Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture
Abstract
1. Introduction
2. Results
2.1. Clinical Signs and Gross Lesions
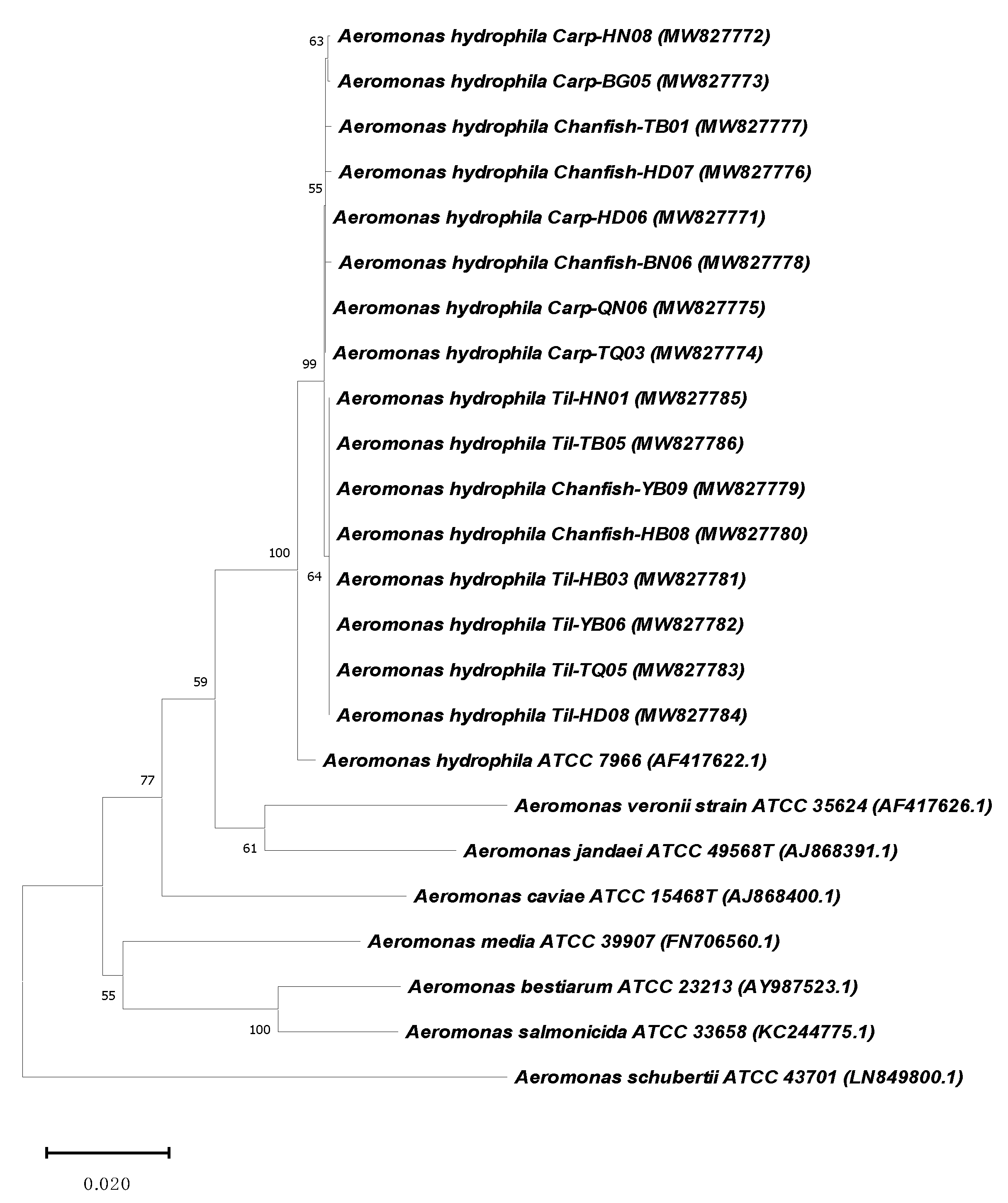
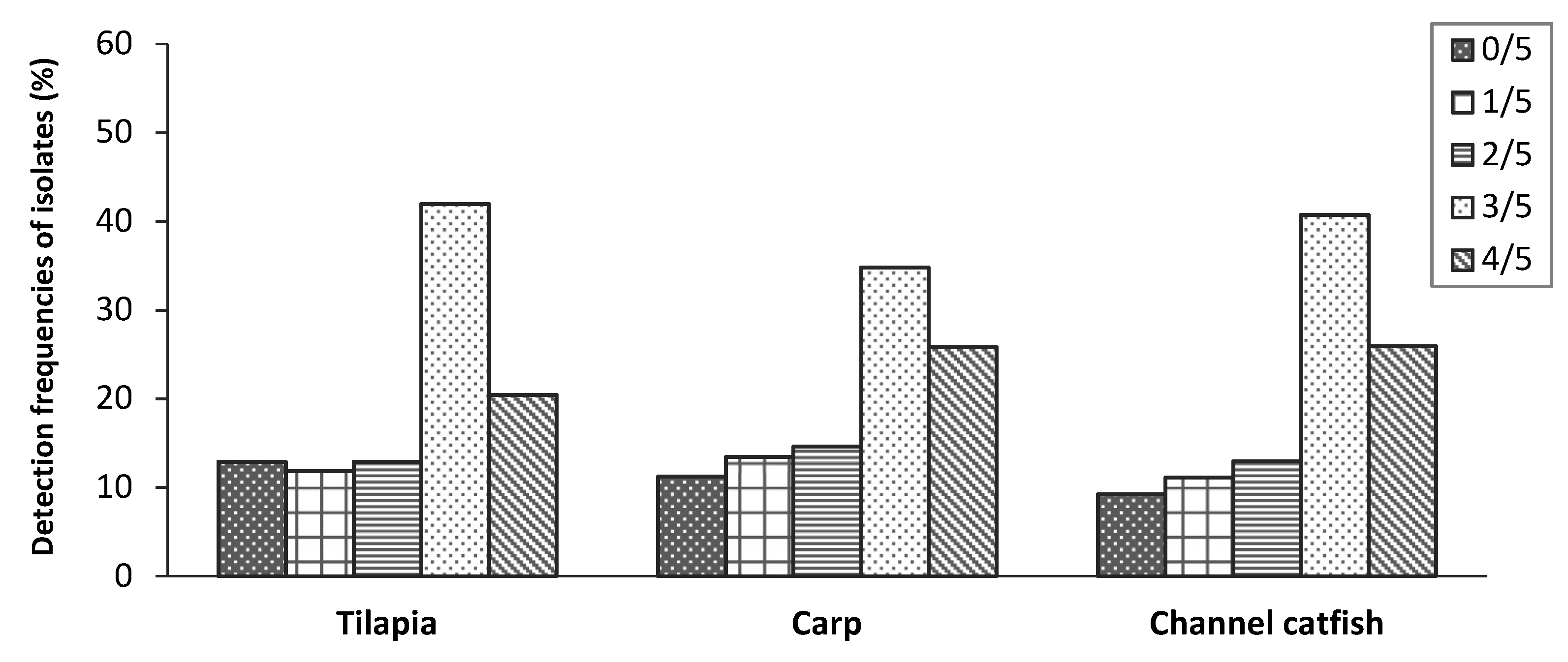
2.2. Aeromonas Hydrophila Identification
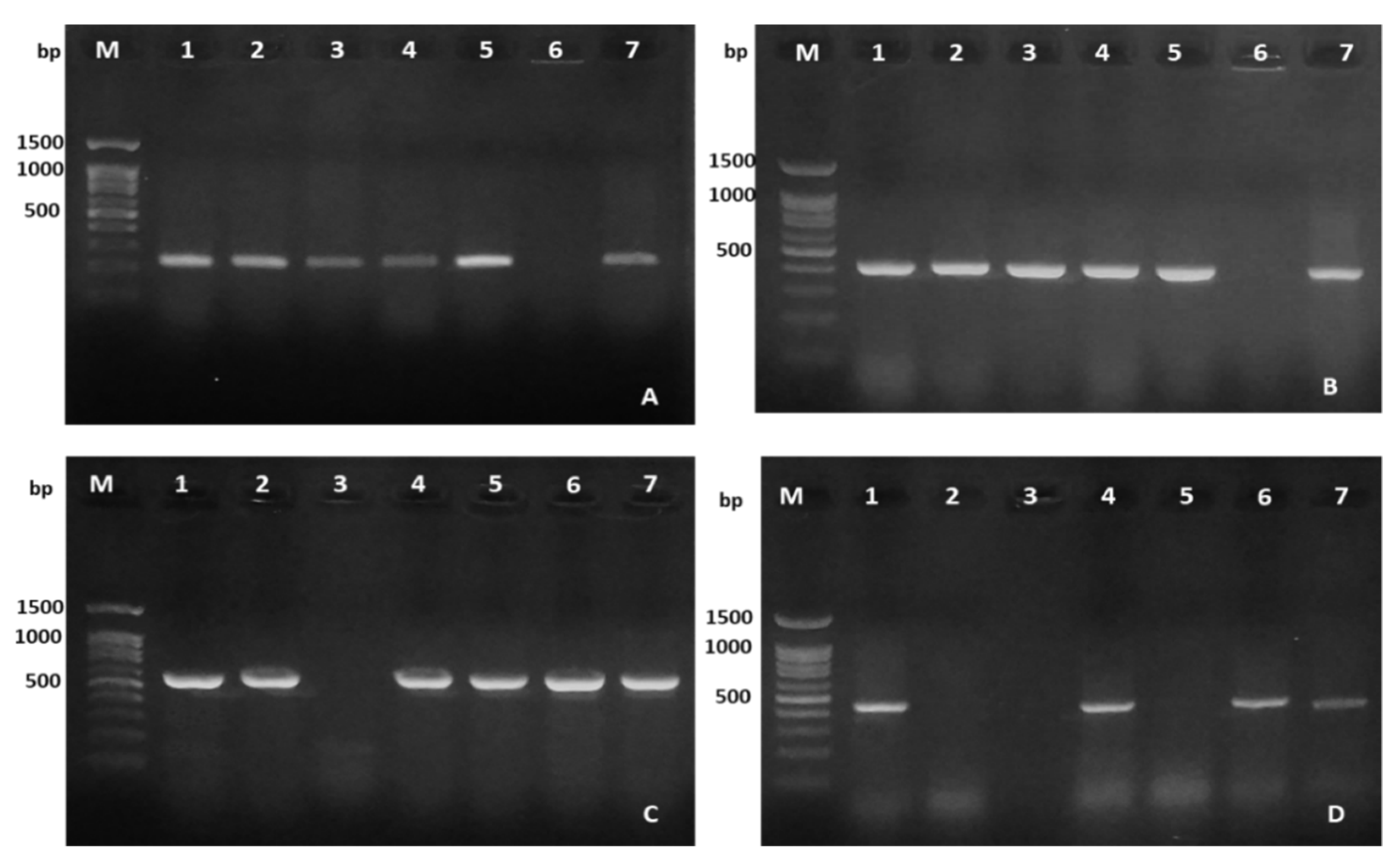
2.3. Virulence Genes Characteristics
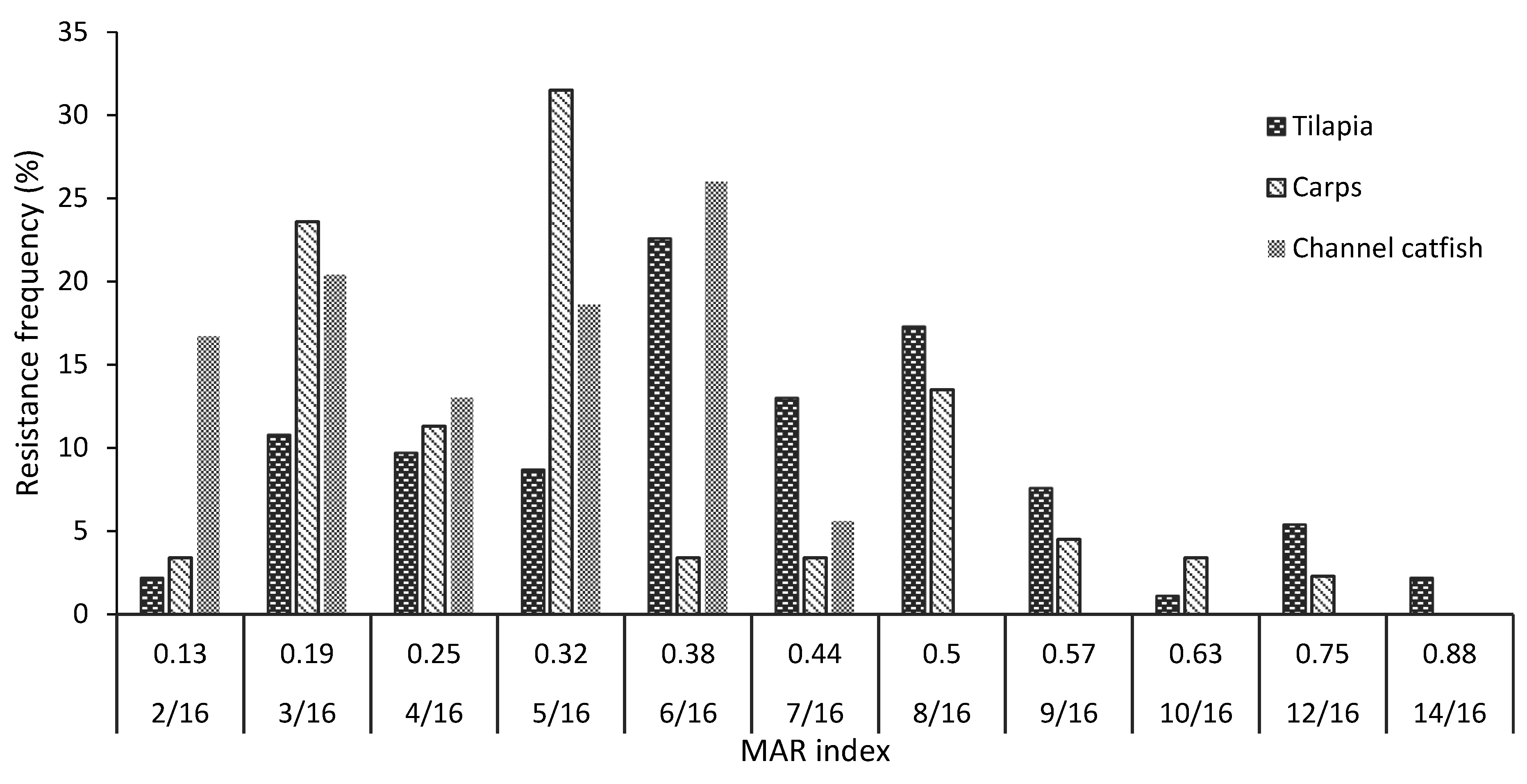
2.4. Antibiotic Susceptibility Patterns
3. Materials and Methods
3.1. Sample Locations and Clinical Examination of Diseased Fish
3.2. Bacterial Culture and Isolation
3.3. Bacterial Identification
3.4. Virulence Genes Detection
3.5. Antimicrobial Susceptibility Testing
3.6. Data Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, D.; Nguyen, T.; Li, M.; Gatlin, D.M.; O’Keefe, T. Advances in aquaculture nutrition: Catfish, tilapia and carp nutrition. In New Technologies in Aquaculture; Burnell, G., Allan, G., Eds.; Woodhead Publishing: Cambridge, UK, 2009; pp. 440–458. [Google Scholar] [CrossRef]
- Eissa, A.; Moustafa, M.; El-Husseiny, I.; Saeid, S.; Saleh, O.; Borhan, T. Identification of some skeletal deformities in freshwater teleosts raised in Egyptian aquaculture. Chemosphere 2009, 77, 419–425. [Google Scholar] [CrossRef]
- Mzula, A.; Wambura, P.N.; Mdegela, R.H.; Shirima, G.M. Present status of aquaculture and the challenge of bacterial diseases in freshwater farmed fish in Tanzania; A call for sustainable strategies. Aquac. Fish. 2020. [Google Scholar] [CrossRef]
