Abstract
The emergence and spread of carbapenem-resistant Enterobacteriaceae (CRE) represent a major clinical problem and raise serious health concerns. The present study aimed to investigate and ascertain the occurrence of CRE among hospitalized patients of Mohamed VI University Hospital, Marrakech, Morocco. Biological samples were collected over a one-year period (2018). The bacterial isolates were identified by MALDI-TOF-MS. Antibiotic susceptibility testing was performed using disc diffusion and Etest. The modified Hodge test and combined disc diffusion test were used for phenotypic detection. CRE hydrolyzing enzyme encoding genes: blaOXA-48, blaKPC, blaIMP, blaVIM, and blaNDM were characterized by PCR and DNA sequencing. In total, 131 non-duplicate CRE clinical strains resistant to Ertapenem were isolated out of 1603 initial Enterobacteriaceae. Klebsiella pneumoniae was the most common species (59%), followed by Enterobacter cloacae (24%), E. coli (10%), Citrobacter freundii (3%), Klebsiella oxycota (2%), Serratia marcescens (1%), and Citrobacter braakii (1%). Of these, 56.49%, 21.37%, 15.27%, 3.38%, and 3.05% were collected from blood, urine, pus, catheters and respiratory samples, respectively. Approximately 85.5% (112/131) of the isolates were carbapenemase producers (40 blaOXA-48, 27 blaNDM, 38 blaOXA-48 + blaNDM and 7 blaVIM). All metallo-β-lactamases isolates were NDM-1 and VIM-1 producers. This is the first documentation of blaOXA-48 genes from C. freundii and C. braakii in Morocco.
1. Introduction
Over the past few years, carbapenem-resistant Enterobacteriaceae (CRE), the most frequent pathogens responsible for nosocomial infections, have emerged and rapidly spread all over the world, posing great challenges to human health [1,2,3,4,5,6,7]. Conventionally, carbapenem antibiotics have been considered the most effective among available antibacterial agents for the treatment of infections caused by Enterobacteriaceae (e.g., E. coli and Klebsiella pneumoniae), and are still being used as the antibiotics of last-resort to date [8,9]. Recently, these last-line therapeutic agents have lost effectiveness in more than half of the people treated, across many countries, leading to treatment failure and reducing clinical treatment options [10,11]. According to the latest data reported by the Chinese Antimicrobial surveillance network (www.chinets.com, accessed on 29 September 2018), the resistance rates of K. pneumoniae to meropenem and imipenem (two parenteral carbapenems) were eight times higher in 2018 compared to 2005 (2.9%–26% and 3%–25%, respectively). Moreover, carbapenem-resistant K. pneumoniae has been recently recognized as life-threatening in Europe and is even listed on the global priority list, established by the World Health Organization, of antibiotic-resistant bacteria for which new antibiotics are urgently needed [12,13].
There are three major mechanisms responsible for carbapenem resistance in Enterobacteriaceae: enzyme production (cephalosporinases, extended-spectrum β-lactamases (ESBLs), metallo-β-lactamases (MBLs) and carbapenemases), efflux pumps and porin mutations [14,15].
The production of carbapenemases, a type of β-lactamase hydrolyzing enzymes, is considered a major mechanism conferring carbapenem resistance for CRE and contributes largely to the development and spread of antibiotic resistance [16]. Carbapenemase encoding genes, often located in plasmids, are associated with mobile elements that facilitate their acquisition (e.g., by clonal and horizontal transfer) and spread from bacteria to bacteria [17]. KPC, VIM, IPM, NDM and OXA-48 are examples of the most common resistant genes widely distributed among Enterobacteriaceae clinical isolates. The blaOXA-48, first identified from carbapenem-resistant K. pneumoniae isolates in Istanbul, Turkey, has since been extensively reported as a source of nosocomial infection outbreaks in many parts of the world, notably from countries surrounding the Mediterranean area [18,19,20]. North African countries (mainly Egypt, Libya, Tunisia and Morocco) are considered the principal reservoirs of OXA-48 producing Enterobacteriaceae [21,22,23,24]. In Morocco, the situation of carbapenem-resistant Enterobacteriaceae has been the subject of only a few and outdated studies [25,26]. Most current and available research is confined to case reports or research studies that have focused on few patient numbers, for specific hospital units, specific bacterial species and specific genes, over a short period of time [27,28,29,30]. Thus, we conducted this one-year surveillance study (2018) to characterize the dissemination and characteristics of carbapenem-resistant Enterobacteriaceae (including blaOXA-48, blaKPC, blaIMP, blaVIM, and blaNDM) among hospitalized patients, from different units, in Mohamed VI University Hospital, Marrakech, Morocco.
