Role of AmpC-Inducing Genes in Modulating Other Serine Beta-Lactamases in Escherichia coli
Abstract
1. Introduction
2. Results and Discussion
2.1. Deletion of AmpC Induction Genes Altered the Beta-Lactam Sensitivity of E. coli
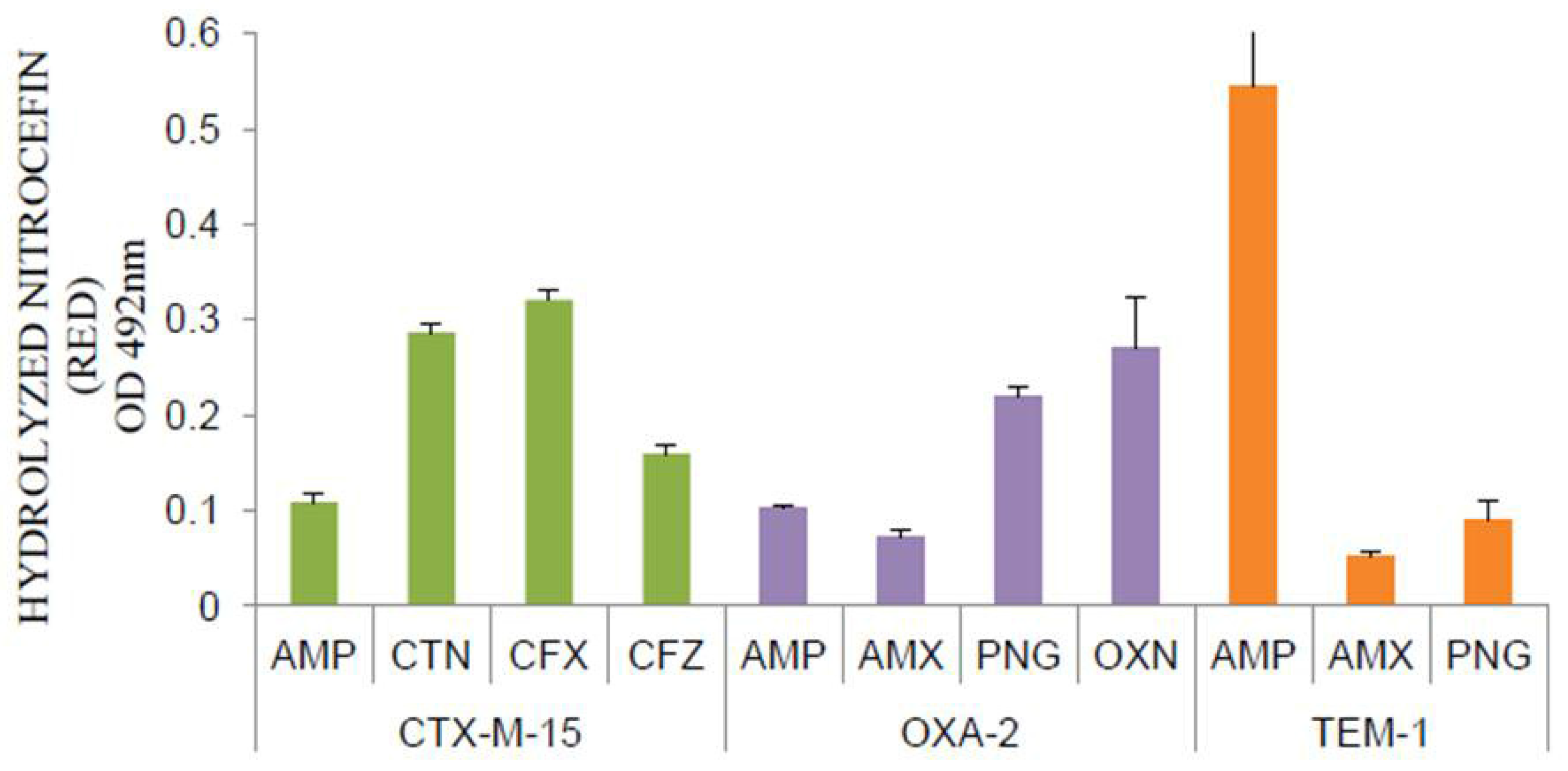
2.2. Each Serine Beta-Lactamase Displayed Its Characteristic Spectrum of Beta-Lactam Hydrolysis
2.3. Induction of Classes A and D Beta-Lactamases in E. coli by Sub-Inhibitory Concentrations of Beta-Lactam Antibiotics
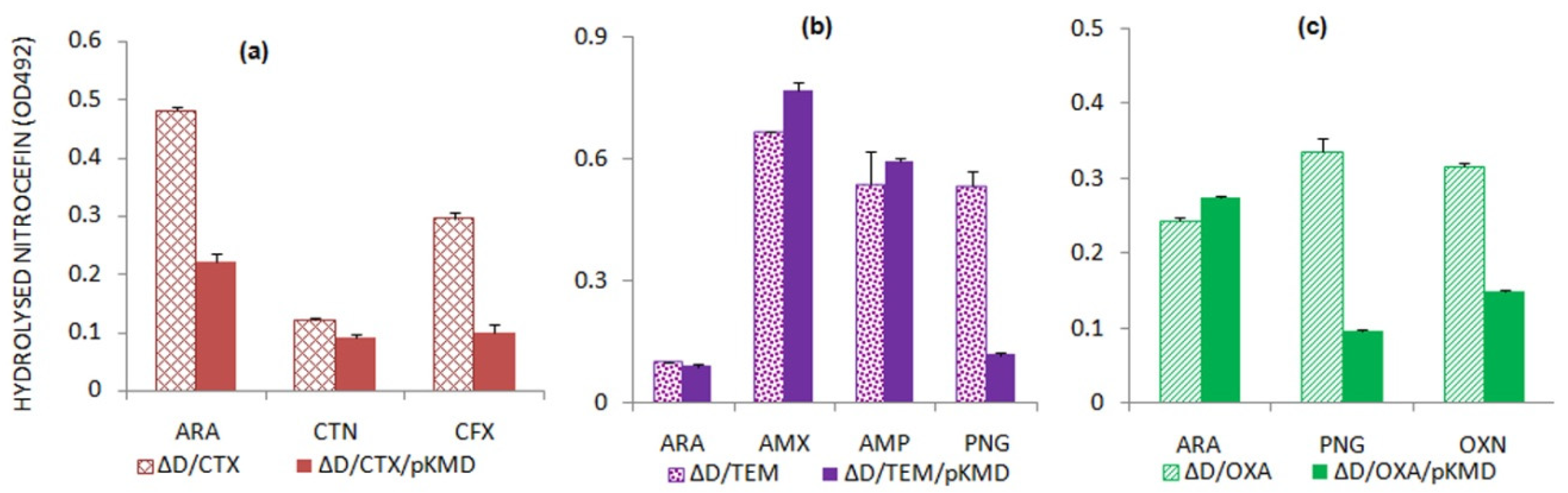
2.4. AmpD Expression Leads to the Alterations in Beta-Lactamase Expression
Variability Exists in the Expression of Beta-Lactamases in ΔampD Mutant
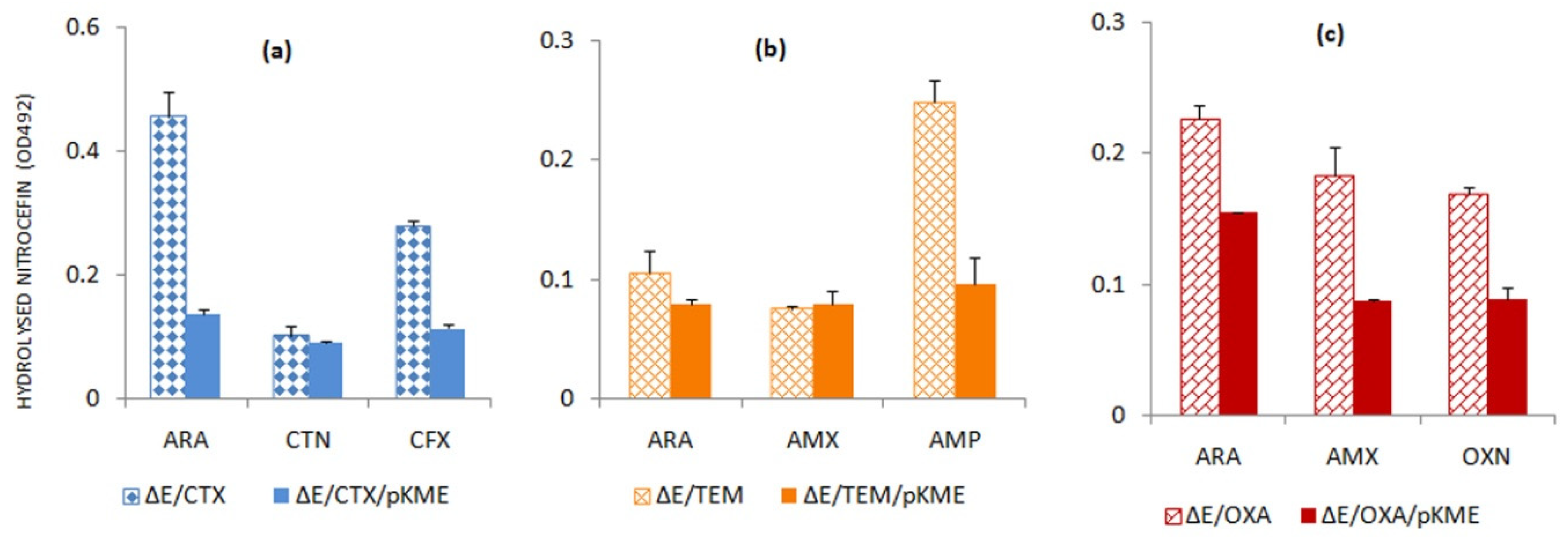
2.5. AmpE Acts as a Negative Regulator of Beta-Lactamase Expression
Presence of AmpE Effect on the Induction of Serine Beta-Lactamases by Beta-Lactams
3. Materials and Methods
3.1. Strains, Plasmids and Chemicals
3.2. Genetic Manipulations in E. coli BW25113
3.3. Cloning of Predicted Beta-Lactamase-Inducing Genes
3.4. Drug Susceptibility Testing (DST)
3.5. Nitrocefin Hydrolysis Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Breukink, E. The membrane steps of bacterial cell wall synthesis as antibiotic targets. Antibiotics 2016, 5, 28. [Google Scholar] [CrossRef]
- Maitra, A.; Munshi, T.; Healy, J.; Martin, L.T.; Vollmer, W.; Keep, N.H.; Bhakta, S. Cell wall peptidoglycan in Mycobacterium tuberculosis: An Achilles’ heel for the TB-causing pathogen. FEMS Microbiol. Rev. 2019, 43, 548–575. [Google Scholar] [CrossRef]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407. [Google Scholar] [CrossRef]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef]
- Johnson, J.W.; Fisher, J.F.; Mobashery, S. Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci. 2013, 1277, 54. [Google Scholar] [CrossRef]
- Van Heijenoort, J. Biosynthesis of the bacterial peptidoglycan unit. New Compr. Biochem. 1994, 27, 39–54. [Google Scholar]