- Heuer, O.E.; Kruse, H.; Grave, K.; Collignon, P.; Karunasagar, I.; Angulo, F.J. Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 2009, 49, 1248–1253. [Google Scholar] [CrossRef]
- Tavares-Dias, M.; Martins, M.L. An overall estimation of losses caused by diseases in the Brazilian fish farms. J. Parasit. Dis. 2017, 41, 913–918. [Google Scholar] [CrossRef]
- Dias, M.K.; Sampaio, L.S.; Proietti-Junior, A.A.; Yoshioka, E.T.; Rodrigues, D.P.; Rodriguez, A.F.; Ribeiro, R.A.; Faria, F.S.; Ozório, R.O.; Tavares-Dias, M. Lethal dose and clinical signs of Aeromonas hydrophila in Arapaima gigas (Arapaimidae), the giant fish from Amazon. Vet. Microbiol. 2016, 188, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, C.; Economou, E.; Zakas, G.; Salamoura, C.; Dontorou, C.; Apostolou, J. Microbiological and pathogenic contaminants of seafood in Greece. J. Food Qual. 2007, 30, 28–42. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Plumb, J.A.; Hanson, L.A. Health Maintenance and Principal Microbial Diseases of Cultured Fishes; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 1–483. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.; Khafaga, A.F. Natural co-infection of cultured Nile tilapia Oreochromis niloticus with Aeromonas hydrophila and Gyrodactylus cichlidarum experiencing high mortality during summer. Aquac. Res. 2020, 51, 1880–1892. [Google Scholar] [CrossRef]