2. Results
A total of 131 non-duplicate CRE resistant to different carbapenems were isolated and identified out of 1603 initial Enterobacteriaceae clinical strains. The majority were isolated from patients in the neonatal unit (40.3%), followed by the plastic surgical unit (14.1%), urology-nephrology unit (11.1%), pediatric reanimation unit (13%), medical-surgical resuscitations (11.1%) and finally, the medicine unit (10.1%). The age of these patients ranged from 0 months to 84 years. Of these, 57% were children (0–15 years) and 43% adults (≥16–80 years), with 58 (44.27%) females and 73 (55.73%) males in total.
Klebsiella pneumoniae was the most common isolated species (59%, 77/131) (Figure 1), followed by Enterobacter cloacae (24%, 31/131), E. coli (10%, 13/131), Citrobacter freundii (3%, 4/131), Klebsiella oxycota (2%, 3/131), Serratia marcescens (1%, 2/131) and Citrobacter braakii (1%, 1/131). The strains were isolated from various clinical specimens, including blood (56.49%, 74/131), urine (21.37%, 28/131), pus (15.27%, 20/131), catheters (3.38%, 5/131) and respiratory samples (3.05%, 4/131).
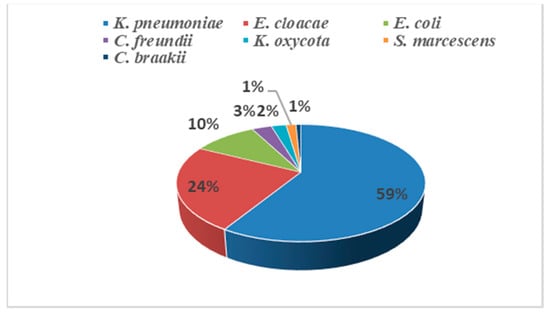
Figure 1.
Distribution of the 131 isolated CRE.
Most of the CRE strains (>83%) were resistant to ertapenem, imipenem, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, and piperacillin-tazobactam (Table 1). When all 131 CRE isolates were considered; 100% were resistant to ETP, GEN and PTZ, 87% were resistant to SXT, 86.4%were resistant to CIP and 82.6% were resistant to IMP. Only 34% of the strains were resistant to colistin. All the strains were resistant to three functional drug classes simultaneously, 100% resistant to ETP, GEN and PTZ, which means they are multidrug-resistant. Approximately 98.7% of K. pneumoniae isolates were multidrug-resistant, while 21 of them were resistant to all tested antibiotics. All isolated strains of E. cloacae and E. coli were found to be multidrug-resistant. Fifteen of the 31 E. cloacae and five out of the 13 E. coli were resistant to all tested antibiotics.

Table 1.
Antimicrobial susceptibility results of the most commonly isolated carbapenem-resistant Enterobacteriaceae strains.
Approximately 85.5% (112/131) of the isolated strains were carbapenemase producers (Table 2). Of these, 40 were positive for blaOXA-48, 27 for blaNDM and 7 for blaVIM, as determined by PCR. In 38 strains, both blaOXA-48 and blaNDM carbapenemase genes were identified. All metallo-β-lactamases isolates were NDM-1 and VIM-1 producers. IMP and KPC genes were not detected in any of our isolates. K. pneumoniae was the most common carbapenemase gene producer (61.6%, 69/112), followed by E. cloacae (21.42%, 24/112) and E. coli (9.82%, 11/112). Over 33.7% (26/77) of the K. pneumoniae isolates possessed blaOXA-48 and blaNDM simultaneously. While in only 19.35% (6/31) of E. cloacae and 23.07% (3/13) of E. coli were the two genes detected alongside each other.

Table 2.
Prevalence of carbapenemase genes among the isolated strains.
Phenotypic detection by MHT correctly identified carbapenemase producers with a sensitivity of 96% and specificity of 100%.
3. Discussion
The emergence and dissemination of carbapenem-resistant Enterobacteriaceae represent a major clinical problem often associated with significant morbidity and mortality rates, due to treatment failure and the limitation of alternative therapeutic options. Early monitoring of CRE emergence and characterization of the types of carbapenemases produced is of great value for preventing and controlling the spread of CRE in hospital settings.
In the present study, a total of 131 non-duplicate CREs resistant to different carbapenems were isolated and identified out of 1603 initial Enterobacteriaceae clinical strains (8.17%). This prevalence is three times higher than that recorded in a similar study, conducted by Wartiti et al. in Rabat, Morocco [31]. In the United States, the prevalence of CRE is generally around 1.4–4.2% [32,33]. In a study in the Asia–Pacific region, CRE isolates rate causing intra-abdominal infections was two times higher (16.5%) compared to our study [34]. On a regional scale, more specifically, in the Mediterranean area, the prevalence of carbapenemases is usually high and variant among countries, partially depending on the population exchange relationship between the regions and the possible reservoirs of each carbapenemase [19].
