Compared with Cotrimoxazole Nitroxoline Seems to Be a Better Option for the Treatment and Prophylaxis of Urinary Tract Infections Caused by Multidrug-Resistant Uropathogens: An In Vitro Study
Abstract
1. Introduction
2. Results
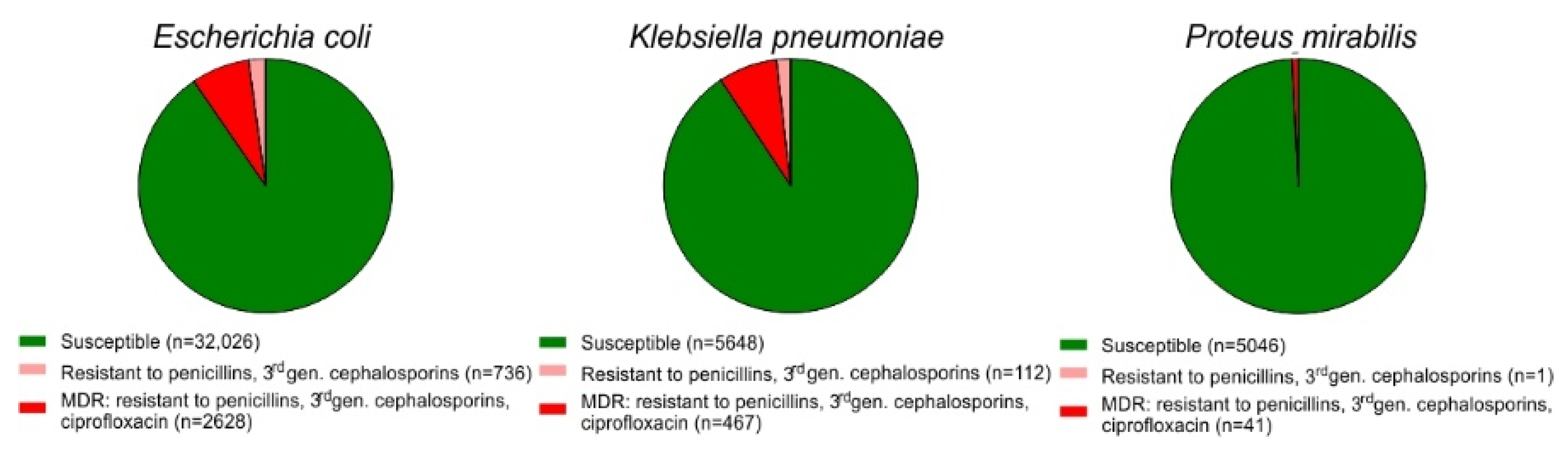
2.1. Prevalence of MDR among E. coli Isolates from Urine Samples
2.2. Overall Activity of Nitroxoline against Multiresistant Uropathogens
2.3. Parallel Resistance to Cotrimoxazole and 3rd Generation Cephalosporins, Ciprofloxacin and MDR Strains of E. coli
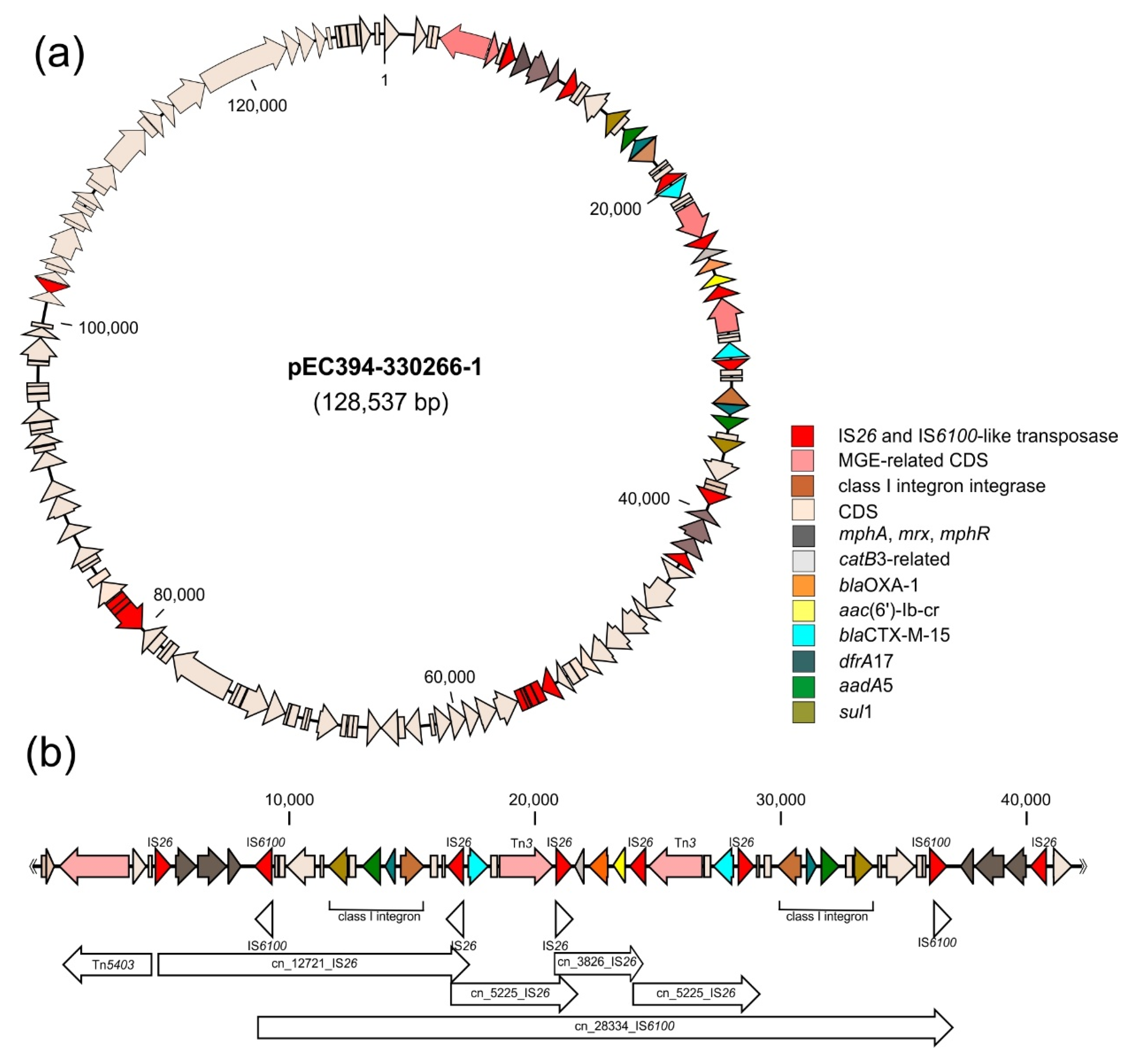
2.4. Co-Localisation of Transferable Resistance to Cotrimoxazole, ESBL and Quinolones
3. Discussion
3.1. Co-Localisation of Transferable Resistance to Cotrimoxazole, ESBL and Quinolones
3.2. Parallel Resistance Due to Accumulation of Resistance Determinants on Mobile Genetic Elements
4. Materials and Methods
4.1. Urine Culture Analysis and Antibiotic Susceptibility Testing
4.2. Genome Sequencing
4.3. Annotation and Strain Typing
4.4. Analysis of Co-Occurrence of Cotrimoxazole and Other Antimicrobial Resistance Genes in Bacterial Genomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Pallett, A.; Hand, K. Complicated urinary tract infections: Practical solutions for the treatment of multiresistant Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65 (Suppl. 3), iii25–iii33. [Google Scholar] [CrossRef]
- Arana, D.M.; Rubio, M.; Alos, J.I. Evolution of antibiotic multiresistance in Escherichia coli and Klebsiella pneumoniae isolates from urinary tract infections: A 12-year analysis (2003–2014). Enferm. Infecc. Microbiol. Clin. 2017, 35, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Cek, M.; Tandogdu, Z.; Wagenlehner, F.; Tenke, P.; Naber, K.; Bjerklund-Johansen, T.E. Healthcare-associated urinary tract infections in hospitalized urological patients-a global perspective: Results from the GPIU studies 2003–2010. World J. Urol. 2014, 32, 1587–1594. [Google Scholar] [CrossRef]
- Esteve-Palau, E.; Grau, S.; Herrera, S.; Sorli, L.; Montero, M.; Segura, C.; Duran, X.; Horcajada, J.P. Impact of an antimicrobial stewardship program on urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Rev. Esp. Quimioter. 2018, 31, 110–117. [Google Scholar] [PubMed]
- Magiorakos, A.P.; Burns, K.; Rodriguez Bano, J.; Borg, M.; Daikos, G.; Dumpis, U.; Lucet, J.C.; Moro, M.L.; Tacconelli, E.; Simonsen, G.S.; et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: Guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control. 2017, 6, 113. [Google Scholar] [CrossRef]
