A Combined Phenotypic-Genotypic Predictive Algorithm for In Vitro Detection of Bicarbonate: β-Lactam Sensitization among Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
2. Results
2.1. Identification of Phenotypic and Genotypic Traits Associated with NaHCO3-Responsiveness
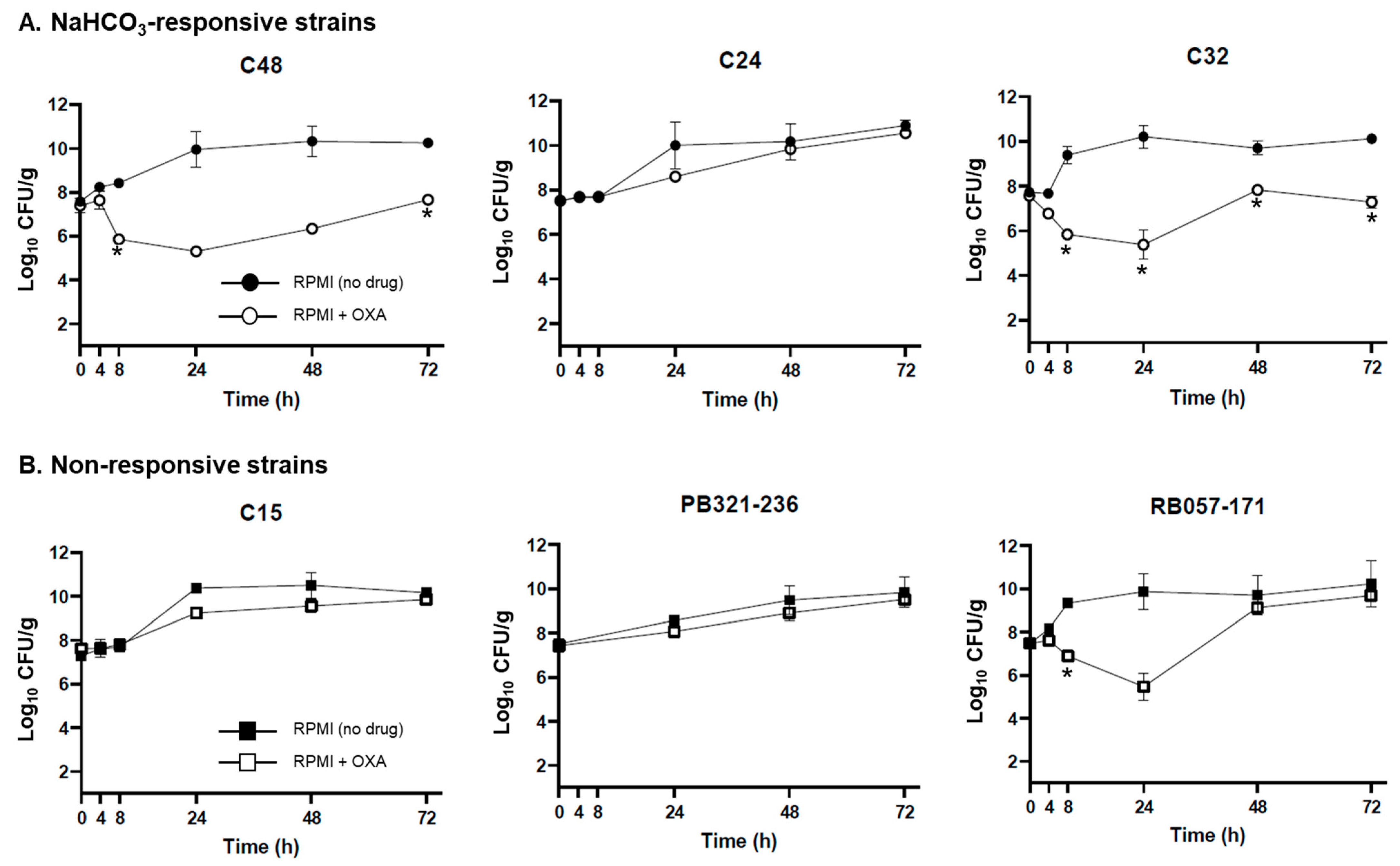
2.2. Ex Vivo Validation of NaHCO3-Responsiveness Screening Criteria
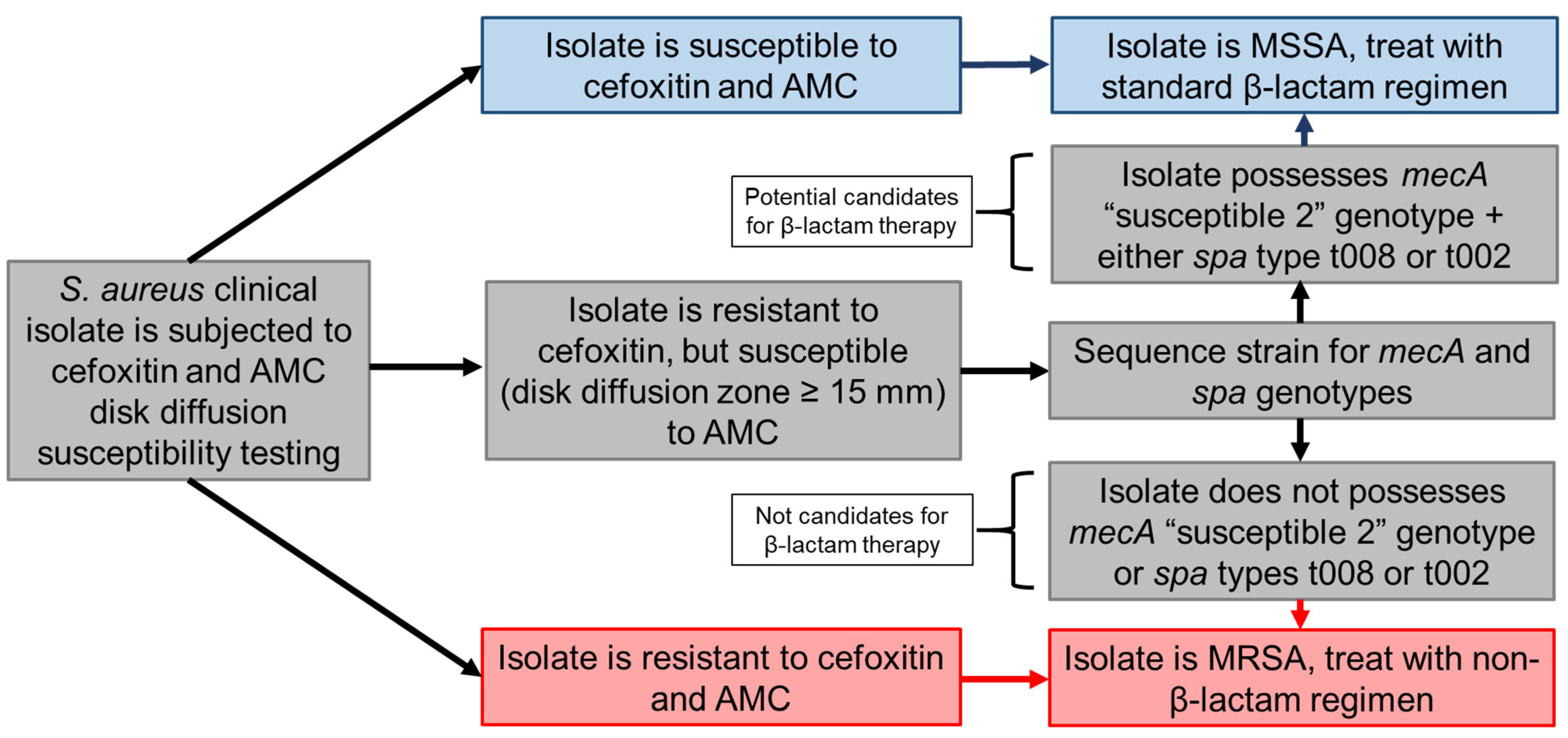
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Conditions, and Susceptibility Testing
4.2. mecA, Clonal Complex (CC), agr, SCCmec, and spa Genotyping
4.3. Statistical Analyses
4.4. Pharmacodynamic Model with Ex Vivo Simulated Endocardial Vegetations (SEVs)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. In Threat Report; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013; pp. 50–52. [Google Scholar]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Abadi, A.T.B.; Rizvanov, A.A.; Haertlé, T.; Blatt, N.L. World Health Organization report: Current crisis of antibiotic resistance. BioNanoScience 2019, 9, 778–788. [Google Scholar] [CrossRef]
- World Health Organization. Standardization of Methods for Conducting Microbic Sensitivity Tests-Second Report of the Expert Committee on Antibiotics; World Health Organization: Geneva, Switzerland, 1961. [Google Scholar]
- Weinstein, M.P.; Patel, J.B.; Campeau, S.; Eliopoulos, G.M.; Galas, M.F.; Humphries, R.M.; Jenkins, S.G.; Limbago, B.; Mathers, A.J.; Mazzulli, T.; et al. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2018. [Google Scholar]
- Tibbetts, R.J. Antimicrobial Susceptibility Testing Paradigms: Current Status and Future Directions. Am. Soc. Clin. Lab. Sci. 2018, 30. [Google Scholar] [CrossRef]
- Nizet, V. The accidental orthodoxy of Drs. Mueller and Hinton. EBioMedicine 2017, 22, 26–27. [Google Scholar] [CrossRef]
- Kubicek-Sutherland, J.Z.; Heithoff, D.M.; Ersoy, S.C.; Shimp, W.R.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Host-dependent induction of transient antibiotic resistance: A prelude to treatment failure. EBioMedicine 2015, 2, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Ersoy, S.C.; Heithoff, D.M.; Barnes, L.T.; Tripp, G.K.; House, J.K.; Marth, J.D.; Smith, J.W.; Mahan, M.J. Correcting a fundamental flaw in the paradigm for antimicrobial susceptibility testing. EBioMedicine 2017, 20, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, F.R. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012.
- Chambers, H.F.; Hackbarth, C.J. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1987, 31, 1982–1988. [Google Scholar] [CrossRef]
- Andrews, J.M.; Baquero, F.; Beltran, J.M.; Canton, E.; Crokaert, F.; Gobernado, M.; Gomez-Ius, R.; Loza, E.; Navarro, M.; Olay, T.; et al. International collaborative study on standardization of bacterial sensitivity to fosfomycin. J. Antimicrob. Chemother. 1983, 12, 357–361. [Google Scholar] [CrossRef]
