Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria
Abstract
1. Introduction
2. Results
2.1. Isolation of Single Bacterial Cultures
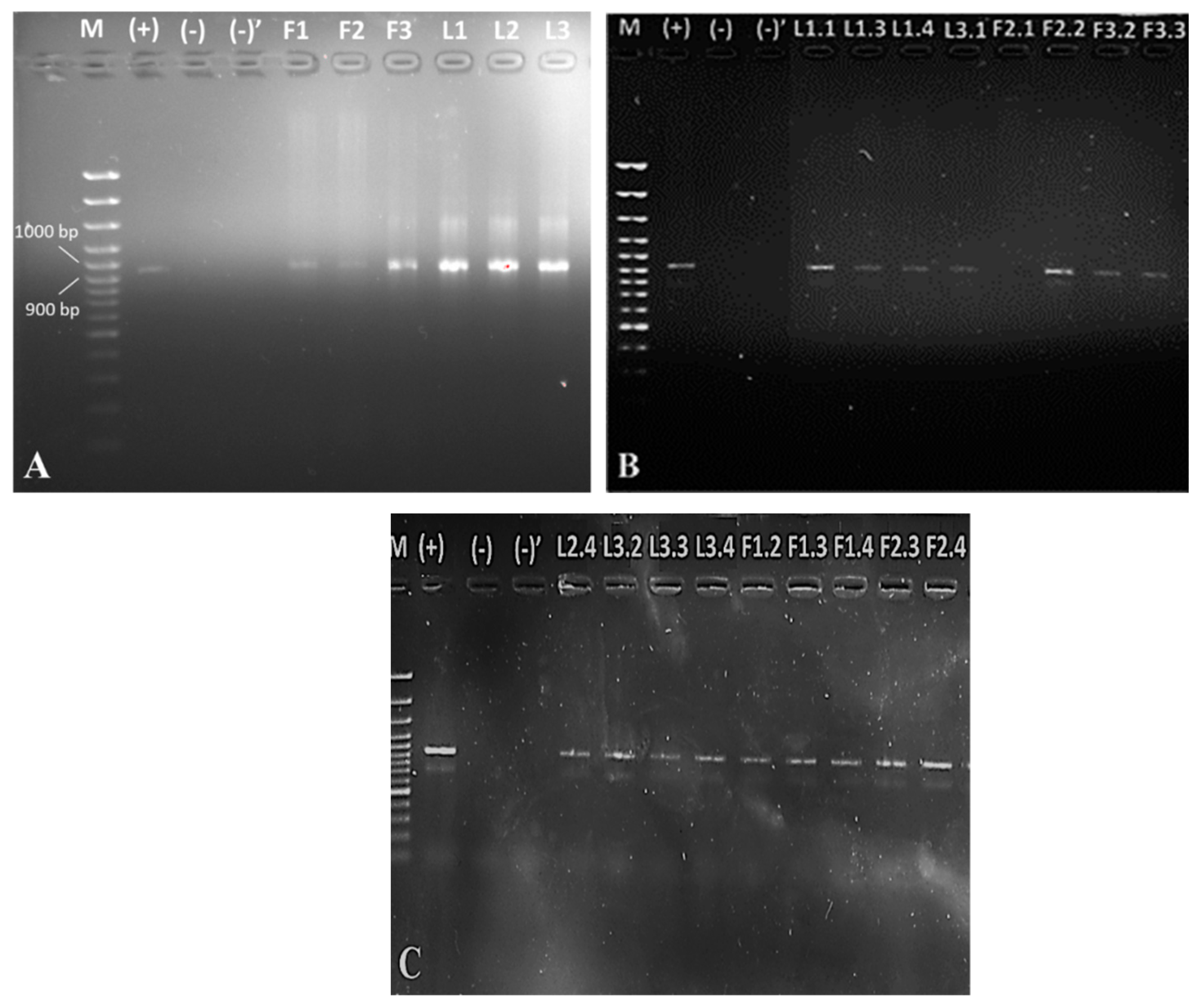
2.2. Biochemical and 16S rDNA Characterization
2.3. Test for Biofilm Formation
2.4. Antibiotic Resistance
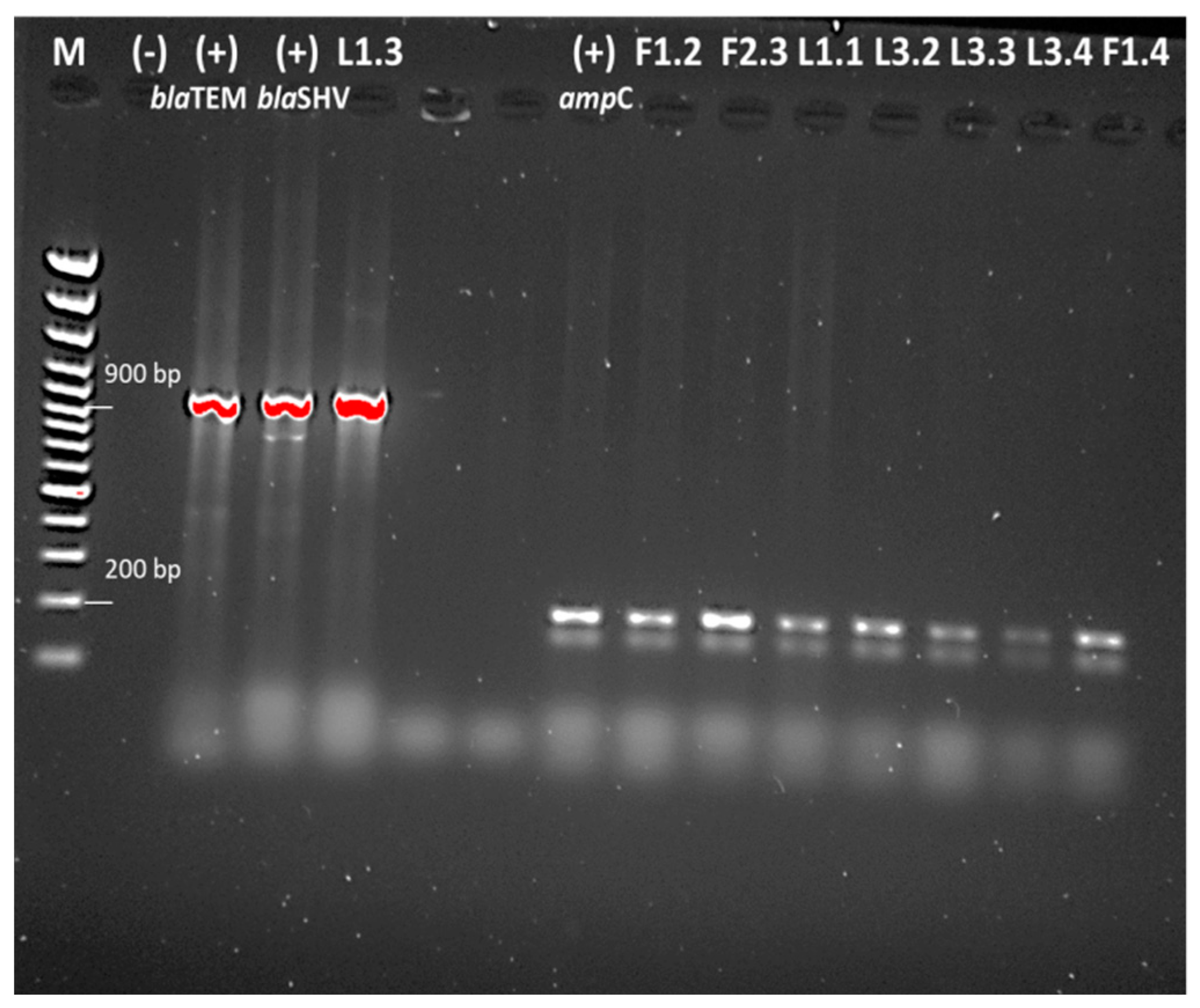
2.5. Detection of Antibiotic Resistance Genes
3. Discussion
4. Materials and Methods
4.1. Swine Farm and Sample Collection
4.2. Isolation of Single Bacterial Cultures
4.3. Biochemical Characterization
4.4. Isolation of 16S rDNA
4.5. PCR Analysis
4.6. Disc Diffusion Method
4.7. Test for Biofilm Formation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’neill, J.I.M. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist 2014, 20, 1–16. [Google Scholar]
- He, L.-Y.; He, L.-K.; Liu, Y.-S.; Zhang, M.; Zhao, J.-L.; Zhang, Q.-Q.; Ying, G.-G. Microbial diversity and antibiotic resistome in swine farm environments. Sci. Total Environ. 2019. [Google Scholar] [CrossRef]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Storteboom, H.; Arabi, M.; Davis, J.; Crimi, B.; Pruden, A. Identification of antibiotic-resistance-gene molecular signatures suitable as tracers of pristine river, urban, and agricultural sources. Environ. Sci. Technol. 2010, 44, 1947–1953. [Google Scholar] [CrossRef] [PubMed]
- Winkworth-Lawrence, C.; Lange, K. Antibiotic resistance genes in freshwater biofilms may reflect influences from high-intensity agriculture. Microb. Ecol. 2016, 72, 763–772. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef]
- Prescott, J.F. The resistance tsunami, antimicrobial stewardship, and the golden age of microbiology. Vet. Microbiol. 2014, 171, 273–278. [Google Scholar] [CrossRef]
- Garner, E.; Benitez, R.; von Wagoner, E.; Sawyer, R.; Schaberg, E.; Hession, W.C.; Krometis, L.-A.H.; Badgley, B.D.; Pruden, A. Stormwater loadings of antibiotic resistance genes in an urban stream. Water Res. 2017, 123, 144–152. [Google Scholar] [CrossRef]