- Mazzariol, A.; Bazaj, A.; Cornaglia, G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: A review. J. Chemother. 2017, 29, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Gomila, A.; Carratala, J.; Eliakim-Raz, N.; Shaw, E.; Tebe, C.; Wolkewitz, M.; Wiegand, I.; Grier, S.; Vank, C.; Cuperus, N.; et al. Clinical outcomes of hospitalised patients with catheter-associated urinary tract infection in countries with a high rate of multidrug-resistance: The COMBACTE-MAGNET RESCUING study. Antimicrob. Resist. Infect. Control 2019, 8, 198. [Google Scholar] [CrossRef]
- Köves, B.; Magyar, A.; Tenke, P. Spectrum and antibiotic resistance of catheter-associated urinary tract infections. GMS Infect. Dis. 2017, 5, Doc06. [Google Scholar] [CrossRef]
- Öztürk, R.; Murt, A. Epidemiology of urological infections: A global burden. World J. Urol. 2020, 38, 2669–2679. [Google Scholar] [CrossRef]
- Parida, S.; Mishra, S.K. Urinary tract infections in the critical care unit: A brief review. Indian J. Crit. Care Med. 2013, 17, 370–374. [Google Scholar] [CrossRef]
- Anesi, J.A.; Lautenbach, E.; Nachamkin, I.; Garrigan, C.; Bilker, W.B.; Omorogbe, J.; Dankwa, L.; Wheeler, M.K.; Tolomeo, P.; Han, J.H. Poor clinical outcomes associated with community-onset urinary tract infections due to extended-spectrum cephalosporin-resistant Enterobacteriaceae. Infect. Control. Hosp. Epidemiol. 2018, 39, 1431–1435. [Google Scholar] [CrossRef]
- European Association of Urologists. EAU Guidelines. Edn. Presented at the EAU Annual Congress Amsterdam 2020; European Association of Urologists: Arnhem, The Netherlands, 2020; ISBN 978-94-92671-07-3. [Google Scholar]
- Huttner, A.; Verhaegh, E.M.; Harbarth, S.; Muller, A.E.; Theuretzbacher, U.; Mouton, J.W. Nitrofurantoin revisited: A systematic review and meta-analysis of controlled trials. J. Antimicrob. Chemother. 2015, 70, 2456–2464. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, F.; Allaert, F. Single-dose treatment of cystitis with fosfomycin trometamol (Monuril): Analysis of 15 comparative trials on 2048 patients. Giorn. Ital. Ost. Gin. 1997, 19, 399–404. [Google Scholar]
- Nicolle, L.E. Pivmecillinam in the treatment of urinary tract infections. J. Antimicrob. Chemother. 2000, 46 (Suppl. 1), 35–39. [Google Scholar] [CrossRef]
- Bouchillon, S.K.; Badal, R.E.; Hoban, D.J.; Hawser, S.P. Antimicrobial susceptibility of inpatient urinary tract isolates of gram-negative bacilli in the United States: Results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin. Ther. 2013, 35, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, P.M.; Warren, R.E.; Livermore, D.M.; McNulty, C.A.M.; Enoch, D.A.; Otter, J.A.; Wilson, A.P.R. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: Report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J. Antimicrob. Chemother. 2018, 73, iii2–iii78. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Durante-Mangoni, E.; Andini, R.; Zampino, R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin. Microbiol. Infect. 2019, 25, 943–950. [Google Scholar] [CrossRef]