- Abd-El-Malek, A.M. Incidence and virulence characteristics of Aeromonas spp. in fish. Vet. World 2017, 10, 34–37. [Google Scholar] [CrossRef]
- Kumar, R.; Pande, V.; Singh, L.; Sharma, L.; Saxena, N.; Thakuria, D.; Singh, A.K.; Sahoo, P.K. Pathological findings of experimental Aeromonas hydrophila infection in golden mahseer (Tor putitora). Fish Aquac. J. 2016, 7, 160. [Google Scholar]
- Zhang, D.; Moreira, G.S.; Shoemaker, C.; Newton, J.C.; Xu, D.-H. Detection and quantification of virulent Aeromonas hydrophila in channel catfish tissues following waterborne challenge. FEMS Microbiol. Lett. 2016, 363, 1–5. [Google Scholar] [CrossRef]
- Palu, A.P.; Gomes, L.M.; Miguel, M.A.L.; Balassiano, I.T.; Queiroz, M.L.P.; Freitas-Almeida, A.C.; de Oliveira, S.S. Antimicrobial resistance in food and clinical Aeromonas isolates. Food Microbiol. 2006, 23, 504–509. [Google Scholar] [CrossRef]
- Lee, H.J.; Hoel, S.; Lunestad, B.T.; Lerfall, J.; Jakobsen, A.N. Aeromonas spp. isolated from ready-to-eat seafood on the Norwegian market: Prevalence, putative virulence factors and antimicrobial resistance. J. Appl. Microbiol. 2020, 130, 1380–1393. [Google Scholar] [CrossRef]
- Albert, M.J.; Ansaruzzaman, M.; Talukder, K.A.; Chopra, A.K.; Kuhn, I.; Rahman, M.; Mollby, R. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 2000, 38, 3785–3790. [Google Scholar]
