1. Introduction
Nontyphoidal
Salmonella (NTS) is a worldwide cause of food-borne illness in humans, causing salmonellosis [
1]. In humans, salmonellosis is transmitted through the ingestion of contaminated food of animal origin (mainly eggs, meat, poultry, and milk). However, other foods, such as green vegetables contaminated by manure, have been involved in its contraction [
2]. The feco-oral route may also cause person-to-person transmission [
3]. The disease is characterized by acute onset of fever, abdominal pain, diarrhea, nausea, and, to a lesser extent, vomiting. The incubation period of the disease is 12–72 h and illness last for 2–7 days [
4].
In most cases, symptoms of salmonellosis are mild, and patients will recover without specific treatment. However, in some cases, mainly children and elderly patients, the resulting dehydration may become severe and fatal [
5]. Although most salmonellosis infections are mild, they account for about 93.8 million foodborne illnesses and 155,000 deaths per year worldwide, making infections a great concern to public health [
6]. More than 2500
Salmonella serovars have been identified, most of which belong to
Salmonella enterica subsp. enterica, which is responsible for most
Salmonella infections in humans [
7].
Salmonella infections could be invasive, requiring efficient antibiotic therapy [
8].
Salmonella virulence is due to presence of chromosomal and plasmid genes. Chromosomal genes are large gene cassettes, called
Salmonella pathogenicity islands (SPIs), which code about 60 genes that account for certain interactions with the human or animal hosts [
9]. Plasmid genes can transfer antimicrobial resistance between bacteria, which can lead to emergence of multidrug resistant (MDR) bacteria and to increasing bacterial virulence, which is a great foodborne risk to human health [
10]. The emergence of multidrug resistant (MDR)
Salmonella serovars exerts a dramatic influence on the efficacy of antibiotic therapy. Increasing prevalence of MDR strains can cause a rise in mortality rates of
Salmonella infections [
11]. The broad prevalence of antimicrobial resistance among nontyphoidal
Salmonella imposes a great threat to public health since these pathogens are mostly food-borne.
Hence, appropriate measures and new guidelines should be established to address the rational use of antibiotics and to prevent their abuse, since they represent a major healthcare challenge. Therefore, the aim of our study was to isolate and identify the most common Salmonella serovars recovered from febrile neutropenic patients suffering from severe gastroenteritis, collected from different microbiological labs in Egypt, followed by phenotypic and molecular analysis of the antimicrobial resistance profiles, and to compare those serovars with those of poultry origin recovered from the same geographical area and during the same period.
3. Discussion
Salmonella is a major cause of food-borne illness in humans, and salmonellosis is mainly contracted through ingestion of contaminated food of poultry origin including, undercooked meat, eggs, and other poultry-derived food [
12]. The
Salmonella organisms can survive in undercooked food and then be transmitted to humans, causing pathological conditions, such as gastroenteritis and fever in humans, which can be associated with significant clinical consequences and complications. Additionally, the antimicrobial resistant genotypes can be carried on transferrable genetic elements such as plasmids or integrons, which can then be transferred from poultry to human serovars and to the bacterial flora—a major consequence of ingestion of undercooked food containing living poultry
Salmonella serovars. In our study, bacteriological examination of 300 human fecal samples collected from febrile neutropenic patients suffering from severe gastroenteritis revealed that NTS represented 16.66% of the recovered isolates (50 isolates), while 6 isolates (2%) were typhoidal
Salmonella. Most of the recovered NTS isolates belonged to
S. Typhimurium (10; 17.85%) and
S. Enteritidis (8; 14.28%), followed by
S. Lumberhurst (8; 14.28%) serovars. These results corroborate those reported by Ahmed et al. [
13], who found that the isolation rate from stool swabs was 18.7% of
Salmonella spp. On the other hand, Diab et al. [
12] isolated
Salmonella from stool samples in a percentage of 4.4% and also serotyped these isolates as
S. Typhimurium and
S. Enteritidis. Adesoji et al. [
14] isolated
Salmonella in a percentage of 60% from stool samples of diarrheic patients.