- Concia, E.; Bragantini, D.; Mazzaferri, F. Clinical evaluation of guidelines and therapeutic approaches in multi drug-resistant urinary tract infections. J. Chemother. 2017, 29, 19–28. [Google Scholar] [CrossRef]
- Amladi, A.U.; Abirami, B.; Devi, S.M.; Sudarsanam, T.D.; Kandasamy, S.; Kekre, N.; Veeraraghavan, B.; Sahni, R.D. Susceptibility profile, resistance mechanisms & efficacy ratios of fosfomycin, nitrofurantoin & colistin for carbapenem-resistant Enterobacteriaceae causing urinary tract infections. Indian J. Med. Res. 2019, 149, 185–191. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Harris, P.N.; Roberts, M.J.; Tambyah, P.A.; Schembri, M.A.; Pezzani, M.D.; Williamson, D.A.; Paterson, D.L. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat. Rev. Urol. 2015, 12, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Wagenlehner, F.; Schmiemann, G. Interdisziplinäre S3 Leitlinie Epidemiologie, Diagnostik, Therapieävention und Management Unkomplizierter, Bakterieller, Ambulanter Erworbener Harnwegsinfektionen bei Erwachsenen Patienten. In Epidemiology, Diagnosis, Treatment Prevention and Management of Uncomplicated, Bacterial, Community-Acquired Urinary Tract Infections in Adult Patients, Langversion 1.1-2 AWMF-Register-Nr. 043/044; AWMF: Düsseldorf, Germany, 2017. [Google Scholar]
- Hof, H. In focus: Pivmecillinam and nitroxoline in the antibiotic treatment. Gynäkologe 2018, 51, 581–589. [Google Scholar] [CrossRef]
- Hof, H.; Bertsch, D.; Passek, D.; Schwarz, R. Nitroxoline—An option for the antibiotic treatment of urinary tract infections. Urologe A 2017, 56, 167–171. [Google Scholar] [CrossRef]
- Sobke, A.; Makarewicz, O.; Baier, M.; Bar, C.; Pfister, W.; Gatermann, S.G.; Pletz, M.W.; Forstner, C. Empirical treatment of lower urinary tract infections in the face of spreading multidrug resistance: In vitro study on the effectiveness of nitroxoline. Int. J. Antimicrob. Agents 2018, 51, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Cherdtrakulkiat, R.; Boonpangrak, S.; Sinthupoom, N.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Derivatives (halogen, nitro and amino) of 8-hydroxyquinoline with highly potent antimicrobial and antioxidant activities. Biochem. Biophys. Rep. 2016, 6, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Molina, J.M.; Llosa, J.; Ventosa, A. In vitro activity of nitroxoline against clinical isolates of Candida species. Mycoses 1991, 34, 323–325. [Google Scholar] [CrossRef]
- Kranz, J.; Schmidt, S.; Lebert, C.; Schneidewind, L.; Mandraka, F.; Kunze, M.; Helbig, S.; Vahlensieck, W.; Naber, K.; Schmiemann, G.; et al. The 2017 Update of the German Clinical Guideline on Epidemiology, Diagnostics, Therapy, Prevention, and Management of Uncomplicated Urinary Tract Infections in Adult Patients. Part II: Therapy and Prevention. Urol. Int. 2018, 100, 271–278. [Google Scholar] [CrossRef]
