Environmental Spread of Antibiotic Resistance
Abstract
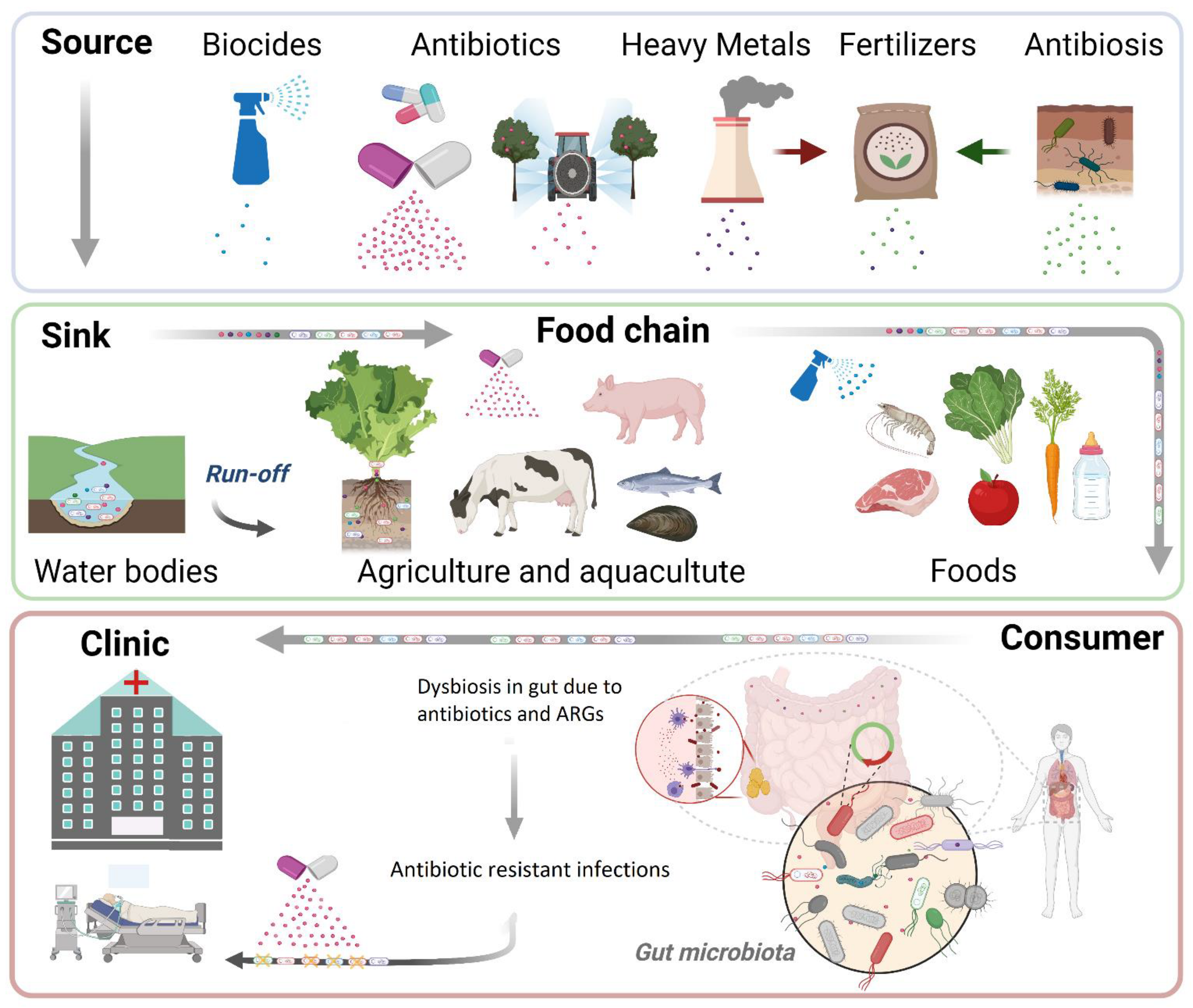
1. Introduction
2. Antibiotics in the Environment
3. Emergence of Antibiotic Resistance in the Environment
4. Co-Selection of ARGs Due to Other Pollutants
5. Transmission of ARGs across the Food Chain
6. Clinical Outcomes
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States, 2013; CDC: Atlanta, GA, USA, 2013. [Google Scholar]
- Spellberg, B.; Barlett, J.G.; Gilbert, D.N. The Future of Antibiotics and Resistance. N. Engl. J. Med. 2013. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Foodborne Antibiotics: Antibiotic Resistance (AR) Solutions Initiative; CDC: Atlanta, GA, USA, 2020. [Google Scholar]
- Liu, C.M.; Stegger, M.; Aziz, M.; Johnson, T.J.; Waits, K.; Nordstrom, L.; Gauld, L.; Weaver, B.; Rolland, D.; Statham, S.; et al. Escherichia Coli ST131-H22 as a Foodborne Uropathogen. MBio 2018, 9. [Google Scholar] [CrossRef]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia Coli O157:H7 Outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.S.; Price, L.B. Recent Research Examining Links among Klebsiella Pneumoniae from Food, Food Animals, and Human Extraintestinal Infections. Curr. Environ. Health Rep. 2016, 3, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kuskowski, M.A.; Smith, K.; O’Bryan, T.T.; Tatini, S. Antimicrobial-Resistant and Extraintestinal Pathogenic Escherichia Coli in Retail Foods. J. Infect. Dis. 2005, 191, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Franklin, A.M.; Williams, C.F.; Watson, J.E. Assessment of Soil to Mitigate Antibiotics in the Environment Due to Release of Wastewater Treatment Plant Effluent. J. Environ. Qual. 2018, 47, 1347–1355. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, Hormones, and Other Organic Wastewater Contaminants in U.S. Streams, 1999–2000: A National Reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar] [CrossRef]
- Faleye, A.C.; Adegoke, A.A.; Ramluckan, K.; Fick, J.; Bux, F.; Stenström, T.A. Concentration and Reduction of Antibiotic Residues in Selected Wastewater Treatment Plants and Receiving Waterbodies in Durban, South Africa. Sci. Total Environ. 2019, 678, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Tolls, J. Sorption of Veterinary Pharmaceuticals in Soils: A Review. Environ. Sci. Technol. 2001, 35, 3397–3406. [Google Scholar] [CrossRef]
- Solomon, E.B.; Yaron, S.; Matthews, K.R. Transmission of Escherichia Coli O157:H7 from Contaminated Manure and Irrigation Water to Lettuce Plant Tissue and Its Subsequent Internalization. Appl. Environ. Microbiol. 2002, 68, 397–400. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Investigation Summary: Factors Potentially Contributing to the Contamination of Romaine Lettuce Implicated in the Fall 2018 Multi-State Outbreak of E. coli O157: H7; US Food and Drug Administration: Silver Spring, MD, USA, 2019. [Google Scholar]
- Wright, G.D. Antibiotic Resistance in the Environment: A Link to the Clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Brandl, M.T. Fitness of Human Enteric Pathogens on Plants and Implications for Food Safety. Annu. Rev. Phytopathol. 2006, 44, 367–392. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Dönhöfer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D.N. Structural Basis for TetM-Mediated Tetracycline Resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 16900–16905. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Cano, M.E.; Velasco, C.; Martínez-Martínez, L.; Pascual, A. Plasmid-Mediated Quinolone Resistance: An Update. J. Infect. Chemother. 2011, 17, 149–182. [Google Scholar] [CrossRef]