- Sha, J.; Kozlova, E.V.; Chopra, A.K. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: Generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 2002, 70, 1924–1935. [Google Scholar] [CrossRef] [PubMed]
- Rico, A.; Phu, T.M.; Satapornvanit, K.; Min, J.; Shahabuddin, A.; Henriksson, P.J.; Murray, F.J.; Little, D.C.; Dalsgaard, A.; Van den Brink, P.J. Use of veterinary medicines, feed additives and probiotics in four major internationally traded aquaculture species farmed in Asia. Aquaculture 2013, 412, 231–243. [Google Scholar] [CrossRef]
- Le, T.X.; Munekage, Y. Residues of selected antibiotics in water and mud from shrimp ponds in mangrove areas in Viet Nam. Mar. Pollut. Bull. 2004, 49, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, G.; Huys, G.; Swings, J.; Mcgann, P.; Hiney, M.; Smith, P.; Pickup, R.W. Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: Implication of Tn1721 in dissemination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol. 2000, 66, 3883–3890. [Google Scholar] [CrossRef]
- Yano, Y.; Hamano, K.; Tsutsui, I.; Aue-umneoy, D.; Ban, M.; Satomi, M. Occurrence, molecular characterization, and antimicrobial susceptibility of Aeromonas spp. in marine species of shrimps cultured at inland low salinity ponds. Food Microbiol. 2015, 47, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Rabobank, World Seafood Trade Map 2019. Available online: https://research.rabobank.com/far/en/sectors/animal-protein/world-seafood-trade-map.html (accessed on 2 May 2019).