- Abouelhassan, Y.; Yang, Q.; Yousaf, H.; Nguyen, M.T.; Rolfe, M.; Schultz, G.S.; Huigens, R.W., 3rd. Nitroxoline: A broad-spectrum biofilm-eradicating agent against pathogenic bacteria. Int. J. Antimicrob. Agents 2017, 49, 247–251. [Google Scholar] [CrossRef]
- Wagenlehner, F.M.; Munch, F.; Pilatz, A.; Barmann, B.; Weidner, W.; Wagenlehner, C.M.; Straubinger, M.; Blenk, H.; Pfister, W.; Kresken, M.; et al. Urinary concentrations and antibacterial activities of nitroxoline at 250 milligrams versus trimethoprim at 200 milligrams against uropathogens in healthy volunteers. Antimicrob. Agents Chemother. 2014, 58, 713–721. [Google Scholar] [CrossRef]
- Wijma, R.A.; Huttner, A.; Koch, B.C.P.; Mouton, J.W.; Muller, A.E. Review of the pharmacokinetic properties of nitrofurantoin and nitroxoline. J. Antimicrob. Chemother. 2018, 73, 2916–2926. [Google Scholar] [CrossRef]
- Naber, K.G.; Niggemann, H.; Stein, G. Review of the literature and individual patients’ data meta-analysis on efficacy and tolerance of nitroxoline in the treatment of uncomplicated urinary tract infections. BMC Infect. Dis. 2014, 14, 628. [Google Scholar] [CrossRef]
- Lambert-Zechovsky, N.; Leveque, B.; Bingen, E.; Pillion, G.; Chapelle, J.; Mathieu, H. Clinical study and effect of nitroxoline on fecal flora in children. Pathol. Biol. 1987, 35, 669–672. [Google Scholar]
- Hassan, A.Y.; Lin, J.T.; Ricker, N.; Anany, H. The Age of Phage: Friend or Foe in the New Dawn of Therapeutic and Biocontrol Applications? Pharmaceuticals 2021, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guerout, A.M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Escudero, J.A.; Loot, C.; Nivina, A.; Mazel, D. The Integron: Adaptation on Demand. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Raman, G.; McMullan, B.; Taylor, P.; Mallitt, K.A.; Kennedy, S.E. Multiresistant E. coli urine infections in children: A case-control study. Arch. Dis. Child. 2018, 103, 336–340. [Google Scholar] [CrossRef]
- Kresken, M.; Körber-Irrgang, B. In vitro activity of nitroxoline against Escherichia coli urine isolates from outpatient departments in Germany. Antimicrob. Agents Chemother. 2014, 58, 7019–7020. [Google Scholar] [CrossRef] [PubMed]
- Hof, H.; Juretschke, C. Nitroxoline: An option for the treatment of urinary tract infection with multi-resistant uropathogenic bacteria. Infection 2019, 47, 493–495. [Google Scholar] [CrossRef]
- Pelletier, C.; Prognon, P.; Bourlioux, P. Roles of divalent cations and pH in mechanism of action of nitroxoline against Escherichia coli strains. Antimicrob. Agents Chemother. 1995, 39, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Sobke, A.; Klinger, M.; Hermann, B.; Sachse, S.; Nietzsche, S.; Makarewicz, O.; Keller, P.M.; Pfister, W.; Straube, E. The urinary antibiotic 5-nitro-8-hydroxyquinoline (Nitroxoline) reduces the formation and induces the dispersal of Pseudomonas aeruginosa biofilms by chelation of iron and zinc. Antimicrob. Agents Chemother. 2012, 56, 6021–6025. [Google Scholar] [CrossRef] [PubMed]