- Floss, H.G.; Yu, T.-W. Rifamycin-Mode of Action, Resistance, and Biosynthesis. Chem. Rev. 2005, 105, 621–632. [Google Scholar] [CrossRef]
- Roberts, M.C. Update on Macrolide-Lincosamide-Streptogramin, Ketolide, and Oxazolidinone Resistance Genes. FEMS Microbiol. Lett. 2008, 282, 147–159. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Brauner, A.; Ronin, I.; Balaban, N.Q. Epistasis between Antibiotic Tolerance, Persistence, and Resistance Mutations. Proc. Natl. Acad. Sci. USA 2019, 116, 14734–14739. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Publisher Correction: Definitions and Guidelines for Research on Antibiotic Persistence. Nat. Rev. Microbiol. 2019, 17, 460. [Google Scholar] [CrossRef]
- Van den Bergh, B.; Michiels, J.E.; Wenseleers, T.; Windels, E.M.; Boer, P.V.; Kestemont, D.; De Meester, L.; Verstrepen, K.J.; Verstraeten, N.; Fauvart, M.; et al. Frequency of Antibiotic Application Drives Rapid Evolutionary Adaptation of Escherichia Coli Persistence. Nat. Microbiol. 2016, 1, 16020. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D.; Poinar, H. Antibiotic Resistance Is Ancient: Implications for Drug Discovery. Trends Microbiol. 2012, 20, 157–159. [Google Scholar] [CrossRef]
- Clardy, J.; Fischbach, M.A.; Currie, C.R. The Natural History of Antibiotics. Curr. Biol. 2009, 19, R437–R441. [Google Scholar] [CrossRef] [PubMed]
- Nasfi, Z.; Busch, H.; Kehraus, S.; Linares-Otoya, L.; König, G.M.; Schäberle, T.F.; Bachoual, R. Soil Bacteria Isolated From Tunisian Arid Areas Show Promising Antimicrobial Activities Against Gram-Negatives. Front. Microbiol. 2018, 9, 2742. [Google Scholar] [CrossRef]
- Mast, Y.; Stegmann, E. Actinomycetes: The Antibiotics Producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a New Resource for Drug Discovery: Marine Actinomycete Bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Kwon, H.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Marinomycins A-D, Antitumor-Antibiotics of a New Structure Class from a Marine Actinomycete of the Recently Discovered Genus “Marinispora”. J. Am. Chem. Soc. 2006, 128, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Hatosy, S.M.; Martiny, A.C. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.; Stedt, J.; Bonnedahl, J.; Molin, Y.; Drobni, M.; Calisto-Ulloa, N.; Gomez-Fuentes, C.; Astorga-España, M.S.; González-Acuña, D.; Waldenström, J.; et al. Human-Associated Extended-Spectrum β-Lactamase in the Antarctic. Appl. Environ. Microbiol. 2012, 78, 2056–2058. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.A.; Gerwick, L. Marine Natural Product Drug Discovery: Leads for Treatment of Inflammation, Cancer, Infections, and Neurological Disorders. Immunopharmacol. Immunotoxicol. 2010, 32, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Cueto, M.; Jensen, P.R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. Pestalone, a New Antibiotic Produced by a Marine Fungus in Response to Bacterial Challenge. J. Nat. Prod. 2001, 64, 1444–1446. [Google Scholar] [CrossRef]
- FDA. Antimicrobials Sold or Distributed for Use in Food-Producing Animals; Food and Drug Administration: Silver Spring, MD, USA, 2016. [Google Scholar]
- Williams-Nguyen, J.; Sallach, J.B.; Bartelt-Hunt, S.; Boxall, A.B.; Durso, L.M.; McLain, J.E.; Singer, R.S.; Snow, D.D.; Zilles, J.L. Antibiotics and Antibiotic Resistance in Agroecosystems: State of the Science. J. Environ. Qual. 2016, 45, 394–406. [Google Scholar] [CrossRef] [PubMed]
- O’neill, C.B.Y.J. The Review on Antimicrobial Resistance. Available online: https://www.wipo.int/edocs/mdocs/mdocs/en/wipo_who_wto_ip_ge_16/wipo_who_wto_ip_ge_16_www_356156.pdf (accessed on 2 February 2020).