- Glauner, B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 1988, 172, 451–464. [Google Scholar] [CrossRef]
- Van Heijenoort, J. Assembly of the monomer unit of bacterial peptidoglycan. Cell. Mol. Life Sci. CMLS 1998, 54, 300–304. [Google Scholar] [CrossRef]
- Ghosh, A.S.; Chowdhury, C.; Nelson, D.E. Physiological functions of D-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 2008, 16, 309–317. [Google Scholar] [CrossRef]
- Tomasz, A. The mechanism of the irreversible antimicrobial effects of penicillins: How the beta-lactam antibiotics kill and lyse bacteria. Annu. Rev. Microbiol. 1979, 33, 113–137. [Google Scholar] [CrossRef]
- Ambler, R. The structure of β-lactamases. Phil. Trans. R. Soc. Lond. B 1980, 289, 321–331. [Google Scholar]
- Zeng, X.; Lin, J. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front. Microbiol. 2013, 4, 128. [Google Scholar] [CrossRef]
- Fisher, J.F.; Mobashery, S. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Biorgan. Chem. 2014, 56, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.; Huang, L.; Bartowsky, E.; Normark, S.; Park, J. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 1994, 13, 4684–4694. [Google Scholar] [CrossRef]
- Park, J.T.; Uehara, T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 2008, 72, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Templin, M.F.; Edwards, D.H.; Höltje, J. A murein hydrolase is the specific target of bulgecin in Escherichia coli. J. Biol. Chem. 1992, 267, 20039–20043. [Google Scholar] [CrossRef]
- Akata, K.; Muratani, T.; Yatera, K.; Naito, K.; Noguchi, S.; Yamasaki, K.; Kawanami, T.; Kido, T.; Mukae, H. Induction of plasmid-mediated AmpC β-lactamase DHA-1 by piperacillin/tazobactam and other β-lactams in Enterobacteriaceae. PLoS ONE 2019, 14, e0218589. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Bennett, P.; Chopra, I. Molecular basis of beta-lactamase induction in bacteria. Antimicrob. Agents Chemother. 1993, 37, 153. [Google Scholar] [CrossRef][Green Version]