- Yu, Z.; He, P.; Shao, L.; Zhang, H.; Lü, F. Co-occurrence of mobile genetic elements and antibiotic resistance genes in municipal solid waste landfill leachates: A preliminary insight into the role of landfill age. Water Res. 2016, 106, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef]
- Luppi, A. Swine enteric colibacillosis: Diagnosis, therapy and antimicrobial resistance. Porc. Health Manag. 2017, 3, 1–18. [Google Scholar] [CrossRef]
- Pereira, D.A.; Vidotto, M.C.; Nascimento, K.A.; Santos, A.C.R.d.; Mechler, M.L.; Oliveira, L.G.d. Virulence factors of Escherichia coli in relation to the importance of vaccination in pigs. Ciência Rural 2016, 46, 1430–1437. [Google Scholar] [CrossRef]
- Loos, M.; Geens, M.; Schauvliege, S.; Gasthuys, F.; van der Meulen, J.; Dubreuil, J.D.; Goddeeris, B.M.; Niewold, T.; Cox, E. Role of heat-stable enterotoxins in the induction of early immune responses in piglets after infection with enterotoxigenic Escherichia coli. PLoS ONE 2012, 7, e41041. [Google Scholar] [CrossRef]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef]
- Fairbrother, J.; Gyles, C. Post-weaning Escherichia coli diarrhea and edema disease. In Diseases of Swine; Straw, B.E., Zimmerman, J.J., D’Allaire, S., Taylor, D.J., Eds.; Blackwell Publishing: Hoboken, NJ, USA, 2006. [Google Scholar]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal enterotoxigenic Escherichia coli. EcoSal Plus 2016, 7, 1–80. [Google Scholar] [CrossRef]
- Blanco, M.; Blanco, J.E.; Dahbi, G.; Mora, A.; Alonso, M.P.; Varela, G.; Gadea, M.P.; Schelotto, F.; Gonzalez, E.A.; Blanco, J. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: Identification of two novel intimin variants (μB and ξR/β2B). J. Med. Microbiol. 2006, 55, 1165–1174. [Google Scholar] [CrossRef][Green Version]
- Beutin, L.; Zimmermann, S.; Gleier, K. Evaluation of the VTEC-Screen “Seiken” test for detection of different types of Shiga toxin (verotoxin)-producing Escherichia coli (STEC) in human stool samples. Diagn. Microbiol. Infect. Dis. 2002, 42, 1–8. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union one health 2018 zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar]