- Hoai, T.D.; Trang, T.T.; Van Tuyen, N.; Giang, N.T.H.; Van Van, K. Aeromonas veronii caused disease and mortality in channel catfish in Vietnam. Aquaculture 2019, 513, 734425. [Google Scholar] [CrossRef]
- Lee, C.; Cho, J.C.; Lee, S.H.; Lee, D.G.; Kim, S.J. Distribution of Aeromonas spp. as identified by 16S rDNA restriction fragment length polymorphism analysis in a trout farm. Appl. Microbiol. 2002, 93, 976–985. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Hoi, L.; Schmidt, A.S.; Qian, D.; Shimada, T.; Shen, J.Y.; Larsen, J.L. Is Aeromonas hydrophila the dominant motile Aeromonas species that causes disease outbreaks in aquaculture production in the Zhejiang Province of China? Dis. Aquat. Organ. 2001, 46, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Samayanpaulraj, V.; Sivaramapillai, M.; Palani, S.N.; Govindaraj, K.; Velu, V.; Ramesh, U. Identification and characterization of virulent Aeromonas hydrophila Ah17 from infected Channa striata in river Cauvery and in vitro evaluation of shrimp chitosan. Food Sci. Nutr. 2020, 8, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Yanez, M.; Catalán, V.; Apráiz, D.; Figueras, M.; Martinez-Murcia, A. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 875–883. [Google Scholar] [CrossRef]
- Korczak, B.; Christensen, H.; Emler, S.; Frey, J.; Kuhnert, P. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int. J. Syst. Evol. Microbiol. 2004, 54, 1393–1399. [Google Scholar] [CrossRef]
- Heuzenroeder, M.W.; Wong, C.Y.; Flower, R.L. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: Correlation with virulence in a suckling mouse model. FEMS Microbiol. Lett. 1999, 174, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Kingombe, C.I.B.; Huys, G.; Tonolla, M.; Albert, M.J.; Swings, J.; Peduzzi, R.; Jemmi, T. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 1999, 65, 5293–5302. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, S.A.; Khan, A.A.; Sung, K.; Tran, Q.; Kerdahi, K.; Steele, R. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010, 27, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.B.; Cockerill, F.R.; Bradfoord, P.A. CLSI M100-S25 performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Clin. Lab. Stand. Inst. 2015, 35, 1–240. [Google Scholar]
- Hedberg, N.; Stenson, I.; Pettersson, M.N.; Warshan, D.; Nguyen-Kim, H.; Tedengren, M.; Kautsky, N. Antibiotic use in Vietnamese fish and lobster sea cage farms; implications for coral reefs and human health. Aquaculture 2018, 495, 366–375. [Google Scholar] [CrossRef]
- Hoa, P.T.P.; Managaki, S.; Nakada, N.; Takada, H.; Shimizu, A.; Anh, D.H.; Viet, P.H.; Suzuki, S. Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci. Total Environ. 2011, 409, 2894–2901. [Google Scholar] [CrossRef]
- Binh, V.N.; Dang, N.; Anh, N.T.K.; Thai, P.K. Antibiotics in the aquatic environment of Vietnam: Sources, concentrations, risk and control strategy. Chemosphere 2018, 197, 438–450. [Google Scholar] [CrossRef]
- Do, T.C.M.V.; Nguyen, D.Q.; Nguyen, T.D.; Le, P.H. Development and validation of a LC-MS/MS method for determination of multi-class antibiotic residues in aquaculture and river waters, and photocatalytic degradation of antibiotics by TiO2 nanomaterials. Catalysts 2020, 10, 356. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Van Huong, N.; Cuong, T.H.; Thu, T.T.N.; Lebailly, P. Efficiency of Different Integrated Agriculture Aquaculture Systems in the Red River Delta of Vietnam. Fish. Aquac. J. 2017, 8, 493. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012, 2012, 1–13. [Google Scholar]
- Pal, M. Is Aeromonas hydrophila a potential pathogen of food safety concern. J. Food Microbiol. 2018, 2, 1–2. [Google Scholar]
- Hoel, S.; Vadstein, O.; Jakobsen, A.N. The significance of mesophilic Aeromonas spp. in minimally processed ready-to-eat seafood. Microorganisms 2019, 7, 1–25. [Google Scholar]
- Parker, J.L.; Shaw, J.G. Aeromonas spp. clinical microbiology and disease. J. Infect. 2011, 62, 109–118. [Google Scholar] [CrossRef]
- Vila, J.; Marco, F.; Soler, L.; Chacon, M.; Figueras, M.J. In vitro antimicrobial susceptibility of clinical isolates of Aeromonas caviae, Aeromonas hydrophila and Aeromonas veronii biotype sobria. J. Antimicrob. Chemothe. 2002, 49, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ni, X.D.; Liu, Y.J.; Lu, C.P. Detection of three virulence genes alt, ahp and aerA in Aeromonas hydrophila and their relationship with actual virulence to zebrafish. J. Appl. Microbiol. 2011, 110, 823–830. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Roos, S.; Edsman, L.; Liu, H.; Söderhäll, K. A highly virulent pathogen, Aeromonas hydrophila, from the freshwater crayfish Pacifastacus leniusculus. J. Invertebr. Pathol. 2009, 101, 56–66. [Google Scholar] [CrossRef]