Indeed, the acquirement of CRE infections is highly related to certain risk factors, such as: being transferred between hospitals or from long-term care centers; a long stay in the hospital, especially in an intensive care unit; having a surgical operation; transplantation; antibiotics usage; central venous catheter presence; having diabetes mellitus [35,36]. In this study, the majority of CRE isolates were from patients in the in the neonatal unit (40.3%), followed by the plastic surgical unit (14.1%), urology-gy-nephrology unit (11.1%), pediatric reanimation unit (13%), medical-surgical resuscitations (11.1%) and finally the medicine unit (10.1%).
Among the isolated strains, K. pneumoniae was the most common species isolated in our study (59%), followed by E. cloacae (24%), then E. coli (10%). This distribution of CRE species is consistent with that of other CRE studies in the United States and Europe [32,37]. Additionally, in a recent study conducted in Tunisian and Libyan hospitals, K. pneumoniae and E. coli were the most isolated strains [23]. This is in concordance with the CDC’s recommendations that K. pneumoniae, E. coli and Enterobacter spp. are the key healthcare-associated pathogens to focus on in the control of the CRE epidemic [38]. In the Asia–Pacific region, a contrary trend was observed in the study conducted by Jean et al. [34], where Enterobacter species, K. pneumoniae and E. coli were the three major pathogens isolated with 77.4%, 40.9% and 11.7%, respectively. At the national level, the study of Wartiti et al. [31] revealed that E. coli isolates accounted for 63.9%, followed by Klebsiella spp. Accounting for 27.9% and Enterobacter spp. for 8.2%.
The majority of CRE isolates in our study were collected from the blood (56.49%), urine (21.37%), pus (15.27%), catheters (3.82%) and respiratory samples (3.05%). In comparison to other studies, urine and blood samples are usually the most common sources of CRE isolates, while other sources such as wound discharge and sputum were the least [1,39].
More than 83% of the CRE isolates were resistant to different classes of antibiotics tested, typically ertapenem, imipenem, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole and piperacillin-tazobactam. A similar antibiotic susceptibility profile was reported by Wartiti et al. [31], except for imipenem found non-susceptible in our case. In contrast, Mathlouthi et al. [23] observed a low resistance for imipenem in Tunisian and Libyan hospitals.
Carbapenemases were detected in 85.5% (112/131) of the CRE isolates (Table 2). Of these, 40 were positive for blaOXA-48, 27 for blaNDM and 7 for blaVIMas, as determined by PCR. In 38 strains, both blaOXA-48 and blaNDM carbapenemase genes were identified. The 19 remaining isolates (14.50%), negative for carbapenemase genes detection, are likely to not be susceptible to carbapenem due to a combination of cephalosporins with a weak carbapenemase activity gene [40], or simply the disposal of other carbapenemases that were not assessed in this study, such as blaGES or blaNMC-A, although these have not been reported to date in the country. OXA-48 is the most prevalent carbapenemase gene detected in Morocco, as is the case in the present work. After its initial identification from a K. pneumoniae isolates in Rabat in 2010, from a 60-year-old man with no previous medical history [24], OXA-48 producers have intensively become the most CRE responsible for nosocomial infections in our country [28,30,41]. Indeed, North African countries (mainly Egypt, Libya, Tunisia and Morocco) are considered the principal reservoirs of OXA-48 producing Enterobacteriaceae [21,22,23,24]. The blaOXA-48 gene identified in Morocco from K. pneumoniae was located inside Tn1999, a composite transposon made of two copies of the insertion sequence IS1999, which is itself always identified on a ca. 62 kb plasmid that is the main vehicle for blaOXA-48 dissemination [41]. It is worth mentioning here, that a closely related or identical OXA-48 gene to the one identified in Morocco was later recognized as a potential source of OXA-48 dissemination in the Netherlands and France [42,43].
Initially, and most frequently, the blaOXA-48 genes were identified in K. pneumoniae. However, many other CRE species have been recently recognized as OXA-48 producers, such as Enterobacter spp., K. oxytoca, E. coli, C. freundii, S. marcescens, etc. In the present study, the majority of blaOXA-48 genes were produced by K. Pneumoniae (25/40), E. cloacae (10/40) and E. coli (4/40). In Morocco, the blaOXA-48 genes were previously identified in different species, such as K. pneumoniae, K. oxytoca, E. coli, E. cloacae and M. morganii [24,25,26,28,41]. According to our best knowledge, this is the first detection of blaOXA-48 genes from C. freundii and C. braakii in the country.
