A Novel Structure Harboring blaCTX-M-27 on IncF Plasmids in Escherichia coli Isolated from Swine in China
Abstract
1. Introduction
2. Results and Discussion
2.1. Phenotypes of blaCTX-M-27-Carrying Isolates
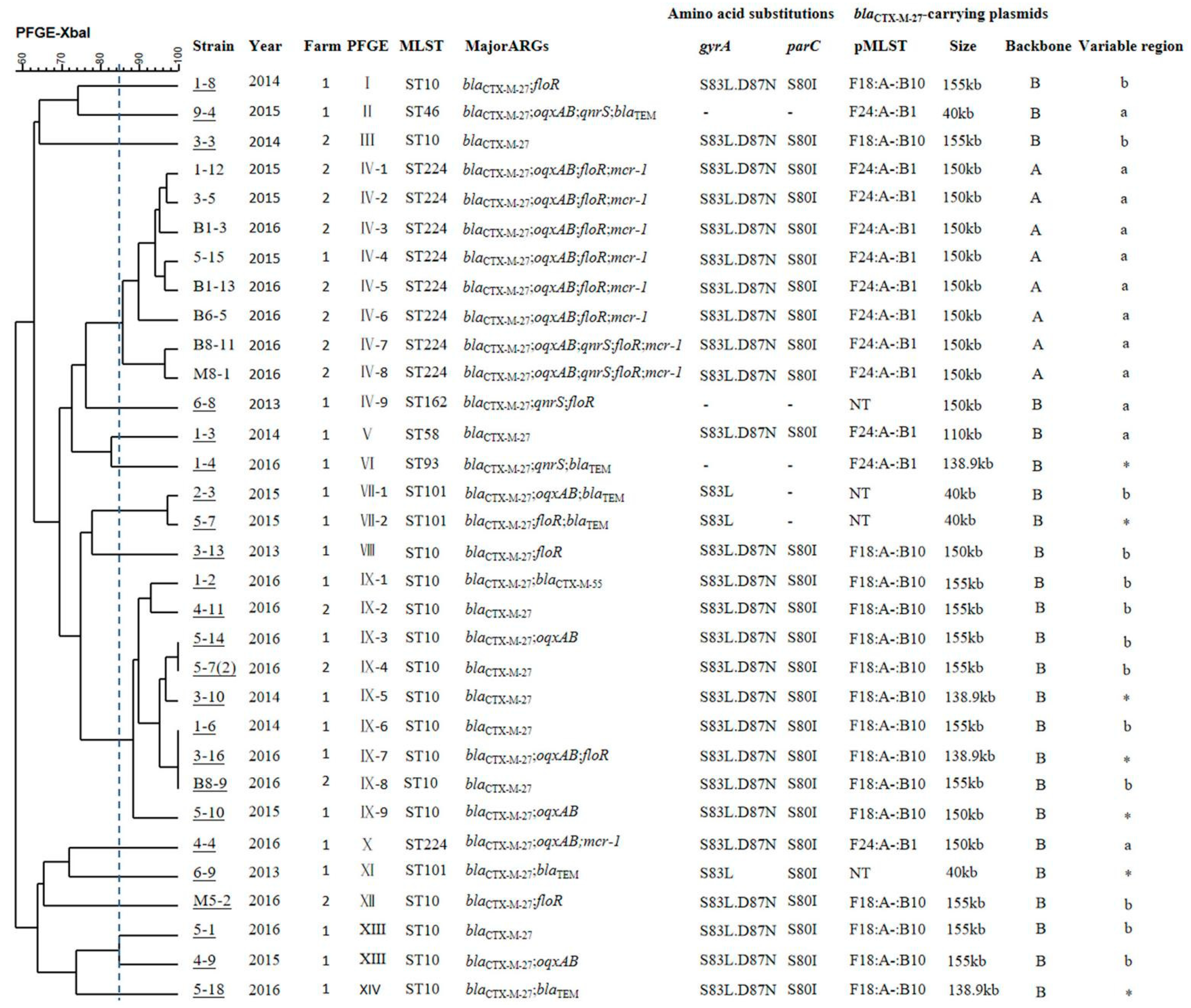
2.2. Molecular Typing Analysis
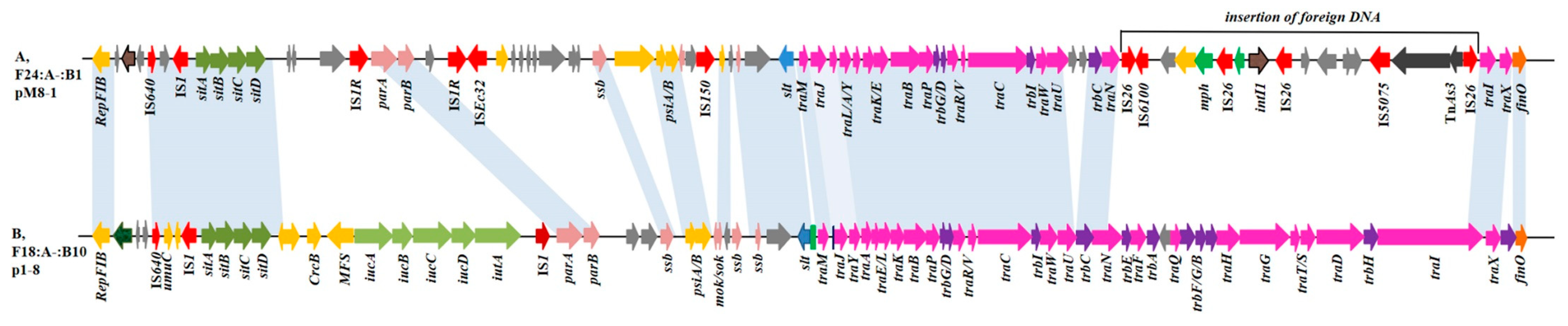
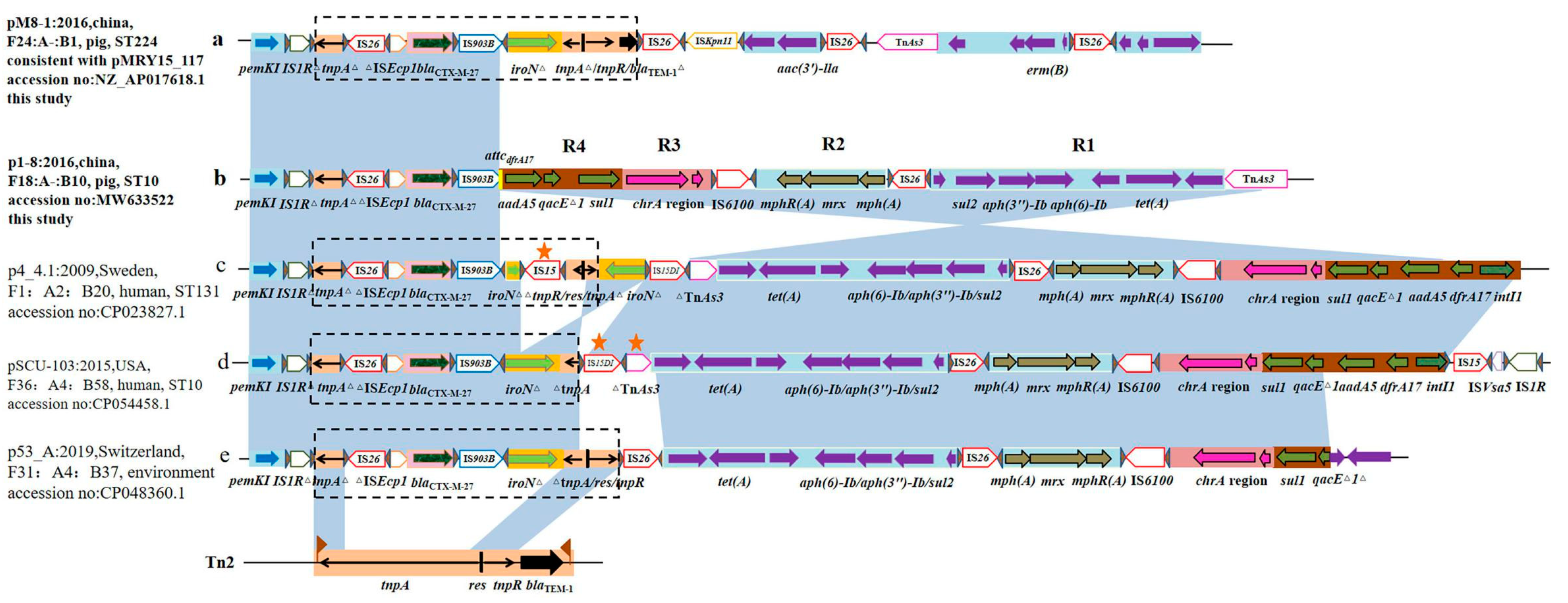
2.3. Location and Genetic Context of the blaCTX-M-27 Gene
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zeng, L.; Doi, Y.; Lv, L.; Liu, J.H. Extended-spectrum beta-lactamase-producing Escherichia coli. Lancet Infect. Dis. 2020, 20, 404–405. [Google Scholar] [CrossRef]
- Matsumura, Y.; Johnson, J.R.; Yamamoto, M.; Nagao, M.; Tanaka, M.; Takakura, S.; Ichiyama, S.; Kyoto, S.C.M.S.; Kyoto-Shiga, C.M.S.G. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J. Antimicrob. Chemother. 2015, 70, 1639. [Google Scholar] [CrossRef] [PubMed]
- Rohde, A.M.; Zweigner, J.; Wiese-Posselt, M.; Schwab, F.; Behnke, M.; Kola, A.; Schröder, W.; Peter, S.; Tacconelli, E.; Wille, T.; et al. Prevalence of third-generation cephalosporin-resistant Enterobacterales colonization on hospital admission and ESBL genotype-specific risk factors: A cross-sectional study in six German university hospitals. J. Antimicrob. Chemother. 2020, 75, 1631–1638. [Google Scholar] [CrossRef] [PubMed]
- Flament-Simon, S.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Nicolas-Chanoine, M.; Blanco, J. High Prevalence of ST131 Subclades C2-H30Rx and C1-M27 Among Extended-Spectrum β-Lactamase-Producing Escherichia coli Causing Human Extraintestinal Infections in Patients from Two Hospitals of Spain and France During 2015. Front. Cell Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of mcr-1 ST10 and ST131 E. coli Isolates. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Kawamura, K.; Sugawara, T.; Matsuo, N.; Hayashi, K.; Norizuki, C.; Tamai, K.; Kondo, T.; Arakawa, Y. Spread of CTX-Type Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates of Epidemic Clone B2-O25-ST131 Among Dogs and Cats in Japan. Microb. Drug Resist. 2017, 23, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Piccirilli, A.; Pompilio, A.; Rossi, L.; Segatore, B.; Amicosante, G.; Rosatelli, G.; Perilli, M.; Di Bonaventura, G. Identification of CTX-M-15 and CTX-M-27 in Antibiotic-Resistant Gram-Negative Bacteria Isolated from Three Rivers Running in Central Italy. Microb. Drug Resist. 2019, 25, 1041–1049. [Google Scholar] [CrossRef]