- Nicholson, P.; Mon-on, N.; Jaemwimol, P.; Tattiyapong, P.; Surachetpong, W. Coinfection of tilapia lake virus and Aeromonas hydrophila synergistically increased mortality and worsened the disease severity in tilapia (Oreochromis spp.). Aquaculture 2020, 520, 734746. [Google Scholar] [CrossRef]
- Xu, X.J.; Ferguson, M.R.; Popov, V.L.; Houston, C.W.; Peterson, J.W.; Chopra, A.K. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: Development of transposon and isogenic mutants. Infect. Immun. 1998, 66, 3501–3509. [Google Scholar] [CrossRef]
- El-Bahar, H.M.; Ali, N.G.; Aboyadak, I.M.; Khalil, S.A.E.S.; Ibrahim, M.S. Virulence genes contributing to Aeromonas hydrophila pathogenicity in Oreochromis niloticus. Int. Microbiol. 2019, 4, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.T.; Veneroni-Gouveia, G.; Costa, M.M. Molecular characterization of virulence factors in Aeromonas hydrophila obtained from fish. Pesquisa Veterinária Brasileira 2012, 32, 701–706. [Google Scholar] [CrossRef]
- Ristow, L.C.; Welch, R.A. Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger? Biochimica et Biophysica Acta (BBA)-Biomembranes 2016, 1858, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Tomás, J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012, 2012, 1–22. [Google Scholar] [CrossRef]
- Hayati, H.R.; Hassan, M.; Ong, B.; Abdelhadi, Y.; Hidayahanum, H.N.; Sharifah, R.; Faten, A.N.; Kuttichantran, S.; Alsaid, M. Virulence genes detection of Aeromonas hydrophila originated from diseased freshwater fishes. Adv. Environ. Biol. 2015, 9, 22–26. [Google Scholar]
- Chopra, A.K.; Houston, C.W. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1999, 1, 1129–1137. [Google Scholar] [CrossRef]
- Rather, M.A.; Willayat, M.M.; Wani, S.A.; Hussain, S.A.; Shah, S.A. Enterotoxin gene profile and molecular epidemiology of Aeromonas species from fish and diverse water sources. J. Appl. Microbiol. 2019, 127, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Abd El Tawab, A.A.; Maarouf, A.A.; El Hofy, F.I.; El Mougy, E.E. Detection of some virulence genes in A. hydrophila and A. caviae isolated from fresh water fishes at Qalubia Governorate. Benha Vet. Med. J. 2017, 33, 489–503. [Google Scholar] [CrossRef]
- Hoel, S.; Vadstein, O.; Jakobsen, A.N. Species distribution and prevalence of putative virulence factors in mesophilic Aeromonas spp. isolated from fresh retail sushi. Front. Microbiol. 2017, 8, 931. [Google Scholar] [CrossRef]
- Aoki, T. Present and future problems concerning the development of resistance in aquaculture. In Chemotherapy in Aquaculture: From Theory to Reality; Michel, C., Alderman, D., Eds.; Office International des EÉpizooties: Paris, France, 1992; pp. 254–262. [Google Scholar]
- Akinbowale, O.L.; Peng, H.; Barton, M. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 2006, 100, 1103–1113. [Google Scholar] [CrossRef]
- WHO. Joint FAO/OIE/WHO Expert Workshop on Non-Human Antimicrobial Usage and Antimicrobial Resistance: Scientific Assessment: Geneva, 1–5 December 2003 (No. WHO/CDS/CPE/ZFK/2004.7); World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Stratev, D.; Stoev, S.; Vashin, I.; Daskalov, H. Some varieties of pathological changes in experimental infection of carps (Cyprinus carpio) with Aeromonas hydrophila. J. Aquac. Eng. Fish. Res. 2015, 1, 191–202. [Google Scholar] [CrossRef]
- Fosse, T.; Giraud-Morin, C.; Madinier, I. Phenotypes of beta-lactam resistance in the genus Aeromonas. Pathologie-Biologie 2003, 51, 290. [Google Scholar] [CrossRef]
- Borella, L.; Salogni, C.; Vitale, N.; Scali, F.; Moretti, V.M.; Pasquali, P.; Alborali, G.L. Motile aeromonads from farmed and wild freshwater fish in northern Italy: An evaluation of antimicrobial activity and multidrug resistance during 2013 and 2016. Acta Vet. Scand. 2020, 62, 6. [Google Scholar] [CrossRef]
- Dahanayake, P.S.; Hossain, S.; Wickramanayake, M.V.K.S.; Heo, G.J. Prevalence of virulence and antimicrobial resistance genes in Aeromonas species isolated from marketed cockles (Tegillarca granosa) in Korea. Lett. Appl. Microbiol. 2020, 71, 94–101. [Google Scholar] [CrossRef]