Citrobacter spp., mostly C. freundii, are known as opportunistic pathogens, accountable for multiple healthcare-associated infections, including respiratory infections and urinary tract infections. In a recent study conducted by Arana et al. [44] in Spain, 57 C. freundii and one C. braakii out of 119 isolates of Citrobacter spp. tested were found to be carbapenemase producers (58.7% VIM-1, 31.7% OXA-48, 19% KPC-2, 3.2% NDM-1 and 1.6% VIM-2). C. freundii produced up to five carbapenemase types simultaneously, including VIM-1 and OXA-48 co-production. The study of Arana et al. also highlighted several remarkable findings, such as clonal and polyclonal dissemination of C. freundii across different geographical areas and hospitals. OXA-48 and VIM-1 enzymes encoded were also detected in C. freundii from patients with acute leukemia in Spain [45].
NDM gene was the second most detected carbapenemase in our study (27/112). This gene (NDM-1, New Delhi metallo-β-lactamase-1), first identified in a K. pneumoniae strain from a Swedish patient previously hospitalized in India, has since been extensively reported as a source of nosocomial infection outbreaks in many parts of the world, including Morocco [46]. Most of the NDM-1 producer isolates in the country were in a hospital setting [28]. In our study, the majority of blaNDM-1 genes were produced by K. pneumoniae (13/27), E. cloacae (7/27) and E. coli (4/27). Over 26 K. pneumoniae, 6 E. cloacae and 3 E. coli isolates, were found to be positive for both the blaNDM-1 and the blaOXA-48 genes. As with the blaOXA-48 gene, NDM-1 was initially and most frequently identified from K. pneumoniae, but has recently been increasingly reported in other species [47]. Being encoded on highly mobile conjugative plasmids, NDM enzymes are easily transferred between bacteria of inter-and intraspecies, especially by horizontal transfer, which facilitates their dissemination [48].
Other carbapenemase genes have been detected in Morocco, such as KPC, IMP, and VIM, but at a low rate compared to OXA-48 and NDM-1 [49,50]. In our study, VIM was identified in only seven isolates (5 K. pneumoniae, one E. cloacae and one C. freundii). No isolate scored positively for KPC and IMP production. In Morocco, IMP carbapenemase producers were essentially isolated from community-acquired infections, while NDM-producers were identified in hospital settings [25,49].
4. Material and Methods
4.1. Bacterial Strains
From January to December 2018, over 131 non-duplicate Gram-negative clinical isolates with reduced susceptibility to carbapenem antibiotics were collected out of 1603 initial Enterobacteriaceae from hospitalized patients in Mohammed VI University Hospital, Marrakech. These CRE strains were isolated from urine, pus, respiratory samples, blood and catheters. Species identification was previously carried out using rapid biochemical and bacterial tests, Phoenix automated microbiology system (Becton–Dickinson Diagnostic Systems, Sparks, MD, USA) and further confirmed by matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (MicroflexTM; Bruker Daltonic, Bremen, Germany).
4.2. Antibiotics Susceptibility Testing
Antibiotic susceptibility was determined on Mueller–Hinton agar using the standard disk diffusion method, as described by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2017 and also using the Phoenix system. Seventeen antibiotics (bioMérieux, Marcy L’Etoile, France) were tested, namely, amoxicillin (AMX), amoxicillin/clavulanate (AMC), piperacillin/tazobactam (PTZ), cephalothin (CEF), ceftriaxone (CRO), cefepime (CPM), ertapenem (ETP), imipenem (IMP), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP), Fosfomycin (FOS), nitrofurantoin (NIT), doxycycline (DCI), trimethoprim/sulfamethoxazole (SXT) and colistin (CS). Sensitivity to imipenem, ertapenem, meropenem (MEM) and colistin was confirmed by the antimicrobial gradient method Etest (bioMérieux, Marcy L’Etoile, France) in collected isolates.
4.3. Phenotypic Detection of Carbapenemases
The detection of carbapenemase production was characterized by two different methods: modified Hodge test (MHT) and combined disc diffusion method (metallo-β-lactamases (MBL), KPC, Ampc and OXA-48). The phenotypic detection of MBL producers by the combined disc diffusion method is based on the specific inhibition of MBLs by chelating agents. Briefly, on Mueller–Hinton agar a carbapenem disc (imipenem 10 µg, Oxoid, Hants, UK) was placed alone and in combination with an MBL inhibitor (EDTA (Sigma-Aldrich, Darmstadt, Germany), 292 µg added), KPC inhibitor (phenylboronic acid, PBA (Sigma-Aldrich), 400 µg added), cloxacillin 250 µg (AmpC inhibitor) and temocillin 30 µg (for OXA-48-like screening). The discs were placed 25 mm apart. Generated inhibition zones around the imipenem-inhibitor discs were compared to that of the imipenem disc alone. Strains showing more than 5 mm zone diameter around the imipenem with EDTA disc and the one with imipenem alone were considered MBL producers. Strains showing more than 5 mm zone diameter around the imipenem disc alone and in combination with cloxacillin were considered positive for AmpC production. If the inhibition zone difference was more than 5 mm with the imipenem plus PBA but there was no difference (<5 mm) with the imipenem plus cloxacillin, then this was an indication of the KPC enzyme. Negative results of all test synergy alongside a zone of inhibition with temocillin (30 µg) were presumptive of an OXA-48-like enzyme.