- Pepin-Puget, L.; El Garch, F.; Bertrand, X.; Valot, B.; Hocquet, D. Genome analysis of enterobacteriaceae with non-wild type susceptibility to third-generation cephalosporins recovered from diseased dogs and cats in Europe. Vet. Microbiol. 2020, 242, 108601. [Google Scholar] [CrossRef]
- Aguirre, L.; Vidal, A.; Seminati, C.; Tello, M.; Redondo, N.; Darwich, L.; Martín, M. Antimicrobial resistance profile and prevalence of extended-spectrum beta-lactamases (ESBL), AmpC beta-lactamases and colistin resistance (mcr) genes in Escherichia coli from swine between 1999 and 2018. Porc. Health Manag. 2020, 6. [Google Scholar] [CrossRef]
- Tamang, M.D.; Nam, H.; Kim, S.; Chae, M.H.; Jang, G.; Jung, S.; Lim, S. Prevalence and Molecular Characterization of CTX-M β-Lactamase–Producing Escherichia coli Isolated from Healthy Swine and Cattle. Foodborne Pathog. Dis. 2013, 10, 13–20. [Google Scholar] [CrossRef]
- Ball, T.A.; Monte, D.F.; Aidara-Kane, A.; Matheu-Alvarez, J.; Ru, H.; Thakur, S.; Horovitz, J.; Ejobi, F.; Lacher, D.W.; Fedorka-Cray, P.J. Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella enterica from Dairy Cattle Farms in the Wakiso District, Uganda: A Cross-Sectional Study. Foodborne Pathog. Dis. 2019, 16, 54–59. [Google Scholar] [CrossRef]
- Afema, J.A.; Ahmed, S.; Besser, T.E.; Jones, L.P.; Sischo, W.M.; Davis, M.A. Molecular Epidemiology of Dairy Cattle-Associated Escherichia coli Carrying blaCTX-M Genes in Washington State. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Matsumura, Y.; Pitout, J.D.D.; Gomi, R.; Matsuda, T.; Noguchi, T.; Yamamoto, M.; Peirano, G.; DeVinney, R.; Bradford, P.A.; Motyl, M.R.; et al. Global Escherichia coli Sequence Type 131 Clade with blaCTX-M-27 Gene. Emerg. Infect. Dis. 2016, 22, 1900–1907. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Li, C.; Mukherjee, S.; Hsu, C.; Bodeis Jones, S.; Gaines, S.A.; Kabera, C.; Loneragan, G.H.; Torrence, M.; Harhay, D.M.; et al. Whole-Genome Sequence Analysis of CTX-M Containing Escherichia coli Isolates from Retail Meats and Cattle in the United States. Microb. Drug Resist. 