- Andersson, D.I.; Hughes, D. Microbiological Effects of Sublethal Levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Kaur, S. Molecular Approaches towards Development of Novel Bacillus Thuringiensis Biopesticides. World J. Microbiol. Biotechnol. 2000, 16, 781–793. [Google Scholar] [CrossRef]
- Fillinger, U.; Lindsay, S.W. Suppression of Exposure to Malaria Vectors by an Order of Magnitude Using Microbial Larvicides in Rural Kenya. Trop. Med. Int. Health 2006, 11, 1629–1642. [Google Scholar] [CrossRef]
- Boyce, R.; Lenhart, A.; Kroeger, A.; Velayudhan, R.; Roberts, B.; Horstick, O. Bacillus Thuringiensis Israelensis (Bti) for the Control of Dengue Vectors: Systematic Literature Review. Trop. Med. Int. Health 2013, 18, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-H.; Koumoutsi, A.; Scholz, R.; Borriss, R. More than Anticipated—Production of Antibiotics and Other Secondary Metabolites by Bacillus Amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 2009, 16, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulou, A.; Theologidis, I.; Liebmann, B.; Kalantidis, K.; Vassilakos, N.; Skandalis, N. Bacillus Amyloliquefaciens MBI600 Differentially Induces Tomato Defense Signaling Pathways Depending on Plant Part and Dose of Application. Sci. Rep. 2019, 9, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Publications Office of the European Union. State of the Art on the Contribution of Water to Antimicrobial Resistance; Publications Office of the European Union: Luxembourg, 2018; ISBN 9789279984785. [Google Scholar]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel Soil Bacteria Possess Diverse Genes for Secondary Metabolite Biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, R.; Hu, J.; Xing, S.; Huang, C.; Mi, J.; Liao, X. Dominant Denitrifying Bacteria Are Important Hosts of Antibiotic Resistance Genes in Pig Farm Anoxic-Oxic Wastewater Treatment Processes. Environ. Int. 2020, 143, 105897. [Google Scholar] [CrossRef]
- Luczkiewicz, A.; Kotlarska, E.; Artichowicz, W.; Tarasewicz, K.; Fudala-Ksiazek, S. Antimicrobial Resistance of Pseudomonas Spp. Isolated from Wastewater and Wastewater-Impacted Marine Coastal Zone. Environ. Sci. Pollut. Res. Int. 2015, 22, 19823–19834. [Google Scholar] [CrossRef]
- Molecular Characterization of Clinical and Environmental Pseudomonas Aeruginosa Isolated in a Burn Center. Saudi J. Biol. Sci. 2019, 26, 1731–1736. [CrossRef] [PubMed]
- Monitoring of Indicator and Multidrug Resistant Bacteria in Agricultural Soils under Different Irrigation Patterns. Agric. Water Manag. 2017, 184, 19–27. [CrossRef]
- Dantas, G.; Sommer, M.O.A.; Oluwasegun, R.D.; Church, G.M. Bacteria Subsisting on Antibiotics. Science 2008, 320, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Maeusli, M.; Lee, B.; Miller, S.; Reyna, Z.; Lu, P.; Yan, J.; Ulhaq, A.; Skandalis, N.; Spellberg, B.; Luna, B. Horizontal Gene Transfer of Antibiotic Resistance from Acinetobacter Baylyi to Escherichia Coli on Lettuce and Subsequent Antibiotic Resistance Transmission to the Gut Microbiome. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Port, J.A.; Wallace, J.C.; Griffith, W.C.; Faustman, E.M. Metagenomic Profiling of Microbial Composition and Antibiotic Resistance Determinants in Puget Sound. PLoS ONE 2012, 7, e48000. [Google Scholar]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial Pharmaceuticals in the Aquatic Environment—Occurrence and Environmental Implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, C.; Parker, D.B.; Snow, D.D.; Zhou, Z.; Li, X. Occurrence of Antimicrobials and Antimicrobial Resistance Genes in Beef Cattle Storage Ponds and Swine Treatment Lagoons. Sci. Total Environ. 2013, 463–464, 631–638. [Google Scholar] [CrossRef]
- Martiny, A.C.; Martiny, J.B.H.; Weihe, C.; Field, A.; Ellis, J.C. Functional Metagenomics Reveals Previously Unrecognized Diversity of Antibiotic Resistance Genes in Gulls. Front. Microbiol. 2011, 2, 238. [Google Scholar] [CrossRef]