- ECDC. Antimicrobial resistance surveillance in Europe 2016. In Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2017. [Google Scholar]
- WHO. Central Asian and Eastern European Surveillance of Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- ECDC. Surveillance of Antimicrobial Resistance in Europe. In Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017; ECDC: Stockholm, Sweden, 2018. [Google Scholar]
- Buranasinsup, S.; Kulpeanprasit, S.; Kong-ngoen, T.; Jangsangthong, A.; Sookrung, N.; Chaicumpa, W.; Indrawattana, N. Prevalence of the Multi-drug Resistance of Shiga Toxin-producing Escherichia coli Isolated from Pigs in Central Thailand. Chiang Mai J. Sci 2018, 45, 21–32. [Google Scholar]
- Urumova, V. Phenotypic and Genotypic Characteristics of Antimicrobial Resistance in Resident Escherichia coli and Enterococcus spp., Isolated from Intensively Farmed Pigs in Bulgaria (in Bulgarian); Trakia University: Stara, Zagora, 2016. [Google Scholar]
- Hu, Y.S.; Shin, S.; Park, Y.H.; Park, K.T. Prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine feces in Korea. J. Food Prot. 2017, 80, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Yang, Y.; Li, F.; Li, X.; Liu, H.; Fazilani, S.A.; Guo, W.; Xu, G.; Zhang, X. The prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine farms in China. BMC Vet. Res. 2020, 16, 258. [Google Scholar] [CrossRef]
- Dimitrova, A.; Yordanov, S.; Petkova, K.; Savova, T.; Bankova, R.; Ivanova, S. Example scheme for antibacterial therapy and metaphylaxis of colibacillosis in the intensive pig production (in Bulgarian). Livest. Science. Agric. Acad. 2016, LIII, 44–53. [Google Scholar]
- Magray, M.S.U.D.; Kumar, A.; Rawat, A.K.; Srivastava, S. Identification of Escherichia coli through analysis of 16S rRNA and 16S-23S rRNA internal transcribed spacer region sequences. Bioinformation 2011, 6, 370. [Google Scholar] [CrossRef]
- Padola, N.L.; Sanz, M.E.; Blanco, J.E.; Blanco, M.; Blanco, J.; Etcheverria, A.I.; Arroyo, G.H.; Usera, M.A.; Parma, A.E. Serotypes and virulence genes of bovine Shigatoxigenic Escherichia coli (STEC) isolated from a feedlot in Argentina. Vet. Microbiol. 2004, 100, 3–9. [Google Scholar] [CrossRef]