- Mallik, D.; Pal, S.; Ghosh, A.S. Involvement of AmpG in mediating a dynamic relationship between serine beta-lactamase induction and biofilm-forming ability of Escherichia coli. FEMS Microbiol. Lett. 2018, 365, fny065. [Google Scholar] [CrossRef]
- Jaurin, B.; Grundström, T.; Edlund, T.; Normark, S. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 1981, 290, 221–225. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Lindquist, S.; Galleni, M.; Lindberg, F.; Normark, S. Signalling proteins in enterobacterial AmpC β-lactamase regulation. Mol. Microbiol. 1989, 3, 1091–1102. [Google Scholar] [CrossRef]
- Höltje, J.-V.; Kopp, U.; Ursinus, A.; Wiedemann, B. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 1994, 122, 159–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bauernfeind, A.; Schweighart, S.; Grimm, H. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 1990, 18, 294–298. [Google Scholar] [CrossRef]
- Then, R. Ability of newer beta-lactam antibiotics to induce beta-lactamase production in Enterobacter cloacae. Eur. J. Clin. Microbiol. 1987, 6, 451–455. [Google Scholar] [CrossRef]
- Zhu, Y.; Englebert, S.; Joris, B.; Ghuysen, J.-M.; Kobayashi, T.; Lampen, J. Structure, function, and fate of the BlaR signal transducer involved in induction of beta-lactamase in Bacillus licheniformis. J. Bacteriol. 1992, 174, 6171–6178. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Curran, I.; Joris, B.; Ghuysen, J.-M.; Lampen, J.O. Identification of BlaR, the signal transducer for beta-lactamase production in Bacillus licheniformis, as a penicillin-binding protein with strong homology to the OXA-2 beta-lactamase (class D) of Salmonella typhimurium. J. Bacteriol. 1990, 172, 1137–1141. [Google Scholar] [CrossRef]
