Google Scholar] [CrossRef]
- Santona, A.; Sumbana, J.J.; Fiamma, M.; Deligios, M.; Taviani, E.; Simbine, S.E.; Zimba, T.; Sacarlal, J.; Rubino, S.; Paglietti, B. High-Risk Lineages among Extended-Spectrum β-Lactamase-Producing Escherichia Coli from Extraintestinal Infections in Maputo Central Hospital, Mozambique. Int. J. Antimicrob. Agents 2022, 60, 106649. [Google Scholar] [CrossRef]
- Silva, K.C.; Moreno, M.; Cabrera, C.; Spira, B.; Cerdeira, L.; Lincopan, N.; Moreno, A.M. First Characterization of CTX-M-15-Producing Escherichia Coli Strains Belonging to Sequence Type (ST) 410, ST224, and ST1284 from Commercial Swine in South America. Antimicrob. Agents Chemother. 2016, 60, 2505–2508. [Google Scholar] [CrossRef]
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST Clonal Complexes, with a Predominance of ST131, of Escherichia Coli Harbouring BlaCTX-M-15 in a Tertiary Hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef] [PubMed]
- Bachiri, T.; Bakour, S.; Ladjouzi, R.; Thongpan, L.; Rolain, J.M.; Touati, A. High Rates of CTX-M-15-Producing Escherichia Coli and Klebsiella Pneumoniae in Wild Boars and Barbary Macaques in Algeria. J. Glob. Antimicrob. Resist. 2017, 8, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kiratisin, P.; Apisarnthanarak, A.; Saifon, P.; Laesripa, C.; Kitphati, R.; Mundy, L.M. The Emergence of a Novel Ceftazidime-Resistant CTX-M Extended-Spectrum Beta-Lactamase, CTX-M-55, in Both Community-Onset and Hospital-Acquired Infections in Thailand. Diagn. Microbiol. Infect. Dis. 2007, 58, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Lv, L.; Zeng, Z.; Chen, S.; He, D.; Chen, X.; Wu, C.; Wang, Y.; Yang, T.; Wu, P.; et al. Increasing Prevalence of Extended-Spectrum Cephalosporin-Resistant Escherichia Coli in Food Animals and the Diversity of CTX-M Genotypes during 2003–2012. Vet. Microbiol. 2014, 172, 534–541. [Google Scholar] [CrossRef]
- Lupo, A.; Saras, E.; Madec, J.-Y.; Haenni, M. Emergence of BlaCTX-M-55 Associated with FosA, RmtB and Mcr Gene Variants in Escherichia Coli from Various Animal Species in France. J. Antimicrob. Chemother. 2018, 73, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic Characterization of ESBL-Producing Escherichia Coli and Klebsiella Pneumoniae Isolated from Wastewater and River Water in Tunisia: Predominance of CTX-M-15 and High Genetic Diversity. Environ. Sci. Pollut. Res. Int. 2020, 27, 44368–44377. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, R.; Recule, C.; Baraduc, R.; Chanal, C.; Sirot, D.; De Champs, C.; Sirot, J. Effect of D240G Substitution in a Novel ESBL CTX-M-27. J. Antimicrob. Chemother. 2003, 52, 29–35. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Mani, Y.; Mansour, W.; Lupo, A.; Saras, E.; Bouallègue, O.; Madec, J.-Y.; Haenni, M. Spread of BlaCTX-M-15-Producing Enterobacteriaceae and OXA-23-Producing Acinetobacter Baumannii Sequence Type 2 in Tunisian Seafood. Antimicrob. Agents Chemother. 2018, 62, e00727-18. [Google Scholar] [CrossRef]
- Matsuo, N.; Nonogaki, R.; Hayashi, M.; Wachino, J.-I.; Suzuki, M.; Arakawa, Y.; Kawamura, K. Characterization of BlaCTX-M-27/F1:A2:B20 Plasmids Harbored by Escherichia Coli Sequence Type 131 Sublineage C1/H30R Isolates Spreading among Elderly Japanese in Nonacute-Care Settings. Antimicrob. Agents Chemother. 2020, 64, e00202-20. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf (accessed on 31 January 2022).
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 September 2022).
- Gdoura-Ben Amor, M.; Siala, M.; Zayani, M.; Grosset, N.; Smaoui, S.; Messadi-Akrout, F.; Baron, F.; Jan, S.; Gautier, M.; Gdoura, R. Isolation, Identification, Prevalence, and Genetic Diversity of Bacillus Cereus Group Bacteria From Different Foodstuffs in Tunisia. Front. Microbiol. 2018, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia Coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and Virulence in Escherichia Coli: An Evolutionary Perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Available online: https://pubmlst.org/bigsdb?db=pubmlst_escherichia_seqdef (accessed on 10 September 2022).
