Abstract
The aim of this study was to determine the prevalence of A. baumannii antibiotic-resistant strains in Kazakhstan and to characterize genotypes related to epidemic “high-risk” clones. Two hundred and twenty four A. baumannii isolates from four cities of Kazakhstan in 2011–2019 were studied. Antibiotic susceptibility testing was performed by using broth microdilutions method according to EUCAST (v 11.0) recommendations. The presence of blaOXA-23-like, blaOXA-24/40-like, blaOXA-58-like, blaVIM, blaIMP, and blaNDM genes was determined by PCR. Genotyping was performed using high-throughput real-time PCR detection of 21 SNPs at 10 chromosomal loci used in existing MLST schemes. Resistance rates to imipenem, meropenem, amikacin, gentamicin, and ciprofloxacin were 81.3%, 78.6%, 79.9%, 65.2%, and 89.3%, respectively. No colistin resistant isolates were detected. The values of the MIC 50% and the MIC 90% of tigecycline were 0.125 mg/L, only four isolates (1.8%) had the ECOFF value >0.5 mg/L. The presence of acquired carbapenemase genes was found in 82.2% strains, including blaOXA-23-like (78.6%) or blaOXA-58-like (3.6%) genes. The spreading of carbapenem resistant A. baumannii strains in Kazakhstan was associated with epidemic “high-risk” clonal groups, predominantly, CG208(92)OXF/CG2PAS (80.8%) and less often CG231(109)OXF/CG1PAS (1.8%).
1. Introduction
Acinetobacter baumannii is an opportunistic pathogen that often causes diseases in immunocompromised patients [1]. A. baumannii is common in hospitals and causes a variety of nosocomial infections and iatrogenic diseases that include bloodstream infections, urinary tract infections, meningitis, wound infections, and more [1,2]. The World Health Organization categorizes included MDR A. baumannii as the “highest priority pathogen” for which antibiotic development is urgently needed [3]. However, carbapenems are considered as effective antibiotics against many drug-resistant microorganisms [4]. At once, the number of carbapenem-resistant A. baumannii isolates has been increasing recently [5].
The prevalence of carbapenem resistance of A. baumannii in etiological structure of nosocomial infections during last decade was heterogeneous. According to the EARS-Net data from 2013 to 2017, the part of carbapenem resistant A. baumannii in West Europe was 35.6%, at this time the prevalence of carbapenem resistant A. baumannii in Greece and Turkey were 79% and 95% respectively. However, according to longitude study in Austria the level of carbapenem resistant A. baumannii was more than 97% [6]. In alignment, the CHINET surveillance system in China from 2004 to 2015 the level of carbapenem resistant A. baumannii increased from 31 to 66.7% [7]. Russian multicenter surveillance study shown the comparable data when carbapenem resistant A. baumannii were detected in 77% [8]. According to multicenter study nosocomial infections caused by A. baumannii in Pakistan the rate of carbapenem resistance in 2017 was 89% [9].
Most isolates are resistant to carbapenems, and also become resistant to other classes of antimicrobial drugs such as aminoglycosides, fluoroquinolones, and to polymyxin in sporadic cases [10,11]. The rapid spread of multidrug-resistant nosocomial strains of Acinetobacter is a global concern. The present situation determines the need for regular monitoring of the sensitivity of nosocomial strains of Acinetobacter spp. and, if necessary, correction of the therapy strategy for infections caused by them [12,13]. The global spread of multidrug resistant strains, including resistant to carbapenems, among nosocomial A. baumannii strains was associated with expansion of two international “high-risk” clonal lineages, called ICL1 and ICL2 [14,15,16,17].