- Byun, J.; Jung, B.; Kim, H.; Fairbrother, J.; Lee, M.; Lee, W. O-serogroups, virulence genes of pathogenic Escherichia coli and Pulsed-field gel electrophoresis (PFGE) patterns of O149 isolates from diarrhoeic piglets in Korea. Vet. Med. 2013, 58, 468–476. [Google Scholar] [CrossRef]
- Memon, J.; Kashif, J.; Hussain, N.; Yaqoob, M.; Ali, A.; Buriro, R.; Soomro, J.; Hassan, M.F.; Sahito, B.; Hongjie, F. Serotypes, Genotypes, Virulence Factors and Antimicrobial Resistance Genes of Escherichia coli Isolated in Bovine Clinical Mastitis from Eastern China. Pak. Vet. J. 2016, 36. [Google Scholar]
- Do, K.-H.; Byun, J.-W.; Lee, W.-K. Virulence genes and antimicrobial resistance of pathogenic Escherichia coli isolated from diarrheic weaned piglets in Korea. J. Anim. Sci. Technol. 2020, 62, 543. [Google Scholar] [CrossRef] [PubMed]
- Annisha, O.D.R.; Li, Z.; Zhou, X.; Stenay Junior, N.M.D.; Donde, O.O. Efficacy of integrated ultraviolet ultrasonic technologies in the removal of erythromycin-and quinolone-resistant Escherichia coli from domestic wastewater through a laboratory-based experiment. J. Water Sanit. Hyg. Dev. 2019, 9, 571–580. [Google Scholar] [CrossRef]
- Pourhossein, Z.; Asadpour, L.; Habibollahi, H.; Shafighi, S.T. Antimicrobial resistance in fecal Escherichia coli isolated from poultry chicks in northern Iran. Gene Rep. 2020, 21, 100926. [Google Scholar] [CrossRef]
- Ranjbar, R.; Dehkordi, F.S.; Shahreza, M.H.S.; Rahimi, E. Prevalence, identification of virulence factors, O-serogroups and antibiotic resistance properties of Shiga-toxin producing Escherichia coli strains isolated from raw milk and traditional dairy products. Antimicrob. Resist. Infect. Control 2018, 7, 53. [Google Scholar] [CrossRef]