- Asempa, T.E.; Abdelraouf, K.; Nicolau, D.P. Metallo-β-lactamase resistance in Enterobacteriaceae is an artefact of currently utilized antimicrobial susceptibility testing methods. J. Antimicrob. Chemother. 2020, 75, 997–1005. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- Schuurmans, J.M.; Hayali, A.S.N.; Koenders, B.B.; ter Kuile, B.H. Variations in MIC value caused by differences in experimental protocol. J. Microbiol. Methods 2009, 79, 44–47. [Google Scholar] [CrossRef]
- Van Belkum, A.; Dunne, W.M., Jr. Next-generation antimicrobial susceptibility testing. J. Clin. Microbiol. 2013, 51, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Puttaswamy, S.; Gupta, S.; Regunath, H.; Smith, L.; Sengupta, S. A comprehensive review of the present and future antibiotic susceptibility testing (AST) systems. Arch. Clin. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Ellington, M.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.; Hopkins, K.L. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef]
- Goldberg, B.; Sichtig, H.; Geyer, C.; Ledeboer, N.; Weinstock, G.M. Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics. MBio 2015, 6, e01888-15. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.; Beisken, S.; Lueftinger, L.; Weinmaier, T.; Klein, M.; Bacher, J.; Patel, R.; von Haeseler, A.; Posch, A.E. Species identification and antibiotic resistance prediction by analysis of whole-genome sequence data by use of ARESdb: An analysis of isolates from the Unyvero lower respiratory tract infection trial. J. Clin. Microbiol. 2020, 58, e00273-20. [Google Scholar] [CrossRef]
- Ersoy, S.C.; Abdelhady, W.; Li, L.; Chambers, H.F.; Xiong, Y.Q.; Bayer, A.S. Bicarbonate resensitization of methicillin-resistant Staphylococcus aureus to β-Lactam antibiotics. Antimicrob. Agents Chemother. 2019, 63, e00496-19. [Google Scholar] [CrossRef]
- Kumaraswamy, M.; Lin, L.; Olson, J.; Sun, C.-F.; Nonejuie, P.; Corriden, R.; Döhrmann, S.; Ali, S.R.; Amaro, D.; Rohde, M.; et al. Standard susceptibility testing overlooks potent azithromycin activity and cationic peptide synergy against MDR Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2016, 71, 1264–1269. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H., Jr.; Corriden, R.; Rohde, M.; et al. Azithromycin synergizes with cationic antimicrobial peptides to exert bactericidal and therapeutic activity against highly multidrug-resistant Gram-negative bacterial pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef]