- Yuan, K.; Wang, X.; Chen, X.; Zhao, Z.; Fang, L.; Chen, B.; Jiang, J.; Luan, T.; Chen, B. Occurrence of antibiotic resistance genes in extracellular and intracellular DNA from sediments collected from two types of aquaculture farms. Chemosphere 2019, 234, 520–527. [Google Scholar] [CrossRef]
- Ottaviani, D.; Santarelli, S.; Bacchiocchi, S.; Masini, L.; Ghittino, C.; Bacchiocchi, I. Occurrence and characterization of Aeromonas spp. in mussels from the Adriatic Sea. Food Microbiol. 2006, 23, 418–422. [Google Scholar] [CrossRef]



| Fish Species | Total no. of Samples | Number of Isolates | ||||
|---|---|---|---|---|---|---|
| Recovered on Rimler–Shotts Medium | Confirmed by Phenotypic Tests | Confirmed by PCR | Confirmed bySequencing | Detection Frequencies (%) | ||
| Tilapia | 198 | 187 | 136 | 102 | 93 | 47.0 |
| Carp | 187 | 156 | 118 | 95 | 89 | 47.6 |
| Channel catfish | 121 | 115 | 84 | 58 | 54 | 44.6 |
| Total/mean | 506 | 458 | 338 | 255 | 236 | 46.4 |
| Virulence Genes | Detection Frequencies % (N) | |||
|---|---|---|---|---|
| Tilapia (n = 93) | Carp (n = 89) | Channel Catfish (n = 54) | Mean (n = 236) | |
| hlyA | 58.1 (54) | 63.0 (56) | 57.5 (31) | 59.7 (141) |
| aerA | 83.9 (78) | 79.8 (71) | 76.0 (41) | 80.5 (190) |
| act | 83.9 (78) | 76.5 (68) | 79.7 (43) | 80.1 (189) |
| alt | 42.0 (39) | 47.2 (42) | 46.3 (25) | 44.9 (106) |
| ast | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Antimicrobial Agents | Antimicrobial Susceptibility of A. hydrophila (N = 236) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tilapia (n = 93) | Carp (n = 89) | Channel Catfish (n = 54) | |||||||
| S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | |
| Penicillins (PNs) | |||||||||
| Oxacillin (Ox) | 0 (0) | 4.4 (4) | 95.7 (89) | 0 (0) | 0 (0) | 100 (89) | 0 (0) | 9.3 (5) | 90.8 (49) |
| Amoxicillin (Ax) | 15.1 (14) | 7.6 (7) | 77.5 (72) | 3.4 (3) | 0(0) | 96.7 (86) | 0 (0) | 0 (0) | 100 (54) |
| Β-Lactam/β-Lactamase inhibitor combination (BL/BLI) | |||||||||
| Amoxicillin-Clavulanic acid (AC) | 76.4 (71) | 15.1 (14) | 8.7 (8) | 73.1 (65) | 18 (16) | 9.0 (8) | 68.6 (37) | 16.7 (9) | 14.9 (8) |
| Cephems (CPs) | |||||||||
| Ceftriaxone (Ct) | 69.9 (65) | 4.4 (4) | 25.9 (24) | 95.6 (85) | 4.5 (4) | 0 (0) | 90.8 (49) | 9.3 (5) | 0(0) |
| Cefuroxime (Cu) | 81.8 (76) | 5.4 (5) | 13.0 (12) | 96.7 (86) | 0 (0) | 3.4 (3) | 100 (54) | 0(0) | 0(0) |
| Cefotaxime (Cx) | 77.5 (72) | 0 (0) | 22.6 (21) | 96.7 (86) | 0 (0) | 3.4 (3) | 100 (54) | 0(0) | 0(0) |
| Macrolides (MCs) | |||||||||
| Erythromycin (Er) | 24.8 (23) | 32.3 (30) | 43.1 (40) | 38.3 (34) | 14.7 (13) | 47.2 (42) | 57.5 (31) | 33.4 (18) | 9.3 (5) |
| Quinolones (QLs) | |||||||||
| Nalidixic (Na) | 54.9 (51) | 0 (0) | 45.2 (42) | 46.1 (41) | 3.4 (3) | 50.6 (45) | 53.8 (29) | 9.3 (5) | 37.1 (20) |
| Sulfonamides (SULs) (Folate pathway inhibitors) | |||||||||
| Sulfamethoxzole- Trimethoprim (SM/TM) | 36.6 (34) | 18.3 (17) | 45.2 (42) | 75.3 (67) | 0 (0) | 24.8 (22) | 68.6 (37) | 9.3 (5) | 22.3 (12) |
| Aminoglycosides (AMGs) | |||||||||
| Neomycin (Ne) | 4.4 (4) | 16.2 (15) | 79.6 (74) | 42.7 (38) | 36.0 (32) | 21.4 (19) | 38.9 (21) | 31.5 (17) | 29.7 (16) |
| Glycopeptide (GLs) | |||||||||
| Vancomycin (Va) | 3.3 (3) | 15.1 (14) | 81.8 (76) | 6.8 (6) | 20.3 (18) | 73.1 (65) | 0 (0) | 16.7 (9) | 83.4 (45) |
| Flouroquinolones (FQNs) | |||||||||
| Ofloxacin (Of) | 49.5 (46) | 8.7 (8) | 42 (39) | 79.8 (71) | 3.4 (3) | 16.9 (15) | 61.2 (33) | 38.9 (21) | 0 (0) |
| Norfloxacin (No) | 63.5 (59) | 24.8 (23) | 11.9 (11) | 83.2 (74) | 3.4 (3) | 13.5 (12) | 77.8 (42) | 22.3 (12) | 0 (0) |
| Tetracyclines (TCs) | |||||||||
| Doxycycline (Dx) | 88.2 (82) | 9.7 (9) | 2.2 (2) | 75.3 (67) | 18 (16) | 6.8 (6) | 100 (54) | 0 (0) | 0(0) |
| Oxytetracycline (OTC) | 59.2 (55) | 4.4 (4) | 36.6 (34) | 55.1 (49) | 0 (0) | 45 (40) | 70.4 (38) | 0 (0) | 29.7 (16) |
| Amphenicols (AMPs) | |||||||||
| Florfenicol (Fl) | 72.1 (67) | 3.3 (3) | 24.8 (23) | 76.5 (68) | 0 (0) | 23.6 (21) | 83.4 (45) | 0 (0) | 16.7 (9) |
| Overall | |||||||||
| Average (%) | 48.5 | 10.6 | 40.9 | 59.0 | 7.6 | 33.4 | 60.6 | 12.3 | 27.1 |
| No of Drugs | Resistance Phenotypes | The Ratio of Isolates—% (N) | ||
|---|---|---|---|---|
| Tilapia n = 93 | Carp n = 89 | Channel Catfish n = 54 | ||
| 2 | Ox + Ax | 2.2 (2) | 3.4 (3) | 16.7 (9) |
| 3 | Ox + Ne + Va | 7.6 (7) | 0 (0) | 0 (0) |
| Ox + Ax + Ne | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Na | 0 (0) | 2.3 (2) | 0 (0) | |
| Ox + Ax + Va | 0 (0) | 21.4 (19) | 20.4 (11) | |
| 4 | Ox + Ax + Na + Va | 4.4 (4) | 3.4 (3) | 13 (7) |