4.4. Molecular Characterization of Carbapenemase Genes
DNA was extracted from the different CRE isolates using the boiling lysis method [51]. Briefly, each strain was streaked and cultured on Luria Bertani agar, before overnight incubation at 37 °C. One colony of bacterial growth was suspended in 200 µL of nuclease-free water (Invitrogen, Paisly, UK). The suspension was boiled at 100 °C for 10 min in a thermal block (Polystat 5, Bioblock Scientific, Illkirch, France), then centrifuged at 19,000× g for 5 min. An aliquot of DNA supernatant (5 µL) was used directly as a template for PCR assay.
All CRE isolates were tested for the presence of carbapenemases genes by real-time PCR (BioRad CFX96 Real-Time System, Marnes-la-Coquette, France). Several carbapenemase genes (blaNDM, blaVIM, blaIMP, blaKPC, and blaOXA-48) were screened with specific primers listed in Table 3.

Table 3.
List of primers and probes.
PCR positive products were purified and sequenced using the Big Dye terminator chemistry on an ABI 3130XL automated sequencer (Thermo Fisher Scientific, Waltham, MA, USA). Obtained sequences were analyzed using Codon Code Aligner software (version 3.7.1.1) and then examined using the BlastN and BlastP compared against the NCBI database (www.ncbi.nlm.nih.gov, accessed on accessed on 29 September 2018) and ARG-ANNOT (Antibiotic Resistance Gene-ANNOTation).
5. Conclusions
In conclusion, in the present study, a total of 131 non-duplicate CREs were isolated and characterized from hospitalized patients of Mohamed VI University Hospital, Marrakech, Morocco. K. pneumoniae, E. cloacae, and E. coli were the most common strains among the different isolated CRE. More than 83% of the isolates were resistant to different classes of antibiotics tested, typically gentamicin, piperacillin-tazobactam, and ertapenem. Carbapenemase genes were detected in 85.5% (112/131) of the CRE isolates. Of these, 40 were positive for blaOXA-48, 27 for blaNDM, and 7 for blaVIM, as determined by PCR. In 38 strains, both blaOXA-48 and blaNDM carbapenemase genes were identified. OXA-48 and NDM were the most prevalent carbapenemase genes among K. pneumoniae, E. cloacae and E. coli isolates. This study reports the occurrence of OXA-48 in C. freundii and C. braakii for the first time in the country. Moreover, it represents the first documentation of the emergence of blaNDM and blaVIM genes in Marrakech hospitals. The alarming spread of these isolates will seriously limit options for clinical treatment in the future. Such findings emphasize the urgent need for control precautions to be taken to prevent the further spread of CRE.
Author Contributions
Conceptualization, S.L., N.S., S.M.D. and J.-M.R.; methodology, S.L., N.S. and J.-M.R.; software, S.L.; validation, S.L. and J.-M.R.; formal analysis, S.L.; investigation, S.L., N.S. and S.M.D.; resources, S.L., N.S. and J.-M.R.; data curation, S.L.; writing—original draft preparation, writing—review and editing, S.L., N.S., S.M.D. and J.-M.R.; visualization, S.L. and J.-M.R.; supervision, J.-M.R.; project administration, S.L.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable Ethical review and approval were waived, due to the retrospectively obtained and anonymized data for this non-interventional study.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Oli, A.N.; Itumo, C.J.; Okam, P.C.; Ezebialu, I.U.; Okeke, K.N.; Ifezulike, C.C.; Ezeobi, I.; Emechebe, G.O.; Okezie, U.M.; Adejumo, S.A.; et al. Carbapenem-Resistant Enterobacteriaceae Posing a Dilemma in Effective Healthcare Delivery. Antibiotics 2019, 8, 156. [Google Scholar] [CrossRef]
- Kim, J.S.; Jin, Y.-H.; Park, S.-H.; Han, S.; Kim, H.S.; Yu, J.K.; Jang, J.I.; Kim, J.; Hong, C.-K.; Lee, J.-H.; et al. Horizontal transfer of blaNDM-1-carrying IncX3 plasmid between carbapenem-resistant Enterobacteriaceae in a single patient. J. Infect. 2020, 81, 816–846. [Google Scholar] [CrossRef]