- Foti, M.; Giacopello, C.; Bottari, T.; Fisichella, V.; Rinaldo, D.; Mammina, C. Antibiotic Resistance of Gram Negatives Isolates from Loggerhead Sea Turtles (Caretta Caretta) in the Central Mediterranean Sea. Mar. Pollut. Bull. 2009, 58, 1363–1366. [Google Scholar] [CrossRef]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; González-Rocha, G. Antibiotic Resistance in Bacterial Isolates from Freshwater Samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef] [PubMed]
- Rabbia, V.; Bello-Toledo, H.; Jiménez, S.; Quezada, M.; Domínguez, M.; Vergara, L.; Gómez-Fuentes, C.; Calisto-Ulloa, N.; González-Acuña, D.; López, J.; et al. Antibiotic Resistance in Escherichia Coli Strains Isolated from Antarctic Bird Feces, Water from inside a Wastewater Treatment Plant, and Seawater Samples Collected in the Antarctic Treaty Area. Polar Sci. 2016, 10, 123–131. [Google Scholar] [CrossRef]
- Miller, R.V.; Gammon, K.; Day, M.J. Antibiotic Resistance among Bacteria Isolated from Seawater and Penguin Fecal Samples Collected near Palmer Station, Antarctica. Can. J. Microbiol. 2009, 55, 37–45. [Google Scholar] [CrossRef]
- Dafforn, K.A.; Lewis, J.A.; Johnston, E.L. Antifouling Strategies: History and Regulation, Ecological Impacts and Mitigation. Mar. Pollut. Bull. 2011, 62, 453–465. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Z.; van der Kuijp, T.J.; Yuan, Z.; Huang, L. A Review of Soil Heavy Metal Pollution from Mines in China: Pollution and Health Risk Assessment. Sci. Total Environ. 2014, 468, 843–853. [Google Scholar] [CrossRef]
- Chowdhury, S.; Mazumder, M.A.J.; Al-Attas, O.; Husain, T. Heavy Metals in Drinking Water: Occurrences, Implications, and Future Needs in Developing Countries. Sci. Total Environ. 2016, 569–570, 476–488. [Google Scholar] [CrossRef]
- Subcommittee on Economic and Consumer Policy, Committee on Oversight and Reform. Baby Foods Are Tainted with Dangerous Levels of Arsenic, Lead, Cadmium, and Mercury; U.S. House of Representatives: Washington, DC, USA, 2021. [Google Scholar]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy Metal Toxicity and the Environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar]
- Nies, D.H. Microbial Heavy-Metal Resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy Metal Driven Co-Selection of Antibiotic Resistance in Soil and Water Bodies Impacted by Agriculture and Aquaculture. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef]
- Li, L.G.; Xia, Y.; Zhang, T. Co-Occurrence of Antibiotic and Metal Resistance Genes Revealed in Complete Genome Collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res. 2014, 42, 737–743. [Google Scholar] [CrossRef]
- Wales, A.D.; Davies, R.H. Co-Selection of Resistance to Antibiotics, Biocides and Heavy Metals, and Its Relevance to Foodborne Pathogens. Antibiotics 2015, 4, 567–604. [Google Scholar] [CrossRef] [PubMed]
- Scientific Committee on Emerging and Newly Identified Health Risks Assessment of the Antibiotic Resistance Effects of Biocides. Available online: https://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf (accessed on 20 March 2021).
- Maillard, J.-Y. Resistance of Bacteria to Biocides. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Paul, D.; Mondal, S.K.; Mandal, S.M. Biologia Futura: Use of Biocides during COVID-19-Global Reshuffling of the Microbiota. Biol. Futur. 2021. [Google Scholar] [CrossRef]
- Kumar, K.S.; Priya, S.M.; Peck, A.M.; Sajwan, K.S. Mass Loadings of Triclosan and Triclocarbon from Four Wastewater Treatment Plants to Three Rivers and Landfill in Savannah, Georgia, USA. Arch. Environ. Contam. Toxicol. 2010, 58, 275–285. [Google Scholar] [CrossRef]
- Juksu, K.; Zhao, J.-L.; Liu, Y.-S.; Yao, L.; Sarin, C.; Sreesai, S.; Klomjek, P.; Jiang, Y.-X.; Ying, G.-G. Occurrence, Fate and Risk Assessment of Biocides in Wastewater Treatment Plants and Aquatic Environments in Thailand. Sci. Total Environ. 2019, 690, 1110–1119. [Google Scholar] [CrossRef]
- Gnanadhas, D.P.; Marathe, S.A.; Chakravortty, D. Biocides--Resistance, Cross-Resistance Mechanisms and Assessment. Expert Opin. Investig. Drugs 2013, 22, 191–206. [Google Scholar] [CrossRef]