| Ox + Ax + Of + Ne | 3.3 (3) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Ne + Va | 2.2 (2) | 0 (0) | 0 (0) | |
| Ox + Ax + Va + Fl | 0 (0) | 4.5 (4) | 0 (0) | |
| 5 | Ox + Ax + Na + Ne + Va | 3.3 (3) | 0 (0) | 11.2 (6) |
| Ox + Ax + Er + Ne + Va | 5.4 (5) | 5.7 (5) | 0 (0) | |
| Ox + Ax + SM/TM + Fl + OTC | 0 (0) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Na + Va + OTC | 0 (0) | 3.4 (3) | 7.5 (4) | |
| Ox + Ax + SM/TM + Va + OTC | 0 (0) | 1.2 (1) | 0 (0) | |
| Ox + Ax + Er + Na + OCT | 0 (0) | 18 (16) | 0 (0) | |
| 6 | Ox + Ct + Cx + Va + Fl + OTC | 5.4 (5) | 0 (0) | 0 (0) |
| Ox + Ax + Er + SM/TM + Ne + Va | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + SM/TM + Ne + Va + OTC | 3.3 (3) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Er + Na + Ne + OTC | 2.2 (2) | 0 (0) | 0 (0) | |
| Ox + Ax + Cu + SM/TM + Ne + Va | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Ac + Er + Ne + Va | 4.4 (4) | 0 (0) | 9.3 (5) | |
| Ox + Ax + Er + Of + Ne + Va | 1.1 (1) | 0 (0) | 0 (0) | |
| Ox + Ax + SM/TM + Va + Fl + OTC | 0 (0) | 0 (0) | 7.5 (4) | |
| Ax + SM/TM + Ne + Va + Fl + OTC | 0 (0) | 0 (0) | 9.3 (5) | |
| 7 | Ox + Ax + Ct + Er + Ne + Va + OTC | 3.3 (3) | 0 (0) | 0 (0) |
| Ox + Ax + SM/TM + Ne + Va + Fl + OTC | 2.2 (2) | 0 (0) | 0 (0) | |
| Ox + Ax + Na + SM/TM + Ne + Va + Fl | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Na + Of + Ne + Va + OTC | 4.4 (4) | 0 (0) | 0 (0) | |
| Ox + Ax + Ac + Na + Of + No + Va | 0 (0) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Ac + Na + SM/TM + Va + OTC | 0 (0) | 0 (0) | 5.6 (3) | |
| 8 | Ox + Ct + Cx + Cu + Of + Ne + Va + OTC | 2.2 (2) | 0 (0) | 0 (0) |
| Ox + Ct + Cx + Na + Of + SM/TM + Ne + OTC | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Er + Na + Of + SM/TM + Ne + Va | 5.4 (5) | 0 (0) | 0 (0) | |
| Ox + Ax + Er + Na + Of + No + SM/TM + Va | 5.4 (5) | 4.5 (4) | 0 (0) | |
| Ox + Ax + Ac + Er + Na + Of + Ne + Va | 1.1 (1) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Er + No + SM/TM + Va + Fl + OTC | 0 (0) | 3.4 (3) | 0 (0) | |
| Ox + Ax + Er + Na + SM/TM + Va + Fl + OTC | 0 (0) | 2.3 (2) | 0 (0) | |
| 9 | Ct + Cx + Na + Of + No + SM/TM + Ne + Fl + OTC | 4.4 (4) | 0 (0) | 0 (0) |
| Ox + Ax + Er + Na + Of + SM/TM + Va + Fl + OTC | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Er + Na + SM/TM + Va + Dx + Fl + OTC | 0 (0) | 4.5 (4) | 0 (0) | |
| 10 | Ox + Ax + Er + Na + Of + SM/TM + Ne + Va + Fl + OTC | 1.1 (1) | 0 (0) | 0 (0) |
| Ox + Ax + Cu + Cx + Er + Na + Ne + Va + Fl + OTC | 0 (0) | 3.4 (3) | 0 (0) | |
| 12 | Ox + Ax + Ct + Cu + Cx + Er + Na + Of + SM/TM + Ne + Va + Fl | 2.2 (2) | 0 (0) | 0 (0) |
| Ox + Ax + Ac + Ct + Cu + Cx + Er + Of + SM/TM + Ne + Va + Fl | 3.3 (3) | 0 (0) | 0 (0) | |
| Ox + Ax + Ac + Er + Na + Of + No + SM/TM + Ne + Va + Fl + OTC | 0 (0) | 2.3 (2) | 0 (0) | |
| 14 | Ox + Ax + Ct + Cu + Cx + Er + Na + Of + No + SM/TM + Ne + Va + Dx + OTC | 2.2 (2) | 0 (0) | 0 (0) |
| Gene | Primers | DNA Sequence (5′→3′) | Product Size (bp) | References |
|---|---|---|---|---|
| 16S rRNA | Aero16S-F | CTACTTTTGCCGGCGAGCGG TGATTCCCGAAGGCACTCCC | 953 | [24] |
| Aero16S-R | ||||
| AeroH | AeroH-F | GAAAGGTTGATGCCTAATACGTA | 625 | [25] |
| AeroH-R | CGTGCTGGCAACAAAGGACAG | |||
| gyrB | gyrB 3F | TCCGGCGGTCTGCACGGCGT | 1110 | [30] |
| gyrB 14R | TTGTCCGGGTTGTACTCGTC | |||
| rpoB | PasrpoB-L | GCAGTGAAAGARTTCTTTGGTTC | 560 | [31] |
| RpoB-R | GTTGCATGTTNGNACCCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nhinh, D.T.; Le, D.V.; Van, K.V.; Huong Giang, N.T.; Dang, L.T.; Hoai, T.D. Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture. Antibiotics 2021, 10, 532. https://doi.org/10.3390/antibiotics10050532
Nhinh DT, Le DV, Van KV, Huong Giang NT, Dang LT, Hoai TD. Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture. Antibiotics. 2021; 10(5):532. https://doi.org/10.3390/antibiotics10050532
Chicago/Turabian StyleNhinh, Doan Thi, Dung Viet Le, Kim Van Van, Nguyen Thi Huong Giang, Lua Thi Dang, and Truong Dinh Hoai. 2021. "Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture" Antibiotics 10, no. 5: 532. https://doi.org/10.3390/antibiotics10050532
APA StyleNhinh, D. T., Le, D. V., Van, K. V., Huong Giang, N. T., Dang, L. T., & Hoai, T. D. (2021). Prevalence, Virulence Gene Distribution and Alarming the Multidrug Resistance of Aeromonas hydrophila Associated with Disease Outbreaks in Freshwater Aquaculture. Antibiotics, 10(5), 532. https://doi.org/10.3390/antibiotics10050532