- Ersoy, S.C.; Otmishi, M.; Milan, V.T.; Li, L.; Pak, Y.; Mediavilla, J.; Chen, L.; Kreiswirth, B.; Chambers, H.F.; Proctor, R.A. Scope and predictive genetic/phenotypic signatures of ‘bicarbonate [NaHCO3]-responsiveness’ and β-Lactam sensitization among methicillin-resistant Staphylococcus aureus (MRSA). Antimicrob. Agents Chemother. 2020, 64, e02445-19. [Google Scholar] [CrossRef]
- Rose, W.E.; Bienvenida, A.M.; Xiong, Y.Q.; Chambers, H.F.; Bayer, A.S.; Ersoy, S.C. Ability of bicarbonate supplementation to sensitize selected methicillin-resistant Staphylococcus aureus (MRSA) strains to β-Lactam antibiotics in an ex vivo simulated endocardial vegetation model. Antimicrob. Agents Chemother. 2019, 64, e0272-19. [Google Scholar]
- Ersoy, S.C.; Chambers, H.F.; Proctor, R.A.; Rosato, A.E.; Mishra, N.N.; Xiong, Y.Q.; Bayer, A.S. Impact of Bicarbonate on PBP2a Production, Maturation, and Functionality in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2021, 65, e02621-20. [Google Scholar] [CrossRef]
- Harrison, E.M.; Ba, X.; Coll, F.; Blane, B.; Restif, O.; Carvell, H.; Köser, C.U.; Jamrozy, D.; Reuter, S.; Lovering, A. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1680–1691. [Google Scholar] [CrossRef]
- Cockerill, F.R. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012; Volume 32. [Google Scholar]
- Patel, J.; Cockerill, F.; Alder, J.; Bradford, P.; Eliopoulos, G.; Hardy, D.; Hindler, J.; Jenkins, S.; Lewis, J.; Miller, L. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2014; Volume 34. [Google Scholar]
- Dien Bard, J.; Hindler, J.A.; Gold, H.S.; Limbago, B. Rationale for eliminating Staphylococcus breakpoints for β-lactam agents other than penicillin, oxacillin or cefoxitin, and ceftaroline. Clin. Infect. Dis. 2014, 58, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, F.; Patel, J.; Alder, J.; Bradford, P.; Dudley, M.; Eliopoulos, G.; Hardy, D.; Hecht, D.; Hindler, J.; Powell, M.; et al. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Third Informational Supplement; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2013; Volume 33. [Google Scholar]
- Bamgbola, O. Review of vancomycin-induced renal toxicity: An update. Ther. Adv. Endocrinol. Metab. 2016, 7, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Finkelberg, D.; Spooner, L.M. Daptomycin-induced acute renal and hepatic toxicity without rhabdomyolysis. Ann. Pharmacother. 2008, 42, 719–721. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.; Reilly, J.; Bunyan, D.; Walker, A. Costs of healthcare-associated methicillin-resistant Staphylococcus aureus and its control. Clin. Microbiol. Infect. 2010, 16, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Prod’hom, G.; Faverjon, F.; Rochais, Y.; Greub, G. Laboratory automation in clinical bacteriology: What system to choose? Clin. Microbiol. Infect. 2016, 22, 217–235. [Google Scholar] [CrossRef]