Microbial genotyping methods is an important tool for molecular epidemiological studies, particularly, for understanding of the population dynamics and transmission of pathogens. Even the ubiquity of whole-genome data, multilocus sequence typing (MLST) is still a “gold standard” for molecular typing of A. baumannii due to standardized landscape about population structure [18,19]. Two available MLST approaches for A. baumannii (Oxford [20] and Pasteur [14] schemes) characterized by different resolution ability but provide concordant discrimination at the level of predominant clonal groups [21,22]. In epidemiological studies, the epidemic “high-risk” clones are typically defined using by MLST nomenclature of sequence types (STs) and clonal complexes (CCs) or clonal groups (CGs) [23] and are characterized as CC231(109)OXF/CC1PAS (for ICL1) and CC208(92)OXF/CC2PAS (for ICL2) [16]. However, such approaches is still expensive and laborious especially for analysis of large sample collections, whereas high-throughput approaches allow to study in detail the entire diversity of A. baumannii strains. A lot of existing high-throughput approaches have been proposed [24,25,26,27,28,29,30] but only single nucleotide typing (SNP)-typing scheme provide direct compare SNP-typing and MLST data due to identify a set of SNPs of MLST loci [30].
In the present study, we aimed to investigate the rates of antibiotic resistance and production of acquired carbapenemase genes and to determine the genotypes and prevalence of “international high-risk clones” among A. baumannii isolates in Kazakhstan.
2. Results and Discussion
Determining the antimicrobial susceptibility of A. baumannii strains results are presented in Table 1. Resistance to carbapenems (imipenem and meropenem) were shown by 81.3% and 78.6% of A. baumannii isolates, respectively. Comparable the resistance phenotype was detected in South-Est Asia: Korea (87.0%), Singapore (95.2%), Hong Kong (50.0%) and Thailand (59.2%) [31]. Fluoroquinolones have high resistance indicators: 89.3% of A. baumannii isolates were resistant to ciprofloxacin. The frequency of resistance to aminoglycosides (amikacin and gentamicin) was 79.9% and 65.2%, respectively. Among non-beta-lactam antibiotics, colistin was highly active in vitro, none resistant isolates were detected. The values of the MIC 50% and the MIC 90% of tigecycline were 0.125 mg/l, only four isolates (1.8%) had the ECOFF value >0.5 mg/L. Combined resistance to imipenem, amikacin, and ciprofloxacin was observed in 73.2% isolates. Extremely profile of resistance make polymyxins (polymyxin B and colistin) as a last-resort treatment option [32].

Table 1.
Antibiotic sensitivity of A. baumannii isolates (n = 224).
The presence of genes for acquired molecular class D carbapenemases belonging to the OXA-23 (78.6%) and OXA-58 (3.6%) groups were revealed in 82.2% of A. baumannii isolates. No MBL genes were found in A. baumannii isolates. Actually, number of isolates harboring carbapenemase genes was higher than carbapenem resistant isolates: eight isolates were susceptible to imipenem and meropenem despite the presence of carbapenemase genes. The OXA-23 carbapenemases are major mechanism of resistance A. baumannii to carbapenems in the Central Asia and early the spreading of OXA-23 produced hospitals acquired strains of A. baumannii was described in Russia [8] and South-East Asia countries [17]. The results of assessing the sensitivity of A. baumannii isolates carrying the genes of acquired OXA-carbapenemases are presented in Table 2. The majority of carbapenemase producers were characterized by associated resistance to ciprofloxacin (97.1%), amikacin (89.7%) and gentamicin (69.5%). No resistant to colistin strains were observed.

Table 2.
Antibiotic sensitivity of A. baumannii isolates (n = 184) producing acquired OXA-carbapenemases.
All isolates were distributed into 20 genotypes (SNP-types) that grouped into 14 clonal groups (genetic clusters combining strains of related SNP-types) (Figure 1). Genotypes that differed in one or two SNPs were considered related. Despite this diversity, most of isolates (n = 181, 80.8%) from four cities were assigned to the same clonal group that combined five related genotypes (SNP-types). Through in silico analysis, nucleotide profiles of these genotypes corresponded to a set of STs that combined into a CG208OXF (formerly known as CC92OXF) and CG2PAS. Strains of clonal group CG208(92)OXF/CG2PAS were characterized by a high frequency of blaOXA-23-like carbapenemase genes (170 of 181 isolates, 93.9%). CG208(92)OXF/CG2PAS is related to international clonal lineage ICL2 that was associated with global dissemination of multidrug resistant A. baumannii strains, including neighboring countries [8,16,34].