- Jafari, E.; Mostaan, S.; Bouzari, S. Characterization of antimicrobial susceptibility, extended-spectrum β-lactamase genes and phylogenetic groups of enteropathogenic Escherichia coli isolated from patients with diarrhea. Osong Public Health Res. Perspect. 2020, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Kuan, N.-L.; Yeh, K.-S. Characteristics of extended-spectrum β-lactamase–producing Escherichia coli from dogs and cats admitted to a Veterinary Teaching Hospital in Taipei, Taiwan From 2014 to 2017. Front. Vet. Sci. 2020, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Abimiku, R.H.; Ngwai, Y.B.; Nkene, I.H.; Bassey, B.E.; Tsaku, P.A.; Ibrahim, T.; Tama, S.C.; Ishaleku, D.; Pennap, G.R.I. Molecular Diversity and extended spectrum beta-lactamase resistance of diarrheagenic Escherichia coli from patients attending selected health care facilities in Nasarawa state, Nigeria. Int. J. Pathog. Res. 2019, 3, 1–18. [Google Scholar] [CrossRef]
- Khoirani, K.; Indrawati, A.; Setiyaningsih, S. Detection of ampicillin resistance encoding gene of Escherichia coli from chickens in Bandung and Purwakarta. J. Ris. Vet. Indones. (J. Indones. Vet. Res.) 2019, 3. [Google Scholar] [CrossRef]
- Kim, Y.W.; Choe, J.C.; Jablonski, P.G. Detection of antibiotic-resistant Escherichia coli from the feces of the Oriental magpie nestlings. J. Korean Soc. Ornithol. 2020, 27, 3–9. [Google Scholar] [CrossRef]
- Böckelmann, U.; Dörries, H.-H.; Ayuso-Gabella, M.N.; de Marçay, M.S.; Tandoi, V.; Levantesi, C.; Masciopinto, C.; Van Houtte, E.; Szewzyk, U.; Wintgens, T. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl. Environ. Microbiol. 2009, 75, 154–163. [Google Scholar] [CrossRef]
- Nguyen, M.C.P.; Woerther, P.-L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 2009, 15, 1648. [Google Scholar] [CrossRef] [PubMed]
- Hassanen, F.; Sabike, I.; Haikel, G.; Elged, E. Screening for Antimicrobial resistance in some pathogens isolated from cold smoked fish marketed in Menofiya government. Benha Vet. Med. J. 2018, 35, 269–283. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 3rd ed.; Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009. [Google Scholar]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: https://eucast.org/clinical_breakpoints/ (accessed on 2 August 2021).