- Matuschek, E.; Brown, D.F.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [PubMed]
- Skov, R.L.; Pallesen, L.V.; Poulsen, R.L.; Espersen, F. Evaluation of a new 3-h hybridization method for detecting the mecA gene in Staphylococcus aureus and comparison with existing genotypic and phenotypic susceptibilty testing methods. J. Antimicrob. Chemother. 1999, 43, 467–475. [Google Scholar] [CrossRef][Green Version]
- Hiramatsu, K.; Asada, K.; Suzuki, E.; Okonogi, K.; Yokota, T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 1992, 298, 133–136. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Chia, N.; Jeraldo, P.R.; Quest, D.J.; Johnson, J.A.; Boxrud, D.J.; Taylor, A.J.; Chen, J.; Jenkins, G.D.; Drucker, T.M.; et al. Comparison of Whole-Genome Sequencing Methods for Analysis of Three Methicillin-Resistant Staphylococcus aureus Outbreaks. J. Clin. Microbiol. 2017, 55, 1946–1953. [Google Scholar] [CrossRef]
- Madigan, T.; Cunningham, S.A.; Patel, R.; Greenwood-Quaintance, K.E.; Barth, J.E.; Sampathkumar, P.; Cole, N.C.; Kohner, P.C.; Colby, C.E.; Asay, G.F.; et al. Whole-genome sequencing for methicillin-resistant Staphylococcus aureus (MRSA) outbreak investigation in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 2018, 39, 1412–1418. [Google Scholar] [CrossRef]
- Cho, H.K.; Yang, J.N.; Cunningham, S.A.; Greenwood-Quaintance, K.E.; Dalton, M.L.; Collura, C.A.; Fang, J.L.; Heinrich, A.L.; Huskins, W.C.; Patel, R. Molecular epidemiology of methicillin-susceptible Staphylococcus aureus in infants in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 2020, 41, 1402–1408. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Jeraldo, P.R.; Schuetz, A.N.; Heitman, A.A.; Patel, R. Staphylococcus aureus whole genome sequence–based susceptibility and resistance prediction using a clinically amenable workflow. Diagn. Microbiol. Infect. Dis. 2020, 97, 115060. [Google Scholar] [CrossRef]
- Patel, R.; (Mayo Clinic, Rochester, MN, USA). Personal communication, 2021.
- Fenn, W.O. The carbon dioxide dissociation curve of nerve and muscle. Am. J. Physiol.-Leg. Content 1928, 85, 207–223. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 1–23. [Google Scholar]
- Mathema, B.; Mediavilla, J.; Kreiswirth, B.N. Sequence analysis of the variable number tandem repeat in Staphylococcus aureus protein A gene. In Bacterial Pathogenesis; Springer: Berlin/Heidelberg, Germany, 2008; pp. 285–305. [Google Scholar]