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]

| Isolate ID # (a) | Location (b) | ESBL Content | Other ß-Lactamase | MIC (µg/mL) (c) | Associated Resistances (c) | PFGE Profile | Phylogroup-ST ST131 Subclade (d) | Inc Plasmid pMLST (e) | |
|---|---|---|---|---|---|---|---|---|---|
| CAZ | CTX | ||||||||
| 35SR | Sfax/CB | CTX-M-15 CTX-M-27 | OXA-1-like | 16 | 2,048 | FQ GEN TOB AMK SXT | E1 | B2-ST131-C2 (d) | F31:A4:B1 (e)/W |
| 17J | Sfax/CB | CTX-M-15 CTX-M-27 | OXA-1-like | 16 | 2,048 | FQ TOB AMK SXT | E1 | B2-ST131-C2 (d) | F31:A4:B1 (e) |
| 17MA | Sfax/CB | CTX-M-27 | - | 8 | 256 | FQ SXT | E3 | B2-ST131-C1-M27 (d) | F1:A-:B20 (e) |
| 151 | Tunis/H2 | CTX-M-27 | - | 8 | 256 | FQ SXT | E4 | B2-ST131-C1-M27 (d) | F1:A-:B20 (e) |
| 307 | Tunis/H2 | CTX-M-15 | TEM- and OXA-1-like | 32 | 1,024 | FQ FOX GEN TOB AMK SXT | E5 | B2-ST131-C2 (d) | FII:FIA |
| 67HR | Tunis/H3 | CTX-M-15 | OXA-1-like | 64 | 4,096 | FQ TOB AMK SXT | E6 | B2-ST131-C2 (d) | FII:FIB/Y |
| 917 | Sfax/CB | CTX-M-15 CTX-M-27 | OXA-1-like | 16 | 1,024 | FQ GEN TOB SXT NI | E2 | A-ST617 | F31:A4:B1 (e)/N |
| 46SR | Sfax/CB | CTX-M-15 CTX-M-27 | OXA-1-like | 32 | 1,024 | FQ GEN TOB SXT NI | E2 | A-ST617 | F31:A4:B1 (e)/N |
| 33.2 | Sfax/H1 | CTX-M-55 | TEM-like | 16 | 4,096 | FQ GEN TOB SXT | E7 | A-ST744 | FII:FIB/W/X |
| 724 | Tunis/H1 | CTX-M-15 | - | 64 | 2,048 | FQ SXT | E8 | A-ST617 | FII: FIB/Y |
| 286 | Tunis/H2 | CTX-M-55 | - | 16 | 4,096 | FQ SXT | E9 | A-ST2973 | FII:FIB/Y |
| 101.1 | Sfax/CA | CTX-M-55 | - | 16 | 4,096 | FQ SXT | E10 | B1-ST6448 | FIB |
| 58 | Sfax/H1 | CTX-M-15 | TEM-like | 8 | 1,024 | SXT | E11 | B1-ST224 | FIB/W/N/Y |
| 17G | Sfax/CB | CTX-M-15 | TEM-like | 8 | 1,024 | FQ SXT | E12 | B1-ST1431 | FIB/W/N/Y |
| 18AF | Sfax/CB | CTX-M-27 | - | 4 | 1,024 | SXT | E13 | D-ST38 | FIB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhaya, A.; Trabelsi, R.; Aillerie, S.; M’Zali, F.; Bégu, D.; Tounsi, S.; Gdoura, R.; Arpin, C. Detection of Clones B2-ST131-C2 and A-ST617 in Escherichia coli Producing Both CTX-M-15 and CTX-M-27 from Tunisian Community Patients. Antibiotics 2022, 11, 1329. https://doi.org/10.3390/antibiotics11101329
Mhaya A, Trabelsi R, Aillerie S, M’Zali F, Bégu D, Tounsi S, Gdoura R, Arpin C. Detection of Clones B2-ST131-C2 and A-ST617 in Escherichia coli Producing Both CTX-M-15 and CTX-M-27 from Tunisian Community Patients. Antibiotics. 2022; 11(10):1329. https://doi.org/10.3390/antibiotics11101329
Chicago/Turabian StyleMhaya, Amel, Rahma Trabelsi, Sabine Aillerie, Fatima M’Zali, Dominique Bégu, Slim Tounsi, Radhouane Gdoura, and Corinne Arpin. 2022. "Detection of Clones B2-ST131-C2 and A-ST617 in Escherichia coli Producing Both CTX-M-15 and CTX-M-27 from Tunisian Community Patients" Antibiotics 11, no. 10: 1329. https://doi.org/10.3390/antibiotics11101329
APA StyleMhaya, A., Trabelsi, R., Aillerie, S., M’Zali, F., Bégu, D., Tounsi, S., Gdoura, R., & Arpin, C. (2022). Detection of Clones B2-ST131-C2 and A-ST617 in Escherichia coli Producing Both CTX-M-15 and CTX-M-27 from Tunisian Community Patients. Antibiotics, 11(10), 1329. https://doi.org/10.3390/antibiotics11101329