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.; Baddour, L.; Barrett, F.; Melton, D.; Beachey, E. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
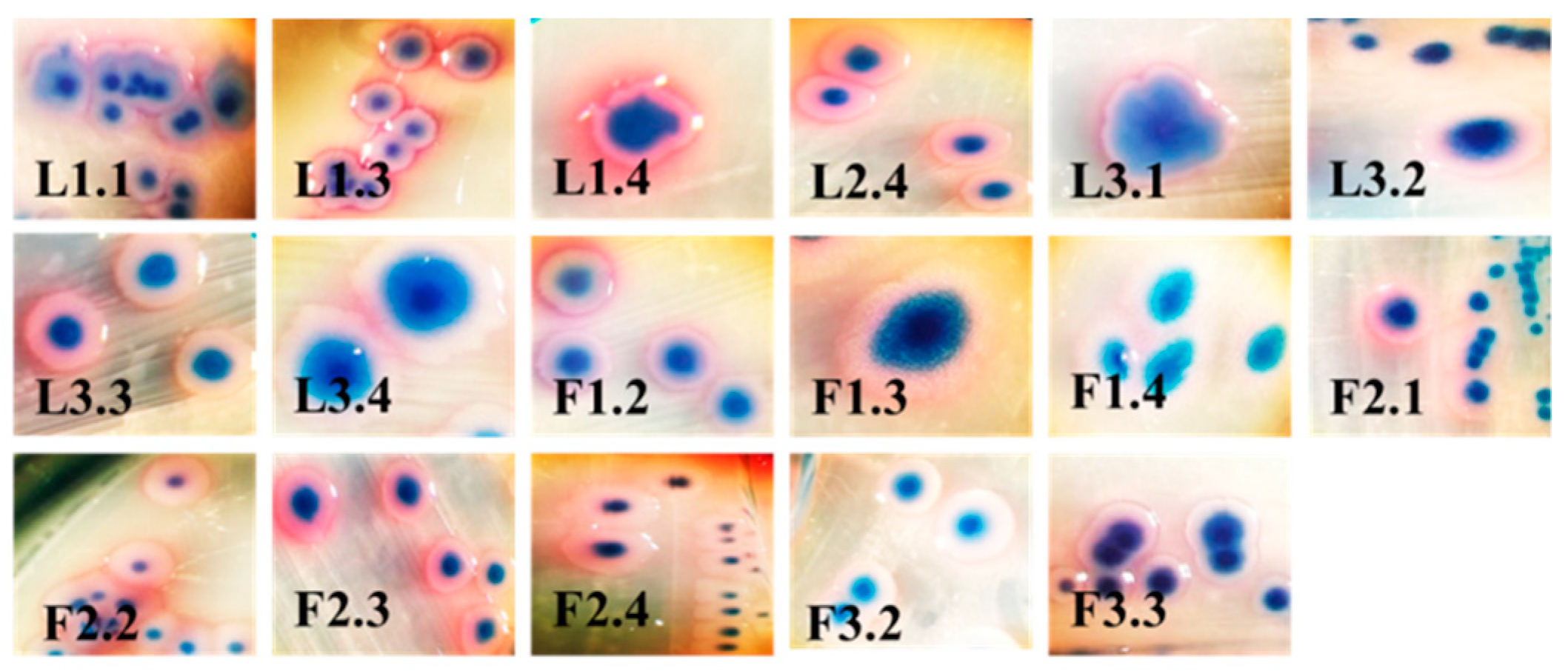
| Strain | OD570 nm | Adherence | Biofilm | Strain | OD570 nm | Adherence | Biofilm |
|---|---|---|---|---|---|---|---|
| ATCC 35218 | 0.463 | MA |  | L3.4 | 0.158 | NA |  |
| O:157 | 1.041 | SA |  | F1.2 | 1.083 | SA |  |
| Blank | 0.162 | --- |  | F1.3 | 0.261 | WA | 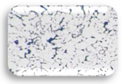 |
| L1.1 | 0.361 | MA |  | F1.4 | 0.261 | WA | 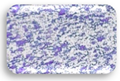 |
| L1.3 | 1.103 | SA | 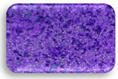 | F2.2 | 0.358 | MA |  |
| L1.4 | 0.364 | MA | 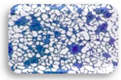 | F2.3 | 1.183 | SA | 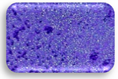 |
| L2.4 | 1.222 | SA |  | F2.4 | 0.378 | MA |  |
| L3.1 | 0.361 | MA | 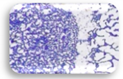 | F3.2 | 0.204 | WA | 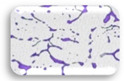 |
| L3.2 | 0.382 | MA | 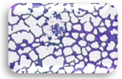 | F3.3 | 0.328 | WA | 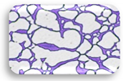 |
| L3.3 | 1.188 | SA |  | ||||
| Drug Class | Antibiotic/Strain | F1.2 | F1.3 | F1.4 | F2.1 | F2.2 | F2.3 | F2.4 | F3.2 | F3.3 | L1.1 | L1.3 | L1.4 | L2.4 | L3.1 | L3.2 | L3.3 | L3.4 | E. coli ATCC 35218 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycosides | Tobramycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Amikacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Gentamicin | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S | |
| Penicillins | Amoxicillin/clavulanic acid | S | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S |
| Ampicillin | S | R | R | S | S | R | S | R | R | R | R | S | R | S | R | R | R | R | |
| Piperacillin/tazobactam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Monobactams | Aztreonam | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Cephalosporins | Cefazolin | I | I | I | I | X | I | I | I | I | I | I | I | I | I | I | I | I | I |
| Cefotaxime | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Ceftazidime | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Cefuroxime | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | |
| Cephalexin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Fluoroquinolones | Ciprofloxacin | S | S | S | S | I | S | S | S | S | S | R | S | S | S | S | S | S | S |
| Carbapenems | Ertapenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Imipenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Meropenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Other agents | Colistin | X | X | X | X | X | X | X | X | X | X | R | X | X | X | X | X | X | X |