- McMurry, L.M.; McDermott, P.F.; Levy, S.B. Genetic Evidence That InhA of Mycobacterium Smegmatis Is a Target for Triclosan. Antimicrob. Agents Chemother. 1999, 43, 711–713. [Google Scholar] [CrossRef]
- Heath, R.J.; Li, J.; Roland, G.E.; Rock, C.O. Inhibition of the Staphylococcus Aureus NADPH-Dependent Enoyl-Acyl Carrier Protein Reductase by Triclosan and Hexachlorophene. J. Biol. Chem. 2000, 275, 4654–4659. [Google Scholar] [CrossRef] [PubMed]
- Tattawasart, U.; Maillard, J.Y.; Furr, J.R.; Russell, A.D. Development of Resistance to Chlorhexidine Diacetate and Cetylpyridinium Chloride in Pseudomonas Stutzeri and Changes in Antibiotic Susceptibility. J. Hosp. Infect. 1999, 42, 219–229. [Google Scholar] [CrossRef]
- Winder, C.L.; Al-Adham, I.S.; Abdel Malek, S.M.; Buultjens, T.E.; Horrocks, A.J.; Collier, P.J. Outer Membrane Protein Shifts in Biocide-Resistant Pseudomonas Aeruginosa PAO1. J. Appl. Microbiol. 2000, 89, 289–295. [Google Scholar] [CrossRef]
- Rouch, D.A.; Cram, D.S.; DiBerardino, D.; Littlejohn, T.G.; Skurray, R.A. Efflux-Mediated Antiseptic Resistance Gene qacA from Staphylococcus Aureus: Common Ancestry with Tetracycline- and Sugar-Transport Proteins. Mol. Microbiol. 1990, 4, 2051–2062. [Google Scholar] [CrossRef]
- Heir, E.; Sundheim, G.; Holck, A.L. The Staphylococcus qacH Gene Product: A New Member of the SMR Family Encoding Multidrug Resistance. FEMS Microbiol. Lett. 1998, 163, 49–56. [Google Scholar] [CrossRef]
- Nishihara, T.; Okamoto, T.; Nishiyama, N. Biodegradation of Didecyldimethylammonium Chloride by Pseudomonas Fluorescens TN4 Isolated from Activated Sludge. J. Appl. Microbiol. 2000, 88, 641–647. [Google Scholar] [CrossRef]
- Meade, M.J.; Waddell, R.L.; Callahan, T.M. Soil Bacteria Pseudomonas Putida and Alcaligenes Xylosoxidans Subsp. Denitrificans Inactivate Triclosan in Liquid and Solid Substrates. FEMS Microbiol. Lett. 2001, 204, 45–48. [Google Scholar] [CrossRef]
- Kampf, G. Biocidal Agents Used for Disinfection Can Enhance Antibiotic Resistance in Gram-Negative Species. Antibiotics 2018, 7, 110. [Google Scholar] [CrossRef]
- Tandukar, M.; Oh, S.; Tezel, U.; Konstantinidis, K.T.; Pavlostathis, S.G. Long-Term Exposure to Benzalkonium Chloride Disinfectants Results in Change of Microbial Community Structure and Increased Antimicrobial Resistance. Environ. Sci. Technol. 2013, 47, 9730–9738. [Google Scholar] [CrossRef]
- Morita, Y.; Murata, T.; Mima, T.; Shiota, S.; Kuroda, T.; Mizushima, T.; Gotoh, N.; Nishino, T.; Tsuchiya, T. Induction of mexCD-oprJ Operon for a Multidrug Efflux Pump by Disinfectants in Wild-Type Pseudomonas Aeruginosa PAO1. J. Antimicrob. Chemother. 2003, 51, 991–994. [Google Scholar] [CrossRef]
- Shigemura, K.; Osawa, K.; Kato, A.; Tokimatsu, I.; Arakawa, S.; Shirakawa, T.; Fujisawa, M. Association of Overexpression of Efflux Pump Genes with Antibiotic Resistance in Pseudomonas Aeruginosa Strains Clinically Isolated from Urinary Tract Infection Patients. J. Antibiot. 2015, 68, 568–572. [Google Scholar] [CrossRef]
- Jutkina, J.; Marathe, N.P.; Flach, C.-F.; Larsson, D.G.J. Antibiotics and Common Antibacterial Biocides Stimulate Horizontal Transfer of Resistance at Low Concentrations. Sci. Total Environ. 2018, 616, 172–178. [Google Scholar] [CrossRef]
- Zeng, W.; Xu, W.; Xu, Y.; Liao, W.; Zhao, Y.; Zheng, X.; Xu, C.; Zhou, T.; Cao, J. The Prevalence and Mechanism of Triclosan Resistance in Escherichia Coli Isolated from Urine Samples in Wenzhou, China. Antimicrob. Resist. Infect. Control. 2020, 9, 1–10. [Google Scholar] [CrossRef]
- Potenski, C.J.; Gandhi, M.; Matthews, K.R. Exposure of Salmonella Enteritidis to Chlorine or Food Preservatives Increases Susceptibility to Antibiotics. FEMS Microbiol. Lett. 2003, 220, 181–186. [Google Scholar] [CrossRef]
- Hardy, K.; Sunnucks, K.; Gil, H.; Shabir, S.; Trampari, E.; Hawkey, P.; Webber, M. Increased Usage of Antiseptics Is Associated with Reduced Susceptibility in Clinical Isolates of Staphylococcus Aureus. MBio 2018, 9. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic Resistance from the Farm to the Table. Available online: https://www.cdc.gov/foodsafety/pdfs/ar-infographic-508c.pdf (accessed on 24 August 2017).