- Kalasseril, S.G.; Krishnan, R.; Vattiringal, R.K.; Paul, R.; Mathew, P.; Pillai, D. Detection of New Delhi Metallo-β-lactamase 1 and Cephalosporin Resistance Genes Among Carbapenem-Resistant Enterobacteriaceae in Water Bodies Adjacent to Hospitals in India. Curr. Microbiol. 2020, 77, 2886–2895. [Google Scholar] [CrossRef] [PubMed]
- Smibert, O.; Satlin, M.J.; Nellore, A.; Peleg, A.Y. Carbapenem-Resistant Enterobacteriaceae in Solid Organ Transplantation: Management Principles. Curr. Infect. Dis. Rep. 2019, 21, 26. [Google Scholar] [CrossRef]
- Zhang, L.; Zhai, W.; Lin, Q.; Zhu, X.; Xiao, Z.; Yang, R.; Zheng, Y.; Zhang, F.; Li, S.; Wang, C.; et al. Carbapenem-resistant Enterobacteriaceae in hematological patients: Outcome of patients with Carbapenem-resistant Enterobacteriaceae infection and risk factors for progression to infection after rectal colonization. Int. J. Antimicrob. Agents 2019, 54, 527–529. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Li, X.; Hu, Z.; Li, Z.; Lv, Y.; Lei, M.; Wu, B.; Chen, H.; Wang, X. Characteristics of Carbapenem-Resistant and Colistin-Resistant Escherichia coli Co-Producing NDM-1 and MCR-1 from Pig Farms in China. Microorganisms 2019, 7, 482. [Google Scholar] [CrossRef]
- Aires-De-Sousa, M.; Fournier, C.; Lopes, E.; De Lencastre, H.; Nordmann, P.; Poirel, L. High Colonization Rate and Heterogeneity of ESBL- and Carbapenemase-Producing Enterobacteriaceae Isolated from Gull Feces in Lisbon, Portugal. Microorganisms 2020, 8, 1487. [Google Scholar] [CrossRef] [PubMed]
- Darville, T. Imipenem and meropenem. Semin. Pediatr. Infect. Dis. 1999, 10, 38–44. [Google Scholar] [CrossRef]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, F.E.; Bukhari, E.E.; Al-Mohizea, M.M.; Hafiz, T.; Essa, E.B.; AlTokhais, Y.I. Emergence of carbapenem-resistant Enterobacteriaceae isolated from patients in a university hospital in Saudi Arabia. Epidemiology, clinical profiles and outcomes. J. Infect. Public Health 2017, 10, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Girmenia, C.; Serrao, A.; Canichella, M. Epidemiology of Carbapenem Resistant Klebsiella Pneumoniae Infections in Mediterranean Countries. Mediterr. J. Hematol. Infect. Dis. 2016, 8, e2016032. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 1 October 2020).
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Suay-García, B.; Pérez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef]
- Martin, J.; Phan, H.T.T.; Findlay, J.; Stoesser, N.; Pankhurst, L.; Navickaite, I.; De Maio, N.; Eyre, D.W.; Toogood, G.; Orsi, N.M.; et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: Long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J. Antimicrob. Chemother. 2017, 72, 3025–3034. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Cox, H.L.; Kitchel, B.; Bonatti, H.; Brassinga, A.K.C.; Carroll, J.; Scheld, W.M.; Hazen, K.C.; Sifri, C.D. Molecular Dissection of an Outbreak of Carbapenem-Resistant Enterobacteriaceae Reveals Intergenus KPC Carbapenemase Transmission through a Promiscuous Plasmid. mBio 2011, 2, e00204-11. [Google Scholar] [CrossRef] [PubMed]
- Carrër, A.; Poirel, L.; Eraksoy, H.; Cagatay, A.A.; Badur, S.; Nordmann, P. Spread of OXA-48-Positive Carbapenem-Resistant Klebsiella pneumoniae Isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 2008, 52, 2950–2954. [Google Scholar] [CrossRef]
- Mathlouthi, N.; Al-Bayssari, C.; Bakour, S.; Rolain, J.M.; Chouchani, C. Retracted Article: Prevalence and emergence of carbapenemases-producing Gram-negative bacteria in Mediterranean basin. Crit. Rev. Microbiol. 2016, 43, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.-P.; Touati, A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 37, 587–604. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.; Harris, P.; Henderson, A.; Paterson, D. Treatment of Infections by OXA-48-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e01195-18. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Mathlouthi, N.; Al-Bayssari, C.; El Salabi, A.; Bakour, S.; Ben Gwierif, S.; Zorgani, A.A.; Jridi, Y.; Ben Slama, K.; Rolain, J.-M.; Chouchani, C. Carbapenemases and extended-spectrum β-lactamases producing Enterobacteriaceae isolated from Tunisian and Libyan hospitals. J. Infect. Dev. Ctries. 2016, 10, 718–727. [Google Scholar] [CrossRef]