The emergence of antibiotic resistance poses a significant public health risk. Antibiotic consumption and the emergence/dissemination of resistant strains in hospitals and intensive care units have been clearly linked in epidemiological studies. Antibiotic therapy is an important factor that induces alteration of gut microbiota composition (called dysbiosis), leading to pathology, including asthma and infectious disease [
15]. Gut microbiota is necessary for proper intestinal tract development as well as immune and nervous system maturation. In fact, an intact, fully developed gastrointestinal (GI) tract microbiota also protects the host from pathogenic microorganism invasion [
16,
17,
18] via a highly complex set of events known as colonization resistance [
19,
20].
Microorganisms coexist in the same ecological niche, i.e., microbiotas, but also a continuum of microorganism exchange occurs between humans, animals, and the environment. For example: yeasts can be found in all environments and have been described as effective antagonists of a variety of plant pathogens [
21]. Additionally, increased “contact” between animals and humans increases the risk of infection and the spread of antimicrobial resistance (AMR) traits. As a result, the possibility of reverse zoonosis, as well as the establishment of animal reservoirs that maintain the infection cycle and AMR spread, is now a growing concern. Antimicrobial resistance of animal origin, which poses a direct and/or indirect threat to human health, especially for extended spectrum beta-lactamase (ESBL) Gram-negative bacteria. [
22].
The antibiogram pattern for human serovars revealed all serovars to be 100% resistant to clindamycin, erythromycin, rifampicin, cefotaxime, amoxicillin, tobramycin, amoxicillin/clavulanic acid, and ampicillin, followed by ampicillin/sulbactam (98%) and lincomycin (92%). For poultry serovars, results reveal that all serovars showed 100% resistance to clindamycin, erythromycin, rifampicin, amoxicillin, lincomycin, and tobramycin, followed by ampicillin and oxytetracycline with 98%, ampicillin/sulbactam and nitrofurantoin with 96%, doxycycline (94%), amoxicillin/clavulanic acid (92%), cefotaxime (90%), and cefradine (82%). Additionally, S. Poona standard strain was resistant to cefradine, clindamycin, erythromycin, rifampicin, streptomycin, cefotaxime, doxycycline, amoxicillin, oxytetracycline, lincomycin, tobramycin, ampicillin/sulbactam, amoxicillin/clavulanic acid, and ampicillin. Interestingly, most of the studies conducted on the recovery of human and poultry Salmonella serovars were found to be resistant to clindamycin, erythromycin, rifampicin, amoxicillin, tobramycin, ampicillin, ampicillin/sulbactam, amoxicillin/clavulanic acid, cefotaxime, lincomycin, and cefradine. The similarity in the resistance patterns between human and poultry serovars suggests that they could be of the same source, pointing out the zoonosis of this microorganism. Moreover, strong evidence was obtained when the standard strain that was used showed the same resistance patterns.