2018, 24, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, W.; Jiang, G.; Zhang, W.; Ding, H.; Liu, Y.; Zeng, Z.; Jiang, H. Characterization of a P1-like bacteriophage carrying CTX-M-27 in Salmonella spp. resistant to third generation cephalosporins isolated from pork in China. Sci. Rep. 2017, 7, 40710. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, W.; Cai, R.; Lu, Y.; Zhang, Y.; Cai, P.; Webber, M.A.; Jiang, H. Mobilization of Tn1721-like structure harboring blaCTX-M-27 between P1-like bacteriophage in Salmonella and plasmids in Escherichia coli in China. Vet. Microbiol. 2021, 253, 108944. [Google Scholar] [CrossRef]
- Jiang, H.X.; Song, L.; Liu, J.; Zhang, X.H.; Ren, Y.N.; Zhang, W.H.; Zhang, J.Y.; Liu, Y.H.; Webber, M.A.; Ogbolu, D.O.; et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 2014, 43, 242–247. [Google Scholar] [CrossRef]
- Cormier, A.; Zhang, P.L.C.; Chalmers, G.; Weese, J.S.; Deckert, A.; Mulvey, M.; McAllister, T.; Boerlin, P. Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 2019, 231, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Kim, D.H.; Ko, K.S. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 2011, 63, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Dhanji, H.; Murphy, N.M.; Akhigbe, C.; Doumith, M.; Hope, R.; Livermore, D.M.; Woodford, N. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum beta-lactamase from UK river water. J. Antimicrob. Chemother. 2011, 66, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Peirano, G.; Lynch, T.; Matsumara, Y.; Nobrega, D.; Finn, T.J.; DeVinney, R.; Pitout, J.D.D. Trends in Population Dynamics of Escherichia coli Sequence Type 131, Calgary, Alberta, Canada, 2006–20161. Emerg. Infect. Dis. 2020, 26, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
- Zogg, A.L.; Simmen, S.; Zurfluh, K.; Stephan, R.; Schmitt, S.N.; Nuesch-Inderbinen, M. High Prevalence of Extended-Spectrum beta-Lactamase Producing Enterobacteriaceae Among Clinical Isolates From Cats and Dogs Admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 2018, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ding, X.; Lin, X.; Sun, R.; Lu, Y.; Cai, R.; Webber, M.A.; Ding, H.; Jiang, H. The Emergence of Chromosomally Located blaCTX-M-55 in Salmonella from Foodborne Animals in China. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Faccone, D.; Moredo, F.A.; Giacoboni, G.I.; Albornoz, E.; Alarcón, L.; Nievas, V.F.; Corso, A. Multidrug-resistant Escherichia coli harbouring mcr-1 and blaCTX-M genes isolated from swine in Argentina. J. Glob. Antimicrob. Resist. 2019, 18, 160–162. [Google Scholar] [CrossRef]