- Benouda, A.; Touzani, O.; Khairallah, M.-T.; Araj, G.F.; Matar, G.M. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann. Trop. Med. Parasitol. 2010, 104, 327–330. [Google Scholar] [CrossRef]
- Barguigua, A.; El Otmani, F.; Talmi, M.; Zerouali, K.; Timinouni, M. Emergence of carbapenem-resistant Enterobacteriaceae isolates in the Moroccan community. Diagn. Microbiol. Infect. Dis. 2012, 73, 290–291. [Google Scholar] [CrossRef]
- Girlich, D.; Bouihat, N.; Poirel, L.; Benouda, A.; Nordmann, P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a University hospital in Morocco. Clin. Microbiol. Infect. 2014, 20, 350–354. [Google Scholar] [CrossRef]
- Maroui, I.; Barguigua, A.; Aboulkacem, A.; Ouarrak, K.; Sbiti, M.; Louzi, H.; Timinouni, M.; Belhaj, A. First report of VIM-2 metallo-β-lactamases producing Pseudomonas aeruginosa isolates in Morocco. J. Infect. Chemother. 2016, 22, 127–132. [Google Scholar] [CrossRef]
- Barguigua, A.; Zerouali, K.; Katfy, K.; El Otmani, F.; Timinouni, M.; Elmdaghri, N. Occurrence of OXA-48 and NDM-1 carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca, Morocco. Infect. Genet. Evol. 2015, 31, 142–148. [Google Scholar] [CrossRef] [PubMed]
- El Hafa, H.; Nayme, K.; El Hamzaoui, N.; Maroui, I.; Sbiti, M.; Zerouali, K.; Timinouni, M.; Belhaj, A. Dissemination of carbapenem-resistant Acinetobacter baumannii strains carrying the blaGES, blaNDM and blaOXA23 in Morocco. Germs 2019, 9, 133–141. [Google Scholar] [CrossRef]
- Taoufik, L.; Hanchi, A.A.; Fatiha, B.; Nissrine, S.; Rabou, M.F.M.; Nabila, S. Emergence of OXA-48 Carbapenemase Producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in Marrakech, Morocco. Clin. Med. Insights Pediatr. 2019, 13, 1179556519834524. [Google Scholar] [CrossRef]
- El Wartiti, M.A.; Bahmani, F.-Z.; Elouennass, M.; Benouda, A. Prevalence of Carbapenemase-Producing Enterobacteriaceae in a University Hospital in Rabat, Morocco: A 19-Months Prospective Study. Int. Arab. J. Antimicrob. Agents 2012, 2, 1–6. [Google Scholar]
- Pollett, S.; Miller, S.; Hindler, J.; Uslan, D.; Carvalho, M.; Humphries, R.M. Phenotypic and Molecular Characteristics of Carbapenem-Resistant Enterobacteriaceae in a Health Care System in Los Angeles, California, from 2011 to 2013. J. Clin. Microbiol. 2014, 52, 4003–4009. [Google Scholar] [CrossRef]
- Morbidity and Mortality Weekly Report. Vital signs: Carbapenem-resistant Enterobacteriaceae. MMWR 2013, 62, 165–170.
- Jean, S.-S.; Lee, W.-S.; Hsueh, P.-R. Ertapenem non-susceptibility and independent predictors of the carbapenemase production among the Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Infect. Drug Resist. 2018, 11, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Sahin, K.; Tekin, A.; Ozdas, S.; Akin, D.; Yapislar, H.; Dilek, A.R.; Sonmez, E. Evaluation of carbapenem resistance using phenotypic and genotypic techniques in Enterobacteriaceae isolates. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.M.; Larsson, M.; Olson, L.; Hoang, N.T.; Le, N.K.; Khu, D.T.; Nguyen, H.D.; Vu, T.V.; Trinh, T.H.; Le, T.Q.; et al. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: Risk factors and burden of disease. J. Infect. 2019, 79, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Brolund, A.; Lagerqvist, N.; Byfors, S.; Struelens, M.J.; Monnet, D.L.; Albiger, B.; Kohlenberg, A.; European Antimicrobial Resistance Genes Surveillance Network (EURGen-Net) Capacity Survey Group. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Eurosurveillance 2019, 24, 1900123. [Google Scholar] [CrossRef] [PubMed]
- National Center for Emerging and Zoonotic Infectious Diseases. Facility Guidance for Control of Carbapenem-resistant Enterobacteriaceae (CRE). Available online: http://www.cdc.gov/hai/pdfs/cre/cre-guidance-508.pdf (accessed on 12 November 2020).