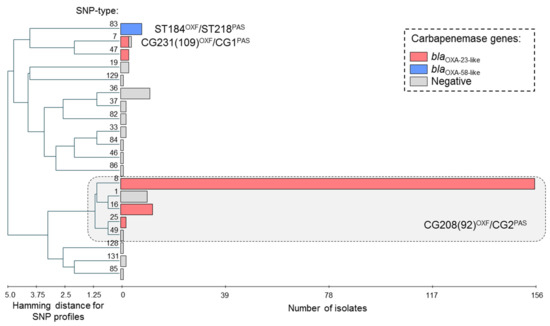
Figure 1.
Genetic diversity and production of carbapenemases in A. baumannii strains in Kazakhstan. UPGMA algorithm was used for hierarchical cluster analysis by PHYLOViZ software [35]. Horizontal rectangles correspond to different genotypes (SNP-types). Length of the rectangles is proportional to the number of isolates. MLST nomenclature was used to characterize of prevalent clonal groups. Types of acquired carbapenemase genes are highlighted.
Among other clonal groups, isolates harboring blaOXA-23-like carbapenemase genes have been also found in SNP-type 7 (3 of 4 isolates, 75.0%) and SNP-type 47 (3 of 3 isolates, 100.0%). Through in silico analysis, nucleotide profiles of SNP-type 7 corresponded to a set of STs that combined into a CG231OXF (formerly known as CC109OXF) and CG1PAS. CG231(109)OXF/CG1PAS was corresponded to another international clonal lineage ICL1 [16,34], however, an incidence of CG231(109)OXF/CG1PAS was sporadic in Kazakhstan—four isolates were collected from Nur-Sultan in 2014 and 2018. Three isolates of SNP-type 47 harboring blaOXA-23-like genes were also collected from Nur-Sultan in 2015. Interestingly enough, the epidemic “high-risk clones” CG231(109)OXF/CG1PAS and CG208(92)OXF/CG2PAS harboring blaOXA-23-like carbapenemases genes were concomitant with spreading of carbapenem resistant A. baumannii strains in Pakistan [9].
Isolates harboring blaOXA-58-like carbapenemase genes (n = 8) have been found in same genotype (SNP-type 83). All isolates of this genotype were carriers of blaOXA-58-like genes and were isolated in 2016 and 2017 from Nur-Sultan. One isolate of SNP-type 83 was investigated by MLST to clarify phylogenetic traits and was assigned to ST184OXF and ST218PAS. Strains of this clonal group CG184OXF/CG218PAS were previously found in South Korea in 2008 and in China in 2009-2010, according to the pubMLST database. Strains of CG184OXF/CG218PAS have not been previously detected in neighboring countries.
Despite the presence of blaOXA-58-like genes, all but one isolates of CG184OXF/CG218PAS were susceptible for imipenem and meropenem. For susceptible isolates, distribution of MIC ranged from 0.5 to 1 mg/L for imipenem and 0.25 to 1 mg/L for meropenem. Expression of blaOXA-58-like gene depends on the presence of insertion sequence, most often ISAba3, in association with carbapenemase gene [36,37], so, presence of blaOXA-58-like gene does not always lead to resistance to carbapenems. Furthermore, another isolate harbored blaOXA-23-like gene was found in CG208(92)OXF/CG2PAS and had a low MIC values (0.5 mg/L) for imipenem and meropenem.