| Fosfomycin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Nitrofurantoin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Trimethoprim | R | S | S | S | R | S | S | R | R | S | R | S | S | S | S | S | S | S | |
| Trimethoprim/sulfamethoxazole | R | S | S | S | R | S | S | S | S | S | R | S | S | S | S | S | S | S |
| Drug Class | Antibiotic/Strain | F1.2 | F1.3 | F1.4 | F2.2 | F2.3 | F2.4 | F3.2 | F3.3 | L1.1 | L1.3 | L1.4 | L2.4 | L3.1 | L3.2 | L3.3 | L3.4 | E. coli O:157 | E. coli ATCC 35218 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tetracycline | Tetracycline | R | S | S | R | I | S | R | S | R | R | R | R | I | R | R | I | R | S |
| Doxycycline hydrochloride | R | I | S | R | R | S | R | R | S | R | S | R | S | S | S | S | S | S | |
| Macrolides | Erythromycin | S | I | S | I | S | S | I | I | I | I | I | S | I | R | S | S | S | S |
| Clarithromycin | S | I | S | S | S | S | I | S | S | S | S | S | S | S | S | S | S | S | |
| Cephalosporins | Cefamandole | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Fluoroquinolones | Nalidixic acid | S | R | S | S | S | S | S | S | R | R | S | S | S | S | S | R | S | R |
| Pefloxacin | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | S | S | |
| Ciprofloxacin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Penicillins | Ampicillin | R | R | S | S | R | S | R | R | R | R | S | R | S | R | R | R | S | R |
| Amoxicillin | S | R | R | S | R | S | R | R | R | R | R | R | S | R | R | R | n.m. | R | |
| Amoxicillin/clavulanic acid | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Penicillin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
| Carbenicillin | S | S | S | S | S | S | S | S | I | I | S | S | I | S | S | S | S | S | |
| Carbapenems | Meropenem | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Aminoglycosides | Streptomycin | S | S | S | S | S | S | S | S | S | I | R | S | S | S | S | R | S | R |
| Other agents | Chloramphenicol | R | R | S | R | R | S | R | R | S | S | S | R | S | S | R | R | R | R |
| Trimethoprim/sulfamethoxazole | R | R | S | R | S | S | R | R | S | R | R | S | S | S | S | S | S | S |
| Primers | Sequences | Reference |
|---|---|---|
| E. coli 16S rDNA F | 5′-AGA GTT TGA TCC TGG CTC AG-3′ | [28] |
| E. coli 16S rDNA R | 5′-CTT GTG CGG GCC CCC GTC AAT TC-3′ | |
| stx1-1 F | 5′-TTA GAC TTC TCG ACT GCA AAG-3′ | [29,30] |
| stx1-1 R | 5′-TGT TGT ACG AAA TCC CCT CTG-3′ | |
| stx2all-1 F | 5′-TTA TAT CTG CGC CGG GTC TG-3′ | [30] |
| stx2all-2 R | 5′-AGA CGA AGA TGG TCA AAA CG-3′ | |
| LT1 F | 5′-TTA CGG CGT TAC TAT CCT CTC TA-3′ | [31] |
| LT2 R | 5′-GGT CTC GGT CAG ATA TGT GAT TC-3′ | |
| STa1 F | 5′-TCC CCT CTT TTA GTC AGT CAA CTG-3′ | [31] |