- Chitto, M.; Berger, M.; Klotz, L.; Dobrindt, U. Sub-Inhibitory concentrations of SOS-Response inducing antibiotics stimulate integrase expression and excision of pathogenicity islands in uropathogenic Escherichia coli strain 536. Int. J. Med. Microbiol. 2020, 310, 151361. [Google Scholar] [CrossRef]
- Puertolas-Balint, F.; Warsi, O.; Linkevicius, M.; Tang, P.C.; Andersson, D.I. Mutations that increase expression of the EmrAB-TolC efflux pump confer increased resistance to nitroxoline in Escherichia coli. J. Antimicrob. Chemother. 2020, 75, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Crellin, E.; Mansfield, K.E.; Leyrat, C.; Nitsch, D.; Douglas, I.J.; Root, A.; Williamson, E.; Smeeth, L.; Tomlinson, L.A. Trimethoprim use for urinary tract infection and risk of adverse outcomes in older patients: Cohort study. BMJ 2018, 360, k341. [Google Scholar] [CrossRef]
- Bandeira, T.D.J.; Moreira, C.A.; Brilhante, R.S.; Castelo-Branco, D.D.S.; Neto, M.P.; de Cordeiro, A.R.; Rodrigues, T.D.J.; Rocha, M.F.; Sidrim, J.J. In vitro activities of amoxicillin-clavulanate, doxycycline, ceftazidime, imipenem, and trimethoprim-sulfamethoxazole against biofilm of Brazilian strains of Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2013, 57, 5771–5773. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Pitout, J.D.; DeVinney, R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000Research 2017, 6. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Riddell, K.; Scholes, D.; Johnson, J.R.; Sokurenko, E.V. The Uropathogenic Escherichia coli Subclone Sequence Type 131-H30 Is Responsible for Most Antibiotic Prescription Errors at an Urgent Care Clinic. Clin. Infect. Dis. 2019, 68, 781–787. [Google Scholar] [CrossRef]
- Norman, A.; Hansen, L.H.; Sorensen, S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009, 364, 2275–2289. [Google Scholar] [CrossRef]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef]
- Gillings, M.R. DNA as a pollutant: The clinical class 1 integron. Curr. Poll. Rep. 2018, 4, 49–55. [Google Scholar] [CrossRef]
- Labbate, M.; Case, R.J.; Stokes, H.W. The integron/gene cassette system: An active player in bacterial adaptation. Methods Mol. Biol. 2009, 532, 103–125. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Littlejohn, T.G.; Radstrom, P.; Sundstrom, L.; Skold, O.; Swedberg, G.; Skurray, R.A. The 3’ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 1993, 37, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Bass, L.; Liebert, C.A.; Lee, M.D.; Summers, A.O.; White, D.G.; Thayer, S.G.; Maurer, J.J. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 1999, 43, 2925–2929. [Google Scholar] [CrossRef]