Moreover, our findings nearly coincide with those obtained by Diab et al. [
12] using 12 antimicrobials: 86.4% were resistant to oxytetracycline and streptomycin, followed by neomycin and erythromycin (77.3%) and norfloxacin and ampicillin (68.2%). On the contrary, 90.9% were sensitive to gentamicin. In addition, Adesoji et al. [
14] mentioned that
Salmonella spp. were 100% resistant to amoxicillin and ampicillin. Furthermore, Voss-Rech et al. [
23] isolated NTS of poultry and human origin in Brazil and found that NTS of poultry origin showed high resistance to sulfonamides (44.3%), nalidixic acid (42.5%), and tetracycline (35.5%), while in those of human origin, the resistance occurred mainly for sulfonamides (46.4%), tetracycline (36.9%), and ampicillin (23.6%). Meanwhile, Singh [
24] stated that
Salmonella isolated from poultry were resistant to ampicillin and tetracycline but not to enrofloxacin. Additionally, Ouali et al. [
25] stated that clinical isolates from a tertiary referring hospital were sensitive to trimethoprim–sulfamethoxazole (72%), chloramphenicol (64%), ampicillin (48%), gentamicin (44%), and ciprofloxacin (2%). Plasmid profile analysis showed that out of the 30 ampicillin-resistant serovars, 19 (63.3%) had plasmid, as did the standard strain. These findings are nearly similar to those of the work of McMillan et al. [
10], which stated that more than 80% of antibiotic resistance genes were located within a plasmid and that many different plasmids were involved in antibiotic resistance in
Salmonella among food animals. In addition, Dos Santos et al. [
26] mentioned that in
S. Typhimurium, most of the virulence factors encoding genes are located in
Salmonella pathogenicity islands (SPI). However, the other virulence genes were located in virulence plasmids (pSLT), particularly in the
spv-operon containing plasmids [