- Ning, J.; Ahmed, S.; Cheng, G.; Chen, T.; Wang, Y.; Peng, D.; Yuan, Z. Analysis of the stability and affinity of BlaR-CTD protein to β-lactam antibiotics based on docking and mutagenesis studies. J. Biol. Eng. 2019, 13, 27. [Google Scholar] [CrossRef]
- Narayanan, N.; Hsieh, M.Y.; Xu, Y.; Chou, C.P. Arabinose-induction of lac-derived promoter systems for penicillin acylase production in Escherichia coli. Biotechnol. Prog. 2006, 22, 617–625. [Google Scholar] [CrossRef]
- Anitha, P.; Bag, S.; Anbarasu, A.; Ramaiah, S. Gene and protein network analysis of AmpC β lactamase. Cell Biochem. Biophys. 2015, 71, 1553–1567. [Google Scholar] [CrossRef] [PubMed]
- Guérin, F.; Isnard, C.; Cattoir, V.; Giard, J.C. Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob. Agents Chemother. 2015, 59, 7753–7761. [Google Scholar] [CrossRef] [PubMed]
- Petrosino, J.F.; Galhardo, R.S.; Morales, L.D.; Rosenberg, S.M. Stress-induced β-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J. Bacteriol. 2009, 191, 5881–5889. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, A.J.; Hanson, N.D. Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob. Agents Chemother. 2006, 50, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.E.; Weiner, J.H. Coordinate regulation of murein peptidase activity and AmpC β-lactamase synthesis in Escherichia coli. FEBS Lett. 1992, 304, 103–108. [Google Scholar] [CrossRef]
- Lindquist, S.; Lindberg, F.; Normark, S. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC beta-lactamase gene. J. Bacteriol. 1989, 171, 3746–3753. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Mao, Y.; Ju, L.; Jin, M.; Sun, Y.; Jin, S.; Gao, H. Distinct roles of major peptidoglycan recycling enzymes in β-lactamase production in Shewanella oneidensis. Antimicrob. Agents Chemother. 2014, 58, 6536–6543. [Google Scholar] [CrossRef][Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Mallik, D.; Kumar, A.; Sarkar, S.K.; Ghosh, A.S. Multiple resistance mechanisms acting in unison in an Escherichia coli clinical isolate. Curr. Microbiol. 2013, 67, 748–753. [Google Scholar] [CrossRef] [PubMed]