| STa2 R | 5′-GCA CAG GCA GGA TTA CAA CAA AGT-3′ | |
| F4-1 F | 5′-ATC GGT GGT AGT ATC ACT GC-3′ | [31] |
| F4-2 R | 5′-AAC CTG CGA CGT CAA CAA GA-3′ | |
| eae (Intimin)-1 F | 5′-CAT TAT GGA ACG GCA GAG GT-3′ | [30,32] |
| eae (Intimin)-2 R | 5′-ATC TTC TGC GTA CTG CGT TCA-3′ | |
| qnrA F | 5′-GGG TAT GGA TAT TAT TGA TAA AG-3′ | [33] |
| qnrA R | 5′-CTA ATC CGG CAG CAC TAT TTA-3′ | |
| qnrB F | 5′-GAT CGT GAA AGC CAG AAA GG-3′ | [34] |
| qnrB R | 5′-ACG ATG CCT GGT AGT TGT CC-3′ | |
| aac(3)-IV F | 5′-CTT CAG GAT GGC AAG TTG GT-3′ | [35] |
| aac(3)-IV R | 5′-TCA TCT CGT TCT CCG CTC AT-3′ | |
| blaSHV F | 5′-TCG CCT GTG TAT TAT CTC CC-3′ | [36] |
| blaSHV R | 5′-CGC AGA TAA ATC ACC ACA ATG-3′ | |
| blaTEM F | 5′-TCG GGG AAA TGT GCG CG-3′ | [37,38] |
| blaTEM R | 5′-TGC TTA ATC AGT GAG GCA CC-3′ | |
| ampC F | 5′-AAT GGG TTT TCT ACG GTC TG-3′ | [39,40] |
| ampC R | 5′-GGG CAG CAA ATG TGG AGC AA-3′ | |
| ampC F | 5′-GTG ACC AGA TAC TGG CCA CA-3′ | [41] |
| ampC R | 5′-TTA CTG TAG CGC CTC GAG GA-3′ | |
| ermB F | 5′-GAA AAA GTA CTC AAC CAA ATA-3′ | [42,43] |
| ermB R | 5′-AAT TTA AGT ACC GTT AC-3′ | |
| ermB F | 5′-GCA TTT AAC GAC GAA ACT GGC T-3′ | [41] |
| ermB R | 5′-GAC AAT ACT TGC TCA TAA GTA ATG GT-3′ |
| Formula | Adherence |
|---|---|
| ODprobe ≤ ODblank | non-adherent |
| ODblank < ODprobe ≤ 2 × ODblank | weakly adherent |
| 2 × ODblank < ODprobe ≤ 4 × ODblank | moderately adherent |
| 4 × ODblank < ODprobe | strongly adherent |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimitrova, L.; Kaleva, M.; Zaharieva, M.M.; Stoykova, C.; Tsvetkova, I.; Angelovska, M.; Ilieva, Y.; Kussovski, V.; Naydenska, S.; Najdenski, H. Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria. Antibiotics 2021, 10, 940. https://doi.org/10.3390/antibiotics10080940
Dimitrova L, Kaleva M, Zaharieva MM, Stoykova C, Tsvetkova I, Angelovska M, Ilieva Y, Kussovski V, Naydenska S, Najdenski H. Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria. Antibiotics. 2021; 10(8):940. https://doi.org/10.3390/antibiotics10080940
Chicago/Turabian StyleDimitrova, Lyudmila, Mila Kaleva, Maya M. Zaharieva, Christina Stoykova, Iva Tsvetkova, Maya Angelovska, Yana Ilieva, Vesselin Kussovski, Sevda Naydenska, and Hristo Najdenski. 2021. "Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria" Antibiotics 10, no. 8: 940. https://doi.org/10.3390/antibiotics10080940
APA StyleDimitrova, L., Kaleva, M., Zaharieva, M. M., Stoykova, C., Tsvetkova, I., Angelovska, M., Ilieva, Y., Kussovski, V., Naydenska, S., & Najdenski, H. (2021). Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria. Antibiotics, 10(8), 940. https://doi.org/10.3390/antibiotics10080940