- Komatsu, Y.; Kasahara, K.; Inoue, T.; Lee, S.; Muratani, T.; Yano, H.; Kirita, T.; Mikasa, K. Molecular epidemiology and clinical features of extended-spectrum beta-lactamase-or carbapenemase-producing Escherichia coli bacteremia in Japan. PLoS ONE 2018, 13, e202276. [Google Scholar] [CrossRef]
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An Overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, N.; Nonogaki, R.; Hayashi, M.; Wachino, J.I.; Suzuki, M.; Arakawa, Y.; Kawamura, K. Characterization of blaCTX-M-27/F1:A2:B20 Plasmids Harbored by Escherichia coli Sequence Type 131 Sublineage C1/H30R Isolates Spreading among Elderly Japanese in Nonacute-Care Settings. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Ghosh, H.; Bunk, B.; Doijad, S.; Schmiedel, J.; Falgenhauer, L.; Spröer, C.; Imirzalioglu, C.; Overmann, J.; Chakraborty, T. Complete Genome Sequence of blaCTX-M-27-Encoding Escherichia coli Strain H105 of Sequence Type 131 Lineage C1/H30R. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Birgy, A.; Levy, C.; Nicolas-Chanoine, M.; Cointe, A.; Hobson, C.A.; Magnan, M.; Bechet, S.; Bidet, P.; Cohen, R.; Bonacorsi, S. Independent Host Factors and Bacterial Genetic Determinants of the Emergence and Dominance of Escherichia coli Sequence Type 131 CTX-M-27 in a Community Pediatric Cohort Study. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Essack, S.Y. Draft genome sequence of an extended-spectrum beta-lactamase (CTX-M-15)-producing Escherichia coli ST10 isolated from a nasal sample of an abattoir worker in Cameroon. J. Glob. Antimicrob. Resist. 2018, 14, 68–69. [Google Scholar] [CrossRef]