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global epidemiology of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mediavilla, J.R.; Oliveira, D.C.; Willey, B.M.; de Lencastre, H.; Kreiswirth, B.N. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec typing. J. Clin. Microbiol. 2009, 47, 3692–3706. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: Role of staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, E.; Coyle, E.A.; Kaatz, G.W.; Zervos, M.J.; Rybak, M.J. Comparison of a rabbit model of bacterial endocarditis and an in vitro infection model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 2000, 44, 1921–1924. [Google Scholar] [CrossRef]
- Rose, W.E.; Leonard, S.N.; Rybak, M.J. Evaluation of daptomycin pharmacodynamics and resistance at various dosage regimens against Staphylococcus aureus isolates with reduced susceptibilities to daptomycin in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 2008, 52, 3061–3067. [Google Scholar] [CrossRef]
- Rose, W.E.; Leonard, S.N.; Sakoulas, G.; Kaatz, G.W.; Zervos, M.J.; Sheth, A.; Carpenter, C.F.; Rybak, M.J. Daptomycin activity against Staphylococcus aureus following vancomycin exposure in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 2008, 52, 831–836. [Google Scholar] [CrossRef]
- Standiford, H.C.; Jordan, M.C.; Kirby, W.M. Clinical pharmacology of carbenicillin compared with other penicillins. J. Infect. Dis. 1970, 122, S9–S13. [Google Scholar] [CrossRef]
- Rao, S.N.; Rhodes, N.J.; Lee, B.J.; Scheetz, M.H.; Hanson, A.P.; Segreti, J.; Crank, C.W.; Wang, S.K. Treatment outcomes with cefazolin versus oxacillin for deep-seated methicillin-susceptible Staphylococcus aureus bloodstream infections. Antimicrob. Agents Chemother. 2015, 59, 5232–5238. [Google Scholar] [CrossRef]
| Responsive Strains (n = 15) | |||||||
|---|---|---|---|---|---|---|---|
| Strain | AMC A | mecA Genotype | Ridom spa Type | agr Type | CC Type | SCCmec Type | β-lactamase (±) |
| MRSA 11/11 | 16 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| MW2 | 20 (S) | susceptible 2 | t128 | agr III | 1 | IV | + |
| BCVA289 | 16 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| PB 031-038 | 11 (R) | resistant 2 | Unknown B | agr I | 8 | IV | + |
| PB 004-193 | 15 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| PB 043-043 | 15 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| PB 077-107 | 13 (R) | susceptible 2 | t002 | agr II | 5 | II | + |
| C48 | 18 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C42 | 15 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C13 | 15 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C32 | 18 (S) | susceptible 2 | t002 | agr II | 5 | IV | + |
| C30 | 19 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C24 | 16 (S) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C38 | 14 (R) | susceptible 2 | t008 | agr I | 8 | IV | + |
| RB 300-087 | 18 (S) | susceptible 2 | t2265 | agr I | 45 | IV | + |
| Nonresponsive Strains (n = 15) | |||||||
| Strain | AMC | mecA Genotype | Ridom spa type | agr type | CC type | SCCmec type | β-lactamase (±) |