26]. Plasmid genes have the ability to transfer antimicrobial resistance between bacteria, which can lead to the presence of multidrug resistant (MDR) bacteria, making them more dangerous to human health [
10]. In our study, plasmids harboring
blaCTX-m,
blaSHV,
blaTEM, and
aac(6′)-Ib were detected in 11 (22%) and 8 (16%) of human and poultry serovars, respectively. Since poultry are usually consumed by humans, the presence of resistant bacteria harboring transmissible genetic elements is of great health and medical importance.
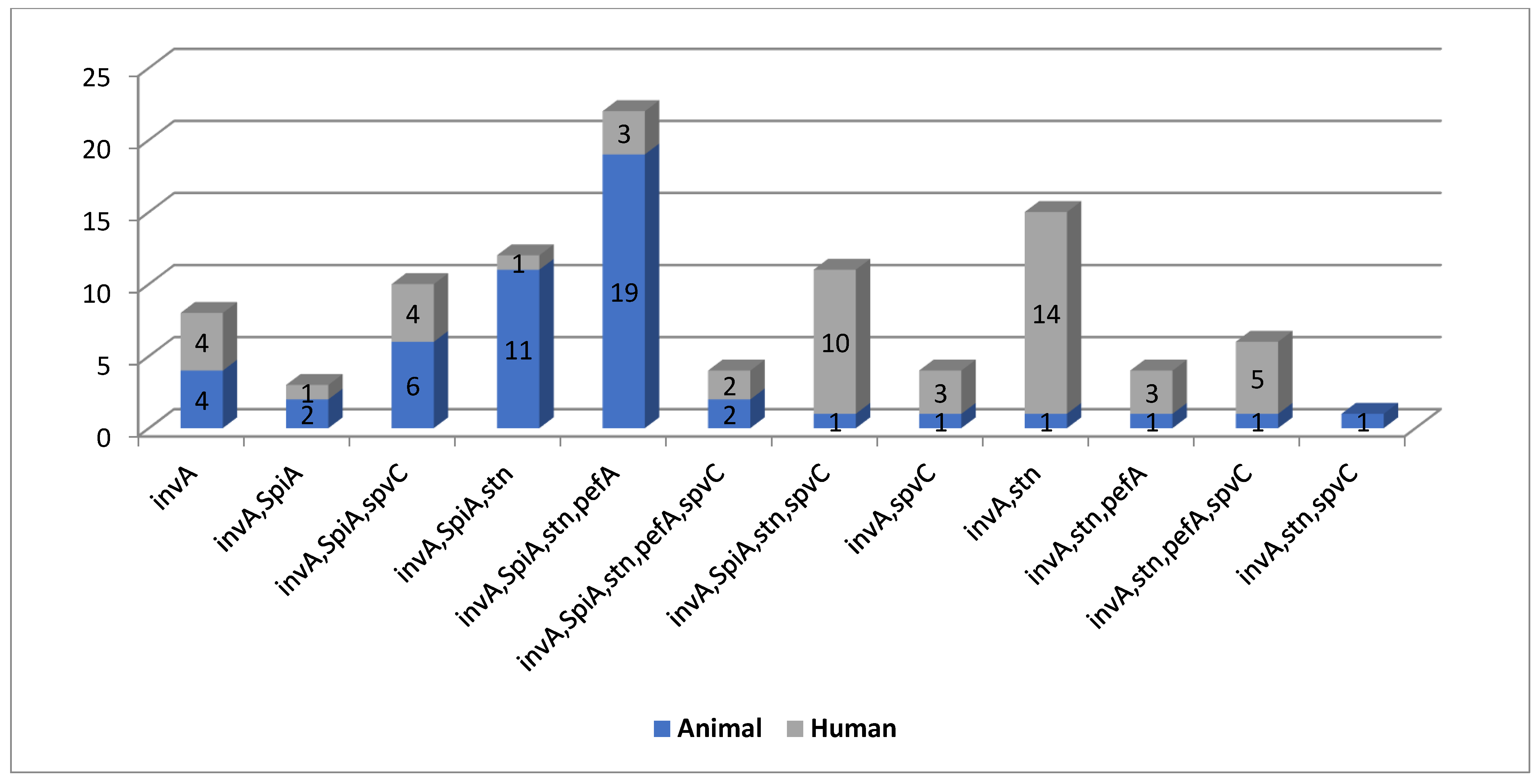
Out of the 50 isolated human serovars, 100, 26, 40, and 42% were positive for
invA gene, both
spiA and
stn,
pefA, and
spvC, respectively. Additionally, out of 50 poultry serovars, 100, 82, 74, 48, and 24% were positive for
invA gene,
spiA,
stn,
pefA, and
spvC, respectively. The
S. Poona standard strain was positive for all tested virulence genes. Molecular detection of the most clinically relevant virulence genes and analysis of the associated virulence genotypes proved that the human (
n = 11) and poultry serovars (
n = 12) shared 11 genotypes. These findings are in agreement with those of a previous study [
27] that reported that, out of 33 NTS,
invA was positive for 28 isolates, of which 89.3% patients were febrile. This study also revealed a positive correlation between the
invA gene and febrile illness, and therefore highlights the importance of
invA as an important marker for bloodstream invasion. Kim et al. [
28] reported that virulence genes profiling showed that
invA and
spiA were found in all antimicrobial-resistant NTS isolates, while
pefA was found in 55% of the resistant NTS isolates. Moreover, Abraham and his coworkers [
29] mentioned that
invA gene was detected in all isolates. Furthermore, 94% of the isolates from sheep showed an abundance of the
spvC gene. However, the