- Spellberg, B.; Hansen, G.R.; Kar, A.; Cordova, C.D.; Price, L.B.; Johnson, J.R. Antibiotic Resistance in Humans and Animals. NAM Perspect. 2016. Discussion paper 6. [Google Scholar] [CrossRef]
- Nadimpalli, M.; Delarocque-Astagneau, E.; Love, D.C.; Price, L.B.; Huynh, B.-T.; Collard, J.-M.; Lay, K.S.; Borand, L.; Ndir, A.; Walsh, T.R.; et al. Combating Global Antibiotic Resistance: Emerging One Health Concerns in Lower- and Middle-Income Countries. Clin. Infect. Dis. 2018, 66, 963–969. [Google Scholar] [CrossRef]
- Food and Drug Administration. Food and Drug Administration 2018 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; Food and Drug Administration: Silver Spring, MD, USA, 2018. [Google Scholar]
- Neyra, R.C.; Frisancho, J.A.; Rinsky, J.L.; Resnick, C.; Carroll, K.C.; Rule, A.M.; Ross, T.; You, Y.; Price, L.B.; Silbergeld, E.K. Multidrug-Resistant and Methicillin-Resistant Staphylococcus Aureus (MRSA) in Hog Slaughter and Processing Plant Workers and Their Community in North Carolina (USA). Environ. Health Perspect. 2014, 122, 471–477. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Tschudin-Sutter, S.; Frei, R.; Stephan, R.; Hächler, H.; Nogarth, D.; Widmer, A.F. Extended-Spectrum β-Lactamase (ESBL)-Producing Enterobacteriaceae: A Threat from the Kitchen. Infect. Control. Hosp. Epidemiol. 2014, 35, 581–584. [Google Scholar] [CrossRef]
- McMahon, M.A.S.; Xu, J.; Moore, J.E.; Blair, I.S.; McDowell, D.A. Environmental Stress and Antibiotic Resistance in Food-Related Pathogens. Appl. Environ. Microbiol. 2007, 73, 211–217. [Google Scholar] [CrossRef]
- Srinivasan, V.; Nguyen, L.T.; Headrick, S.I.; Murinda, S.E.; Oliver, S.P. Antimicrobial Resistance Patterns of Shiga Toxin-Producing Escherichia Coli O157:H7 and O157:H7- from Different Origins. Microb. Drug Resist. 2007, 13, 44–51. [Google Scholar] [CrossRef]
- Mayrhofer, S.; Paulsen, P.; Smulders, F.J.M.; Hilbert, F. Antimicrobial Resistance in Commensal Escherichia Coli Isolated from Muscle Foods as Related to the Veterinary Use of Antimicrobial Agents in Food-Producing Animals in Austria. Microb. Drug Resist. 2006, 12, 278–283. [Google Scholar] [CrossRef]
- Rahman, M.M.; Husna, A.; Elshabrawy, H.A.; Alam, J.; Runa, N.Y.; Badruzzaman, A.T.M.; Banu, N.A.; Al Mamun, M.; Paul, B.; Das, S.; et al. Isolation and Molecular Characterization of Multidrug-Resistant Escherichia Coli from Chicken Meat. Sci. Rep. 2020, 10, 21999. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. 2018 The State of World Fisheries and Aquaculture: Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018; ISBN 9789251305621. [Google Scholar]
- Lewis, G.; Wang, B.; Shafiei Jahani, P.; Hurrell, B.P.; Banie, H.; Aleman Muench, G.R.; Maazi, H.; Helou, D.G.; Howard, E.; Galle-Treger, L.; et al. Dietary Fiber-Induced Microbial Short Chain Fatty Acids Suppress ILC2-Dependent Airway Inflammation. Front. Immunol. 2019, 10, 2051. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet Another Environmental Gateway to the Development and Globalisation of Antimicrobial Resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef]
- Ryu, S.-H.; Park, S.-G.; Choi, S.-M.; Hwang, Y.-O.; Ham, H.-J.; Kim, S.-U.; Lee, Y.-K.; Kim, M.-S.; Park, G.-Y.; Kim, K.-S.; et al. Antimicrobial Resistance and Resistance Genes in Escherichia Coli Strains Isolated from Commercial Fish and Seafood. Int. J. Food Microbiol. 2012, 152, 14–18. [Google Scholar] [CrossRef]
- Furushita, M.; Shiba, T.; Maeda, T.; Yahata, M.; Kaneoka, A.; Takahashi, Y.; Torii, K.; Hasegawa, T.; Ohta, M. Similarity of Tetracycline Resistance Genes Isolated from Fish Farm Bacteria to Those from Clinical Isolates. Appl. Environ. Microbiol. 2003, 69, 5336–5342. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jin, L.; Sun, F.; Hu, Q.; Chen, L. Antibiotic and Heavy-Metal Resistance of Vibrio Parahaemolyticus Isolated from Fresh Shrimps in Shanghai Fish Markets, China. Environ. Sci. Pollut. Res. Int. 2016, 23, 15033–15040. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial Resistance in Aquaculture: Current Knowledge and Alternatives to Tackle the Problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic/Antimicrobial Resistance (AR/AMR). Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 11 November 2020).