- Laroche, E.; Pawlak, B.; Berthe, T.; Skurnik, D.; Petit, F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France). FEMS Microbiol. Ecol. 2009, 68, 118–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leverstein-van Hall, M.A.; Blok, H.E.M.; Donders, A.R.T.; Paauw, A.; Fluit, A.C.; Verhoef, J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 2003, 187, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Nijssen, S.; Florijn, A.; Top, J.; Willems, R.; Fluit, A.; Bonten, M. Unnoticed spread of integron-carrying Enterobacteriaceae in intensive care units. Clin. Infect. Dis. 2005, 41, 1–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, B.; Hu, Y.; Wang, Q.; Yi, Y.; Woo, P.C.; Jing, H.; Zhu, B.; Liu, C.H. Structural diversity of class 1 integrons and their associated gene cassettes in Klebsiella pneumoniae isolates from a hospital in China. PLoS ONE 2013, 8, e75805. [Google Scholar] [CrossRef]
- Fernandez-Lopez, R.; de la Cruz, F. Rebooting the genome: The role of negative feedback in horizontal gene transfer. Mob. Genet. Elem. 2014, 4, 1–6. [Google Scholar] [CrossRef][Green Version]
- Naseer, U.; Sundsfjord, A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 2011, 17, 83–97. [Google Scholar] [CrossRef]
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; de Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. mBio 2016, 7, e02162. [Google Scholar] [CrossRef]
- Beyrouthy, R.; Robin, F.; Hamze, M.; Bonnet, R. IncFIIk plasmid harbouring an amplification of 16S rRNA methyltransferase-encoding gene rmtH associated with mobile element ISCR2. J. Antimicrob. Chemother. 2017, 72, 402–406. [Google Scholar] [CrossRef][Green Version]
- Chen, C.M.; Yu, W.L.; Huang, M.; Liu, J.J.; Chen, I.C.; Chen, H.F.; Wu, L.T. Characterization of IS26-composite transposons and multidrug resistance in conjugative plasmids from Enterobacter cloacae. Microbiol. Immunol. 2015, 59, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hall, R.M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Moran, R.A.; Hall, R.M. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 2014, 5, e01801–e01814. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, L.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirtliff, M.E. Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2018, 51, 145–150. [Google Scholar] [CrossRef]
- Riccobono, E.; Di Pilato, V.; Di Maggio, T.; Revollo, C.; Bartoloni, A.; Pallecchi, L.; Rossolini, G.M. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum beta-lactamase in the Bolivian Chaco region. Antimicrob. Agents Chemother. 2015, 59, 5340–5347. [Google Scholar] [CrossRef]