A. baumannii strains harboring blaOXA-58-like genes were appeared quite recently in Kazakhstan although that strains were a serious problem since the dawn of the worldwide spreading of carbapenem resistant strains of A. baumannii [16]. For the first mention, OXA-58 carbapenemase was found in A. baumannii isolate in France in 2003 [38]. Afterwards, A. baumannii strains harboring blaOXA-58-like genes became widespread in European (Greece, France, Belgium, Italy and Turkey [39,40,41,42]), American (Argentina and USA [43,44,45]), and Asian regions (China, Taiwan and Singapore [46,47,48]). Patient transportation appears to be one of decisive factors in the spread of OXA-58-positive strains in global population of A. baumannii [40,44]. In neighboring countries, OXA-58-positive strains were found in Russia and that strains were noticed in cities bordering Kazakhstan, including Novosibirsk and Yekaterinburg (https://amrmap.net/, last accessed on 12 March 2021 [49]) [8]. According some epidemiological studies, several countries (China, Austria and Iran) remains the endemic regions of A. baumannii strains harboring blaOXA-58-like genes [6,50,51]. Thus, spread of OXA-58-producing strains in the A. baumannii population fell on 2005–2010, whereas now there has been a shift towards carbapenemase of the OXA-23-like and OXA-24/40-like [48,52,53].
Comparison of resistance among isolates of different clonal groups is presented in Table 3. CG208(92)OXF/CG2PAS isolates were significantly more resistant to multiple drugs compared to minor genotypes: to imipenem (93.9% vs 22.6%, p < 0.0001), meropenem (93.4% vs 12.9%, p < 0.0001), amikacin (91.7% vs 25.8%, p < 0.0001), gentamicin (68.5% vs 32.3%, p = 0.0002), and ciprofloxacin (100.0% vs 38.7%, p < 0.0001). It was impossible to compare resistance for CG231(109)OXF/CG1PAS or CG184OXF/CG218PAS due to small number of strains. The values of MIC 90% for tigecycline were 0.125 mg/L for each clonal groups while isolates with ECOFF value >0.5 mg/L were found in CG208(92)OXF/CG2PAS (n = 3) and SNP-type 37 (n = 1). Combined resistance to imipenem, amikacin, and ciprofloxacin among CG208(92)OXF/CG2PAS isolates was also significantly (p > 0.0001) higher compared to minor genotypes: 87.3% (95% CI: 81.7.91.4%) vs 12.9% (95% CI: 5.1–28.9%). Two of four isolates of CG231(109)OXF/CG1PAS and none of CG184OXF/CG218PAS also had combined resistance to imipenem, amikacin and ciprofloxacin.

Table 3.
Antibiotic resistance among isolates of different clonal groups.
Thus, the results of this study indicate the advisability in the routine microbiological diagnostic the detection of carbapenemases genes and genetic profiles for phylogenetic and epidemiological surveillance.
3. Materials and Methods
3.1. Sources of Bacterial Isolates
Acinetobacter baumannii (n = 224) were collected from inpatients of four cities of Kazakhstan (Karaganda, Nur-Sultan, Almaty, and Jezkazgan) in 2011–2019. Isolation and primary identification of bacterial isolates were carried out in local clinical microbiological laboratories by using standard microbiological methods. The final identification of all bacterial isolates was made in the share resource laboratory of the Karaganda Medical University (Kazakhstan). Molecular genetic studies were performed in the Research Institute of Antimicrobial Chemotherapy (Smolensk, Russia).
Material obtained from patients after 48 h of admission to the dispensary for confirmation of nosocomial infection [54,55]. Collected isolates were recovered from respiratory tract of intensive care unit patients (n = 126, 56.7%), surgical intra-abdominal infection (n = 35, 15.6%), blood of intensive care patients (n = 18, 8.0%), urinary tract infections (n = 17, 7.6%), surgical soft tissue infections (n = 17, 7.6%), bones and joints infections (n = 3, 1.3%), central nervous system lesions (n = 3, 1.3%), and other sources (n = 4, 1.8%).