- Carvalheira, A.; Silva, J.; Teixeira, P. Lettuce and Fruits as a Source of Multidrug Resistant Acinetobacter spp. Food Microbiol. 2017, 64, 119–125. [Google Scholar] [CrossRef]
- Bezanson, G.S.; MacInnis, R.; Potter, G.; Hughes, T. Presence and Potential for Horizontal Transfer of Antibiotic Resistance in Oxidase-Positive Bacteria Populating Raw Salad Vegetables. Int. J. Food Microbiol. 2008, 127, 37–42. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the Role of Vegetables in Spreading Antimicrobial-Resistant Bacteria: A Need for Quantitative Risk Assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- O’Flaherty, E.; Solimini, A.G.; Pantanella, F.; De Giusti, M.; Cummins, E. Human Exposure to Antibiotic Resistant-Escherichia Coli through Irrigated Lettuce. Environ. Int. 2019, 122, 270–280. [Google Scholar] [CrossRef]
- Munther, D.S.; Carter, M.Q.; Aldric, C.V.; Ivanek, R.; Brandl, M.T. Formation of Escherichia Coli O157:H7 Persister Cells in the Lettuce Phyllosphere and Application of Differential Equation Models To Predict Their Prevalence on Lettuce Plants in the Field. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella Persisters Promote the Spread of Antibiotic Resistance Plasmids in the Gut. Nature 2019, 573, 270–280. [Google Scholar] [CrossRef]
- Sundin, G.W.; Wang, N. Antibiotic Resistance in Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 2018, 56, 161–180. [Google Scholar] [CrossRef]
- Catara, V. Pseudomonas Corrugata: Plant Pathogen and/or Biological Resource? Mol. Plant. Pathol. 2007, 8, 233–244. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards, USA. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement; CLSI: Annapolis Junction, MD, USA, 2013. [Google Scholar]
- Pérombelon, M.C.M. Potato Diseases Caused by Soft Rot Erwinias: An Overview of Pathogenesis. Plant. Pathol. 2002, 51, 1–12. [Google Scholar] [CrossRef]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus Thuringiensis and Its Pesticidal Crystal Proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards, USA. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. Available online: https://clsi.org/standards/products/microbiology/documents/m45/ (accessed on 2 September 2020).
- Varympopi, A.; Dimopoulou, A.; Theologidis, I.; Karamanidou, T.; Kaldeli Kerou, A.; Vlachou, A.; Karfaridis, D.; Papafotis, D.; Hatzinikolaou, D.G.; Tsouknidas, A.; et al. Bactericides Based on Copper Nanoparticles Restrain Growth of Important Plant Pathogens. Pathogens 2020, 9, 1024. [Google Scholar] [CrossRef]
- Skandalis, N.; Dimopoulou, A.; Georgopoulou, A.; Gallios, N.; Papadopoulos, D.; Tsipas, D.; Theologidis, I.; Michailidis, N.; Chatzinikolaidou, M. The Effect of Silver Nanoparticles Size, Produced Using Plant Extract from Arbutus Unedo, on Their Antibacterial Efficacy. Nanomaterials 2017, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Gurnee, E.A.; Ndao, I.M.; Johnson, J.R.; Johnston, B.D.; Gonzalez, M.D.; Burnham, C.-A.D.; Hall-Moore, C.M.; McGhee, J.E.; Mellmann, A.; Warner, B.B.; et al. Gut Colonization of Healthy Children and Their Mothers With Pathogenic Ciprofloxacin-Resistant Escherichia coli. J. Infect. Dis. 2015, 212, 1862–1868. [Google Scholar] [CrossRef]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Stadler, T.; Meinel, D.; Aguilar-Bultet, L.; Huisman, J.S.; Schindler, R.; Egli, A.; Seth-Smith, H.M.B.; Eichenberger, L.; Brodmann, P.; Hübner, P.; et al. Transmission of ESBL-Producing Enterobacteriaceae and Their Mobile Genetic Elements-Identification of Sources by Whole Genome Sequencing: Study Protocol for an Observational Study in Switzerland. BMJ Open 2018, 8, e021823. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet Infect. Dis. 2018. [Google Scholar] [CrossRef]
- Weingarten, R.A.; Johnson, R.C.; Conlan, S.; Ramsburg, A.M.; Dekker, J.P.; Lau, A.F.; Khil, P.; Odom, R.T.; Deming, C.; Park, M.; et al. Genomic Analysis of Hospital Plumbing Reveals Diverse Reservoir of Bacterial Plasmids Conferring Carbapenem Resistance. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, A.W.; Potter, R.F.; Wallace, M.; Shupe, A.; Patel, S.; Sun, X.; Gul, D.; Kwon, J.H.; Andleeb, S.; Burnham, C.-A.D.; et al. Spatiotemporal Dynamics of Multidrug Resistant Bacteria on Intensive Care Unit Surfaces. Nat. Commun. 2019, 10, 4569. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Johani, K.; Gosbell, I.B.; Jacombs, A.S.W.; Almatroudi, A.; Whiteley, G.S.; Deva, A.K.; Jensen, S.; Vickery, K. Intensive Care Unit Environmental Surfaces Are Contaminated by Multidrug-Resistant Bacteria in Biofilms: Combined Results of Conventional Culture, Pyrosequencing, Scanning Electron Microscopy, and Confocal Laser Microscopy. J. Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef]