| Antibiotics | MIC (mg/L) | ||||||
|---|---|---|---|---|---|---|---|
| BW | ΔampD | ΔampE | ΔampG | ΔampDC | ΔampCE | ΔampGC | |
| Ampicillin | 4 | 4 | 4 | 2 | 4 | 8 | 4 |
| Amoxicillin | 8 | 4 | 4 | 2 | 4 | 8 | 4 |
| Cefalothin | 8 | 4 | 4 | 2 | 8 | 16 | 8 |
| Cefoxitin | 8 | 8 | 4 | 4 | 2 | 4 | 1 |
| Beta-Lactamases | Antibiotics | MIC (mg/L) | |
|---|---|---|---|
| BW | BW/BLA | ||
| CTX-M-15 | Ampicillin | 4 | 62.5 |
| Amoxicillin | 8 | 125 | |
| Cefotaxime | 0.08 | 2.5 | |
| TEM-1 | Amoxicillin | 8 | 62.5 |
| Penicillin G | 16 | 32 | |
| OXA-2 | Amoxicillin | 8 | 125 |
| Ampicillin | 4 | 62 | |
| Oxacillin | 62 | 250 | |
| Strain | Relevant Characteristic(s) | Remarks |
|---|---|---|
| XL1-Blue | F’::Tn10 proA + B + lacIq Δ(lacZ)M15/recA1 endA1 gyrA96 (NalR) thi hsdR17 (rK–mK+) glnV44 relA1 lac | Stratagene, La Jolla, CA, USA |
| BW25113 | F-::Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | The Coli Genetic Stock Center (CGSC) |
| ΔampC | ampC deleted from BW25113 | This work |
| ΔampD | ampD deleted from BW25113 | This work |
| ΔampE | ampE deleted from BW25113 | This work |
| ΔampG | ampG deleted from BW25113 | This work |
| ΔampGampC | ampG and ampC deleted from BW25113 | This work |
| ΔampDampC | ampD and ampC deleted from BW25113 | This work |
| ΔampEampC | ampE and ampC deleted from BW25113 | This work |
| Plasmid | Relevant characteristic(s) | |
| pGEM-T Easy | PCR cloning vector | Promega Corp., Madison, WI, USA |
| pKD46 | Recombinase gene expressed by PBAD promoter, Ampr | CGSC |
| pCP20 | Flippase gene cloned, Camr | CGSC |
| pBM15 | blaCTX-M15 from NGM9 cloned in pBAD18-Cam | This work |
| pBT1 | blaTEM-1 from U-84 cloned in pBAD18-Cam | This work |
| pBO2 | blaOXA-2 from W-12 cloned in pBAD18-Cam | This work |
| pKMD | ampD gene from BW25113 cloned in pBAD18-Kan | This work |