| BMC1001 | 15 (S) | resistant 2 | t064 | agr I | 8 | IV | + |
| C5 | 26 (S) | resistant 2 | t242 | agr II | 5 | II | + |
| RB 067-227 | 18 (S) | susceptible 2 | t128 | agr III | 1 | IV | + |
| RB 010-016 | 11 (R) | resistant 2 | t002 | agr II | 5 | II | + |
| PB 027-133 | 13 (R) | resistant 2 | t002 | agr II | 5 | II | + |
| PB 088-180 | 23 (S) | resistant 2 | t002 | agr II | 5 | II | + |
| RB 034-221 | 23 (S) | resistant 2 | t002 | agr II | 5 | II | + |
| C7 | 14 (R) | susceptible 2 | t008 | agr I | 8 | IV | + |
| C36 | 14 (R) | resistant 2 | t002 | agr II | 5 | II | + |
| C15 | 16 (S) | resistant 2 | t064 | agr I | 8 | IV | + |
| PB 300-111 | 17 (S) | susceptible 2 | t051 | agr I | 8 | IV | + |
| PB 321-236 | 23 (S) | resistant 2 | t003 | agr II | 5 | II | + |
| C3 | 14 (R) | susceptible 2 | t008 | agr I | 8 | IV | + |
| PB 017-037 | 24 (S) | resistant 2 | t002 | agr II | 5 | II | + |
| RB 057-171 | 15 (S) | susceptible 2 | t9878 | agr III | 1 | IV | + |
| Algorithm Criteria Met? | NaHCO3-Responsive | Nonresponsive |
|---|---|---|
| Criteria met | 10 | 0 |
| Criteria not met | 5 | 15 |
| Statistic | Value | 95% CI |
| Sensitivity | 66.67% | 38.38% to 88.18% |
| Specificity | 100.00% | 78.20% to 100.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ersoy, S.C.; Rose, W.E.; Patel, R.; Proctor, R.A.; Chambers, H.F.; Harrison, E.M.; Pak, Y.; Bayer, A.S. A Combined Phenotypic-Genotypic Predictive Algorithm for In Vitro Detection of Bicarbonate: β-Lactam Sensitization among Methicillin-Resistant Staphylococcus aureus (MRSA). Antibiotics 2021, 10, 1089. https://doi.org/10.3390/antibiotics10091089
Ersoy SC, Rose WE, Patel R, Proctor RA, Chambers HF, Harrison EM, Pak Y, Bayer AS. A Combined Phenotypic-Genotypic Predictive Algorithm for In Vitro Detection of Bicarbonate: β-Lactam Sensitization among Methicillin-Resistant Staphylococcus aureus (MRSA). Antibiotics. 2021; 10(9):1089. https://doi.org/10.3390/antibiotics10091089
Chicago/Turabian StyleErsoy, Selvi C., Warren E. Rose, Robin Patel, Richard A. Proctor, Henry F. Chambers, Ewan M. Harrison, Youngju Pak, and Arnold S. Bayer. 2021. "A Combined Phenotypic-Genotypic Predictive Algorithm for In Vitro Detection of Bicarbonate: β-Lactam Sensitization among Methicillin-Resistant Staphylococcus aureus (MRSA)" Antibiotics 10, no. 9: 1089. https://doi.org/10.3390/antibiotics10091089
APA StyleErsoy, S. C., Rose, W. E., Patel, R., Proctor, R. A., Chambers, H. F., Harrison, E. M., Pak, Y., & Bayer, A. S. (2021). A Combined Phenotypic-Genotypic Predictive Algorithm for In Vitro Detection of Bicarbonate: β-Lactam Sensitization among Methicillin-Resistant Staphylococcus aureus (MRSA). Antibiotics, 10(9), 1089. https://doi.org/10.3390/antibiotics10091089