- Roy Chowdhury, P.; Charles, I.G.; Djordjevic, S.P. A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS ONE 2015, 10, e0115781. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wang, X.M.; Dai, L.; Hua, X.; Dong, Z.; Schwarz, S.; Liu, S. Novel conjugative plasmid from Escherichia coli of swine origin that coharbors the multiresistance gene cfr and the extended-spectrum-beta-lactamase gene blaCTX-M-14b. Antimicrob. Agents Chemother. 2015, 59, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Ginn, A.N.; Dobiasova, H.; Iredell, J.R.; Partridge, S.R. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 2015, 80, 118–126. [Google Scholar] [CrossRef]
- Zurfluh, K.; Poirel, L.; Nordmann, P.; Klumpp, J.; Stephan, R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob. Resist. Infect. Control. 2015, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef]
- Cain, A.K.; Hall, R.M. Evolution of a multiple antibiotic resistance region in IncHI1 plasmids: Reshaping resistance regions in situ. J. Antimicrob. Chemother. 2012, 67, 2848–2853. [Google Scholar] [CrossRef]
- Venturini, C.; Hassan, K.A.; Roy Chowdhury, P.; Paulsen, I.T.; Walker, M.J.; Djordjevic, S.P. Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS ONE 2013, 8, e78862. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, S.; Gilbert, C.; Cordaux, R. Cargo capacity of phages and plasmids and other factors influencing horizontal transfers of prokaryote transposable elements. Mob. Genet. Elem. 2012, 2, 115–118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghosh, H.; Doijad, S.; Bunk, B.; Falgenhauer, L.; Yao, Y.; Sproer, C.; Gentil, K.; Schmiedel, J.; Imirzalioglu, C.; Overmann, J.; et al. Detection of translocatable units in a blaCTX-M-15 extended-spectrum beta-lactamase-producing ST131 Escherichia coli isolate using a hybrid sequencing approach. Int. J. Antimicrob. Agents 2016, 47, 245–247. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Feldman, C.; Osei Sekyere, J.; Maningi, N.E.; Modipane, L.; Essack, S.Y. The Resistome, Mobilome, Virulome and Phylogenomics of Multidrug-Resistant Escherichia coli Clinical Isolates from Pretoria, South Africa. Sci. Rep. 2019, 9, 16457. [Google Scholar] [CrossRef]
- Roy Chowdhury, P.; McKinnon, J.; Liu, M.; Djordjevic, S.P. Multidrug Resistant Uropathogenic Escherichia coli ST405 with a Novel, Composite IS26 Transposon in a Unique Chromosomal Location. Front. Microbiol. 2018, 9, 3212. [Google Scholar] [CrossRef]
- Aldeyab, M.A.; Harbarth, S.; Vernaz, N.; Kearney, M.P.; Scott, M.G.; Darwish Elhajji, F.W.; Aldiab, M.A.; McElnay, J.C. The impact of antibiotic use on the incidence and resistance pattern of extended-spectrum beta-lactamase-producing bacteria in primary and secondary healthcare settings. Br. J. Clin. Pharmacol. 2012, 74, 171–179. [Google Scholar] [CrossRef]