- Zahra, R.; Javeed, S.; Malala, B.; Babenko, D.; Toleman, M.A. Analysis of Escherichia coli STs and resistance mechanisms in sewage from Islamabad, Pakistan indicates a difference in E. coli carriage types between South Asia and Europe. J. Antimicrob. Chemother. 2018, 73, 1781–1785. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum beta-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Song, J.; Oh, S.; Kim, J.; Shin, J. Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-spectrum β-lactamase-producing Enterobacteriaceae from animal origin and wastewater in Tunisia: First detection of O25b-B23-CTX-M-27-ST131 Escherichia coli and CTX-M-15/OXA-204-producing Citrobacter freundii from wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.M.; Sellera, F.P.; Fernandes, M.R.; Moura, Q.; Garino, F.; Azevedo, S.S.; Lincopan, N. Genomic features of a highly virulent, ceftiofur-resistant, CTX-M-8-producing Escherichia coli ST224 causing fatal infection in a domestic cat. J. Glob. Antimicrob. Resist. 2018, 15, 252–253. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.C.; Moreno, M.; Cabrera, C.; Spira, B.; Cerdeira, L.; Lincopan, N.; Moreno, A.M. First Characterization of CTX-M-15-Producing Escherichia coli Strains Belonging to Sequence Type (ST) 410, ST224, and ST1284 from Commercial Swine in South America. Antimicrob. Agents Chemother. 2016, 60, 2505–2508. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef]
- Colmenarejo, C.; Hernández-García, M.; Muñoz-Rodríguez, J.R.; Huertas, N.; Navarro, F.J.; Mateo, A.B.; Pellejero, E.M.; Illescas, S.; Vidal, M.D.; Del Campo, R. Prevalence and risks factors associated with ESBL-producing faecal carriage in a single long-term-care facility in Spain: Emergence of CTX-M-24- and CTX-M-27-producing Escherichia coli ST131-H30R. J. Antimicrob. Chemother. 2020, 75, 2480–2484. [Google Scholar] [CrossRef]
- Lucas, P.; Jouy, E.; Le Devendec, L.; de Boisseson, C.; Perrin-Guyomard, A.; Jove, T.; Blanchard, Y.; Touzain, F.; Kempf, I. Characterization of plasmids harboring blaCTX-M genes in Escherichia coli from French pigs. Vet. Microbiol. 2018, 224, 100–106. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, Z.L.; Huang, X.Y.; Ma, Z.B.; Guo, Z.W.; Lv, L.C.; Xia, Y.B.; Zeng, L.; Song, Q.H.; Liu, J.H. Evolution and Comparative Genomics of F33:A-:B- Plasmids Carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae Isolated from Animals, Food Products, and Humans in China. Msphere 2018, 3. [Google Scholar] [CrossRef]