- Harish, B.; Mohamudha, P.R.; Parija, S.C. Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J. Med Res. 2012, 135, 114–119. [Google Scholar] [CrossRef]
- Kim, S.Y.; Shin, J.; Shin, S.Y.; Ko, K.S. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn. Microbiol. Infect. Dis. 2013, 76, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Hays, C.; Benouda, A.; Poirel, L.; Elouennass, M.; Nordmann, P. Nosocomial occurrence of OXA-48-producing enterobacterial isolates in a Moroccan hospital. Int. J. Antimicrob. Agents 2012, 39, 545–547. [Google Scholar] [CrossRef]
- Potron, A.; Kalpoe, J.; Poirel, L.; Nordmann, P. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin. Microbiol. Infect. 2011, 17, E24–E26. [Google Scholar] [CrossRef]
- Poirel, L.; Ros, A.; Carrër, A.; Fortineau, N.; Carricajo, A.; Berthelot, P.; Nordmann, P. Cross-border transmission of OXA-48-producing Enterobacter cloacae from Morocco to France. J. Antimicrob. Chemother. 2011, 66, 1181–1182. [Google Scholar] [CrossRef]
- Arana, D.M.; Ortega, A.; González-Barberá, E.; Lara, N.; Bautista, V.; Gómez-Ruíz, D.; Sáez, D.; Fernández-Romero, S.; Aracil, B.; Pérez-Vázquez, M.; et al. Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 2017, 72, 3283–3287. [Google Scholar] [CrossRef] [PubMed]
- Lalaoui, R.; Djukovic, A.; Bakour, S.; Hadjadj, L.; Sanz, J.; Salavert, M.; López-Hontangas, J.L.; Sanz, M.A.; Ubeda, C.; Rolain, J.-M. Genomic characterization of Citrobacter freundii strains coproducing OXA-48 and VIM-1 carbapenemase enzymes isolated in leukemic patient in Spain. Antimicrob. Resist. Infect. Control. 2019, 8, 167. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, L.; Nordmann, P. Worldwide Dissemination of the NDM-Type Carbapenemases in Gram-Negative Bacteria. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Nahid, F.; Khan, A.A.; Rehman, S.; Zahra, R. Prevalence of metallo-β-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health. J. Infect. Public Health 2013, 6, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, G.; Wu, X.; Wang, L.; Cai, J.; Chan, E.W.; Chen, S.; Zhang, R. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 2015, 6, 595. [Google Scholar] [CrossRef]
- Barguigua, A.; El Otmani, F.; El Yaagoubi, F.L.; Talmi, M.; Zerouali, K.; Timinouni, M. First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. APMIS 2012, 121, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Mahrach, Y.; Mourabit, N.; Arakrak, A.; Bakkali, M.; Laglaoui, A. Phenotypic and Molecular Study of Car-bapenemase-Producing Enterobacteriaceae in a Regional Hospital in Northern Morocco. J. Clin. Med. Sci. 2019, 3, 113. [Google Scholar] [CrossRef]
- Mentasti, M.; Prime, K.; Sands, K.; Khan, S.; Wootton, M. Rapid detection of IMP, NDM, VIM, KPC and OXA-48-like carbapenemases from Enterobacteriales and Gram-negative non-fermenter bacteria by real-time PCR and melt-curve analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2029–2036. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic Features of the Widespread Plasmid Coding for the Carbapenemase OXA-48. Antimicrob. Agents Chemother. 2011, 56, 559–562. [Google Scholar] [CrossRef]
- Al Bayssari, C.; Diene, S.M.; Loucif, L.; Gupta, S.K.; Dabboussi, F.; Mallat, H.; Hamze, M.; Rolain, J.-M. Emergence of VIM-2 and IMP-15 Carbapenemases and Inactivation ofoprDGene in Carbapenem-Resistant Pseudomonas aeruginosa Clinical Isolates from Lebanon. Antimicrob. Agents Chemother. 2014, 58, 4966–4970. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Diene, S.M.; Bruder, N.; Raoult, D.; Rolain, J.-M. Real-time PCR assay allows detection of the New Delhi metallo-β-lactamase (NDM-1)-encoding gene in France. Int. J. Antimicrob. Agents 2011, 37, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Touati, M.; Diene, S.M.; Dekhil, M.; Djahoudi, A.; Racherache, A.; Rolain, J.-M. Dissemination of a Class I Integron Carrying VIM-2 Carbapenemase in Pseudomonas aeruginosa Clinical Isolates from a Hospital Intensive Care Unit in Annaba, Algeria. Antimicrob. Agents Chemother. 2013, 57, 2426–2427. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Dai, W.; Sun, S.; Zhang, X.; Zhang, L. Prevalence of Plasmid-Mediated Quinolone Resistance and Aminoglycoside Resistance Determinants among Carbapeneme Non-Susceptible Enterobacter cloacae. PLoS ONE 2012, 7, e47636. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).