- Strand, L.; Jenkins, A.; Henriksen, I.H.; Allum, A.G.; Grude, N.; Kristiansen, B.E. High levels of multiresistance in quinolone resistant urinary tract isolates of Escherichia coli from Norway: A non clonal phenomen? BMC Res. Notes 2014, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Anesi, J.A.; Lautenbach, E.; Nachamkin, I.; Garrigan, C.; Bilker, W.B.; Wheeler, M.; Tolomeo, P.; Han, J.H. Clinical and Molecular Characterization of Community-Onset Urinary Tract Infections Due to Extended-Spectrum Cephalosporin-Resistant Enterobacteriaceae. Infect. Control. Hosp. Epidemiol. 2016, 37, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Mauch, H.; Podbielski, A.; Hermann, M. Mikrobiologisch-Infektiologische Qualitätsstandards (MiQ): Harnwegsinfektionen; Urban & Fischer: München, Germany, 2005. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy in Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: Mobile Element Finder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Cury, J.; Jove, T.; Touchon, M.; Neron, B.; Rocha, E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef] [PubMed]


| E. coli Urine Isolates | Cotrimoxazole Resistant Isolates [%] |
|---|---|
| Susceptible (n = 29,094) | 20 |
| 3rd generation cephalosporin resistant (n = 763) | 42 |
| Ciprofloxacin resistant (n = 4450) | 46 |
| MDR (n = 2624) | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrindt, U.; Wami, H.T.; Schmidt-Wieland, T.; Bertsch, D.; Oberdorfer, K.; Hof, H. Compared with Cotrimoxazole Nitroxoline Seems to Be a Better Option for the Treatment and Prophylaxis of Urinary Tract Infections Caused by Multidrug-Resistant Uropathogens: An In Vitro Study. Antibiotics 2021, 10, 645. https://doi.org/10.3390/antibiotics10060645
Dobrindt U, Wami HT, Schmidt-Wieland T, Bertsch D, Oberdorfer K, Hof H. Compared with Cotrimoxazole Nitroxoline Seems to Be a Better Option for the Treatment and Prophylaxis of Urinary Tract Infections Caused by Multidrug-Resistant Uropathogens: An In Vitro Study. Antibiotics. 2021; 10(6):645. https://doi.org/10.3390/antibiotics10060645
Chicago/Turabian StyleDobrindt, Ulrich, Haleluya T. Wami, Torsten Schmidt-Wieland, Daniela Bertsch, Klaus Oberdorfer, and Herbert Hof. 2021. "Compared with Cotrimoxazole Nitroxoline Seems to Be a Better Option for the Treatment and Prophylaxis of Urinary Tract Infections Caused by Multidrug-Resistant Uropathogens: An In Vitro Study" Antibiotics 10, no. 6: 645. https://doi.org/10.3390/antibiotics10060645
APA StyleDobrindt, U., Wami, H. T., Schmidt-Wieland, T., Bertsch, D., Oberdorfer, K., & Hof, H. (2021). Compared with Cotrimoxazole Nitroxoline Seems to Be a Better Option for the Treatment and Prophylaxis of Urinary Tract Infections Caused by Multidrug-Resistant Uropathogens: An In Vitro Study. Antibiotics, 10(6), 645. https://doi.org/10.3390/antibiotics10060645