- Hayashi, M.; Matsui, M.; Sekizuka, T.; Shima, A.; Segawa, T.; Kuroda, M.; Kawamura, K.; Suzuki, S. Dissemination of IncF group F1:A2:B20 plasmid-harbouring multidrug-resistant Escherichia coli ST131 before the acquisition of blaCTX-M in Japan. J. Glob. Antimicrob. Resist. 2020, 23, 456–465. [Google Scholar] [CrossRef]
- Zong, Z.; Ginn, A.N.; Dobiasova, H.; Iredell, J.R.; Partridge, S.R. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid 2015, 80, 118–126. [Google Scholar] [CrossRef]
- Canton, R.; Coque, T.M. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Jiang, H.X.; Tang, D.; Liu, Y.H.; Zhang, X.H.; Zeng, Z.L.; Xu, L.; Hawkey, P.M. Prevalence and characteristics of beta-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 2012, 67, 2350–2353. [Google Scholar] [CrossRef]
- Hibbert-Rogers, L.C.; Heritage, J.; Gascoyne-Binzi, D.M.; Hawkey, P.M.; Todd, N.; Lewis, I.J.; Bailey, C. Molecular epidemiology of ceftazidime resistant Enterobacteriaceae from patients on a paediatric oncology ward. J. Antimicrob. Chemother. 1995, 36, 65–82. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Meth. 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.F.; Mullikin, J.C.; Biesecker, L.G. Systematic Evaluation of Sanger Validation of Next-Generation Sequencing Variants. Clin. Chem. 2016, 62, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Sun, Y.-H.; Wang, J.-Y.; Chang, M.-X.; Zhao, Q.-Y.; Jiang, H.-X. A Novel Structure Harboring blaCTX-M-27 on IncF Plasmids in Escherichia coli Isolated from Swine in China. Antibiotics 2021, 10, 387. https://doi.org/10.3390/antibiotics10040387
Zhang Y, Sun Y-H, Wang J-Y, Chang M-X, Zhao Q-Y, Jiang H-X. A Novel Structure Harboring blaCTX-M-27 on IncF Plasmids in Escherichia coli Isolated from Swine in China. Antibiotics. 2021; 10(4):387. https://doi.org/10.3390/antibiotics10040387
Chicago/Turabian StyleZhang, Yan, Yin-Huan Sun, Jiang-Yang Wang, Man-Xia Chang, Qiu-Yun Zhao, and Hong-Xia Jiang. 2021. "A Novel Structure Harboring blaCTX-M-27 on IncF Plasmids in Escherichia coli Isolated from Swine in China" Antibiotics 10, no. 4: 387. https://doi.org/10.3390/antibiotics10040387
APA StyleZhang, Y., Sun, Y.-H., Wang, J.-Y., Chang, M.-X., Zhao, Q.-Y., & Jiang, H.-X. (2021). A Novel Structure Harboring blaCTX-M-27 on IncF Plasmids in Escherichia coli Isolated from Swine in China. Antibiotics, 10(4), 387. https://doi.org/10.3390/antibiotics10040387