pefA gene was detected in 18 isolates from chicken and 1 isolate from sheep. Moreover, a previous study [
30] detected the presence of the
stn gene in 85.7% isolates of
S. Enteritidis among 41 collected isolates.
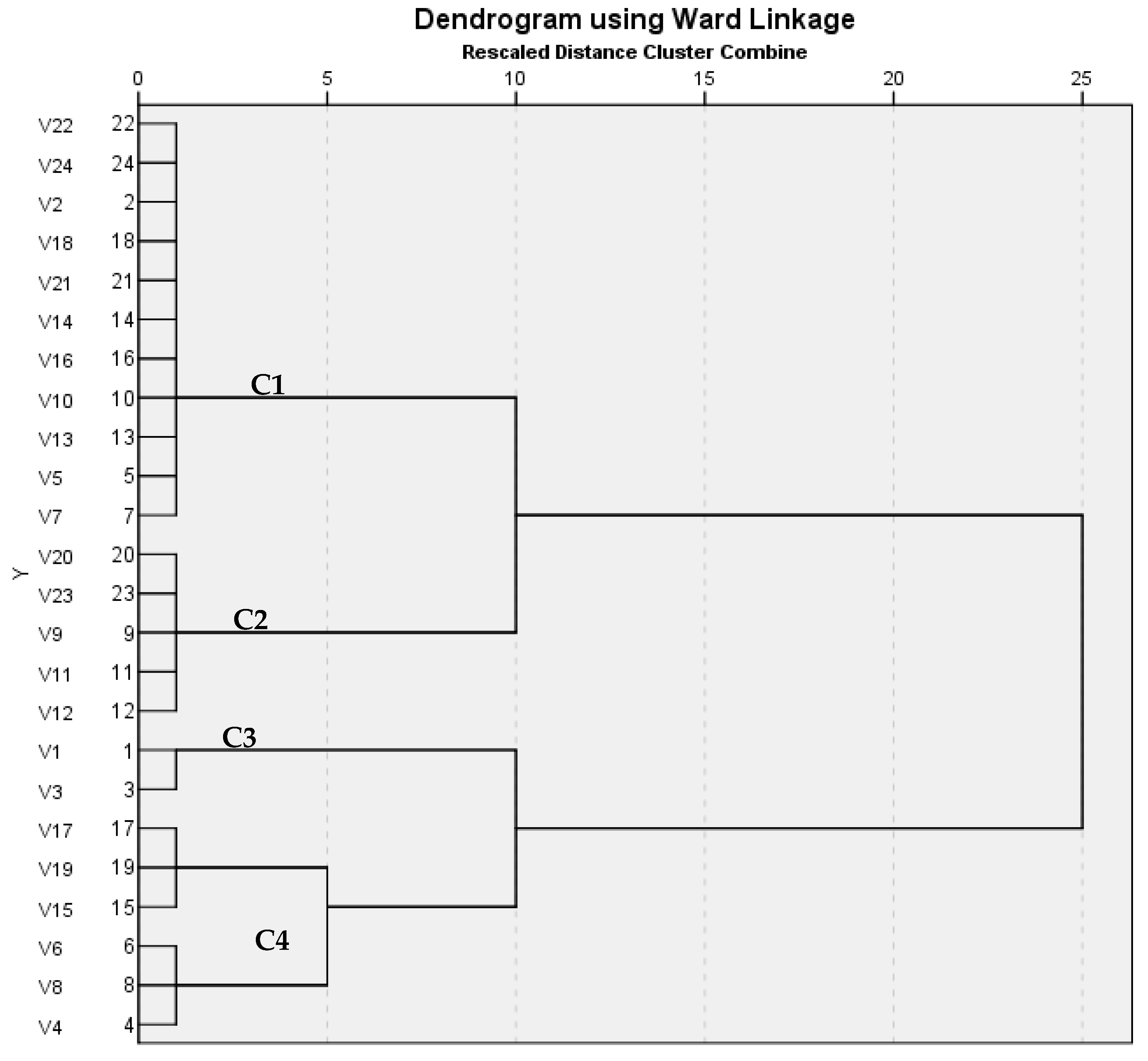
ERIC-PCR is a highly effective DNA-based typing method for fingerprinting. Epidemiology of
Salmonella Enteritidis can be studied through using ERIC-PCR analysis [
31]. In this study, ERIC PCR was performed on 12 different human serovars, which were compared with 12 different poultry serovars; the analysis showed different patterns with 1–6 band, a dendrogram revealed four clusters, and the similarity index for them showed similarity that ranged from 0.15 to 1. Similar findings were reported by Kim et al. [
28], which stated that distribution of profiles among serotypes revealed that different serotypes had similar fingerprinting patterns. Additionally, Khakzad et al. [
32] stated that most isolated strains were relevant to
Salmonella Enteritidis, and a dendrogram study showed that the bacteria were grouped in one cluster in a dendrogram that all 37 strains were put into—a large cluster of
Salmonella’s type which was divided into two clusters:
Salmonella Enterica and Bongori. Furthermore, Saravanan et al. [
33] mentioned that ERIC-PCR was used to determine the degree of variation between the isolates. All the isolates produced 8 distinct banding patterns (ERIC 1–8) ranging from 1 to 6 bands, with the fragments ranging from 150 to 2000 bp. Molecular detection of the most clinically relevant virulence genes and analysis of the associated virulence genotypes proved that the human (
n = 11) and poultry serovars (
n = 12) shared 11 genotypes. ERIC-PCR analysis revealed that human and poultry serovars clustered in 3 out of the 4 clusters using Jaccard/Tanimoto Coefficient with a similarity index ranged from 0.15 to 1.