| pKME | ampE gene from BW25113 cloned in pBAD18-Kan | This work |
| pKMG | ampG gene from BW25113 cloned in pBAD18-Kan | This work |
| Beta-Lactamases | Antibiotics | MIC (mg/L) | ||
|---|---|---|---|---|
| ΔampD | ΔampD/BLA | ΔampD/pKMD/BLA | ||
| CTX-M-15 | Ampicillin | 4 | 125 | 250 |
| Amoxicillin | 4 | 32 | 62.5 | |
| Cefalothin | 4 | 32 | 125 | |
| TEM-1 | Ampicillin | 4 | 32 | 8 |
| Amoxicillin | 4 | 125 | 16 | |
| Cefoxitin | 4 | 4 | 4 | |
| OXA-2 | Ampicillin | 4 | 8 | 500 |
| Amoxicillin | 4 | 32 | 500 | |
| Oxacillin | 250 | 62 | >500 | |
| Beta-Lactamases | Antibiotics | MIC (mg/L) | ||
|---|---|---|---|---|
| ΔampE | ΔampE/BLA | ΔampE/pKME/BLA | ||
| CTX-M-15 | Amoxicillin | 4 | 62.5 | 62.5 |
| Ampicillin | 4 | 62.5 | 31.25 | |
| Cefotaxime | 0.1 | 1.6 | 0.4 | |
| TEM-1 | Amoxicillin | 4 | 125 | 4 |
| Ampicillin | 4 | 125 | 2 | |
| Cefoxitin | 4 | 4 | 4 | |
| OXA-2 | Amoxicillin | 4 | 62.5 | 62.5 |
| Cefoxitin | 4 | 4 | 2 | |
| Oxacillin | 250 | 500 | 250 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallik, D.; Jain, D.; Bhakta, S.; Ghosh, A.S. Role of AmpC-Inducing Genes in Modulating Other Serine Beta-Lactamases in Escherichia coli. Antibiotics 2022, 11, 67. https://doi.org/10.3390/antibiotics11010067
Mallik D, Jain D, Bhakta S, Ghosh AS. Role of AmpC-Inducing Genes in Modulating Other Serine Beta-Lactamases in Escherichia coli. Antibiotics. 2022; 11(1):67. https://doi.org/10.3390/antibiotics11010067
Chicago/Turabian StyleMallik, Dhriti, Diamond Jain, Sanjib Bhakta, and Anindya Sundar Ghosh. 2022. "Role of AmpC-Inducing Genes in Modulating Other Serine Beta-Lactamases in Escherichia coli" Antibiotics 11, no. 1: 67. https://doi.org/10.3390/antibiotics11010067
APA StyleMallik, D., Jain, D., Bhakta, S., & Ghosh, A. S. (2022). Role of AmpC-Inducing Genes in Modulating Other Serine Beta-Lactamases in Escherichia coli. Antibiotics, 11(1), 67. https://doi.org/10.3390/antibiotics11010067