- Muloi, D.; Ward, M.J.; Pedersen, A.B.; Fèvre, E.M.; Woolhouse, M.E.J.; van Bunnik, B.A.D. Are Food Animals Responsible for Transfer of Antimicrobial-Resistant Escherichia Coli or Their Resistance Determinants to Human Populations? A Systematic Review. Foodborne Pathog. Dis. 2018, 15, 467–474. [Google Scholar] [CrossRef]
- Peter, S.; Bosio, M.; Gross, C.; Bezdan, D.; Gutierrez, J.; Oberhettinger, P.; Liese, J.; Vogel, W.; Dörfel, D.; Berger, L.; et al. Tracking of Antibiotic Resistance Transfer and Rapid Plasmid Evolution in a Hospital Setting by Nanopore Sequencing. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Evans, D.R.; Griffith, M.P.; Sundermann, A.J.; Shutt, K.A.; Saul, M.I.; Mustapha, M.M.; Marsh, J.W.; Cooper, V.S.; Harrison, L.H.; Van Tyne, D. Systematic Detection of Horizontal Gene Transfer across Genera among Multidrug-Resistant Bacteria in a Single Hospital. eLife 2020, 9, e53886. [Google Scholar] [CrossRef] [PubMed]
- Hurdle, J.G.; O’Neill, A.J.; Mody, L.; Chopra, I.; Bradley, S.F. In Vivo Transfer of High-Level Mupirocin Resistance from Staphylococcus Epidermidis to Methicillin-Resistant Staphylococcus Aureus Associated with Failure of Mupirocin Prophylaxis. J. Antimicrob. Chemother. 2005, 56, 1166–1168. [Google Scholar] [CrossRef]
- Conlan, S.; Lau, A.F.; Deming, C.; Spalding, C.D.; Lee-Lin, S.; Thomas, P.J.; Park, M.; Dekker, J.P.; Frank, K.M.; Palmore, T.N.; et al. Plasmid Dissemination and Selection of a Multidrug-Resistant Klebsiella Pneumoniae Strain during Transplant-Associated Antibiotic Therapy. MBio 2019, 10. [Google Scholar] [CrossRef]
- Conlan, S.; Park, M.; Deming, C.; Thomas, P.J.; Young, A.C.; Coleman, H.; Sison, C.; Weingarten, R.A.; Lau, A.F.; NISC Comparative Sequencing Program. Plasmid Dynamics in KPC-Positive Klebsiella Pneumoniae during Long-Term Patient Colonization. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- León-Sampedro, R.; DelaFuente, J.; Díaz-Agero, C.; Crellen, T.; Musicha, P.; Rodríguez-Beltrán, J.; de la Vega, C.; Hernández-García, M.; López-Fresneña, N.; R-GNOSIS WP5 Study Group. Pervasive Transmission of a Carbapenem Resistance Plasmid in the Gut Microbiota of Hospitalized Patients. Nat. Microbiol. 2021, 6, 606–616. [Google Scholar] [CrossRef] [PubMed]
| Pseudomonas corrugata | |||||||||||||||
| Antibiotic | AMP | PEN | FEP | VAN | FOF | ERY | CLI | GEN | MEM | TET | PMB | CHL | CIP | RIF | LCM |
| MIC (mg/L) | >32 | >64 | 32 | >64 | 256 | >64 | >8 | 16 | 8 | 1 | >16 | 32 | <0.5 | 32 | >32 |
| Breakpoint | R1 ≥ 32 ≤ R2 | R1 ≥ 32 ≤ R2 | I1 = 8 ≤ R2 | S1 ≤ 4 | R2 ≥ 4 | R1 ≥ 32 | S1 > 0.5 = S2 | ||||||||
| Pectobacterium carotovorum sbsp. carotovorum | |||||||||||||||
| Antibiotic | AMP | PEN | FEP | VAN | FOF | ERY | CLI | GEN | MEM | TET | PMB | CHL | CIP | RIF | LCM |
| MIC (mg/L) | >32 | >64 | 8 | >32 | 128 | 32 | >8 | 32 | <1 | 1 | <1 | 128 | <0.5 | 4 | >32 |
| Breakpoint | R ≥ 32 | R ≥ 16 | R ≥ 256 | R ≥ 16 | R ≥ 4 | R ≥ 16 | R ≥ 4 | R ≥ 32 | R ≥ 1 | ||||||
| Bacillus thuringiensis sbsp. kurstaki | |||||||||||||||
| Antibiotic | AMP | PEN | FEP | VAN | FOF | ERY | CLI | GEN | MEM | TET | PMB | CHL | CIP | RIF | LCM |
| MIC (mg/L) | 32 | 16 | >64 | <4 | 64 | >8 | 1 | <2 | <1 | <2 | >16 | <4 | <0.5 | <0.5 | 16 |
| Breakpoint | R ≥ 0.5 | R ≥ 0.25 | S ≤ 4 | R ≥ 8 | R ≥ 4 | R ≥ 16 | R ≥ 16 | R ≥ 16 | R ≥ 32 | R ≥ 4 | R ≥ 4 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skandalis, N.; Maeusli, M.; Papafotis, D.; Miller, S.; Lee, B.; Theologidis, I.; Luna, B. Environmental Spread of Antibiotic Resistance. Antibiotics 2021, 10, 640. https://doi.org/10.3390/antibiotics10060640
Skandalis N, Maeusli M, Papafotis D, Miller S, Lee B, Theologidis I, Luna B. Environmental Spread of Antibiotic Resistance. Antibiotics. 2021; 10(6):640. https://doi.org/10.3390/antibiotics10060640
Chicago/Turabian StyleSkandalis, Nicholas, Marlène Maeusli, Dimitris Papafotis, Sarah Miller, Bosul Lee, Ioannis Theologidis, and Brian Luna. 2021. "Environmental Spread of Antibiotic Resistance" Antibiotics 10, no. 6: 640. https://doi.org/10.3390/antibiotics10060640
APA StyleSkandalis, N., Maeusli, M., Papafotis, D., Miller, S., Lee, B., Theologidis, I., & Luna, B. (2021). Environmental Spread of Antibiotic Resistance. Antibiotics, 10(6), 640. https://doi.org/10.3390/antibiotics10060640