3.2. Species Identification and Storage of Isolates
The isolates were identified by matrix-assisted laser desorption/ionization—time-of-flight mass spectrometry (MALDI-TOF MS) using the Microflex LT system and the MALDI Biotyper Compass v.4.1.80 software (Bruker Daltonics, Hamburg, Germany). The values known for Acinetobacter representatives were used as a criterion for reliable species identification score ≥ 2.0 according to manual. The species identification of A. baumanii isolates was confirmed by detection of species-specific blaOXA-51-like genes using real-time PCR with commercial kits “AmpliSensR MDR Ab-OXA-FL” (Central Research Institute of Epidemiology, Moscow, Russia) and the DTPrime 5X1 system (DNA-Technology, Moscow, Russia). Prior to analysis, the isolates were stored at −70 °C in trypticase-soy broth (BD, Sparks, MD, USA) supplemented with 30% glycerol. The obtained strains from local laboratories were revived on Columbia blood agar (BD, USA) aerobically at 36 ± 1 °C.
3.3. Determination of Sensitivity to Antibiotics
Determination of sensitivity to antimicrobial drugs (amikacin, gentamicin, imipenem, meropenem, netilmicin, ciprofloxacin, tigecycline, colistin) was carried out by the microdilution method in Mueller-Hinton broth. Interpretation of susceptibility testing results was performed according to recommendation of EUCAST v 11.0 [33]. To control the quality of the sensitivity determination, we used Escherichia coli ATCC®25922, Escherichia coli ATCC®35218 strains, Pseudomonas aeruginosa ATCC®27853.
3.4. Identification of Carbapenemase Genes
The presence of acquired class D carbapenemases genes common for Acinetobacter spp. (groups OXA-23, OXA-24/40, and OXA-58), as well as class B carbapenemases (metallo-beta-lactamases (MBL) of VIM, IMP, and NDM groups) were determined by real-time PCR using commercial kits “AmpliSensR MDR Acinetobacter-OXA-FL” and “AmpliSensR MDR MBL-FL” (Central Research Institute of Epidemiology, Moscow, Russia). For amplification, a DTPrime 5X1 real-time PCR system (DNA Technology, Moscow, Russia) was used. Strains A. baumannii, A. pittii, and P. aeruginosa, carring the known carbapenemases genes of the listed groupswere used as positive controls. DNA extraction was performed by express method using InstaGeneTM matrix (Bio-Rad, Hercules, CA, USA). Samples of extracted DNA were stored at −20 °C before testing. The results of assessing the sensitivity to antibiotics and determining the genes of various types of carbapenemases have been deposited to the AMRmap website database [49].
3.5. Molecular Genotyping of A. baumannii
Genetic diversity of A. baumannii was explored by single nucleotide polymorphism (SNP)-typing method based on the analysis of 21 informative SNPs at 10 chromosomal loci (gltA, recA, cpn60, gyrB, gdhB, rpoD, fusA, pyrG, rplB, and rpoB) used in the University of Oxford and the Institute Pasteur multilocus sequencing-typing (MLST) schemes [30]. Detection of each SNP was performed by allele-specific real-time PCR according to the high-throughput approach proposed Myakishev et al. [56]. Obtained sequences of 21 nucleotide bases for each A. baumannii isolate were used to assign a certain genotype (SNP-type) and to assess genetic relatedness between genotypes. The QIAgility system (QIAGEN, Hilden, Germany) and DTPrime 5X1 (DNA-Technology, Moscow, Russia) were used to prepare and conduct PCR in 384-well format that ensured high-throughput genotyping of isolates. A. baumannii strains of known STs submitted to pubMLST database (https://pubmlst.org/organisms/acinetobacter-baumannii, last accessed on 12 March 2021): id4785, id4790, id4793, id4798, id4816, id4825, id4841, id4932, id4985, id5036 were used as controls.
The selected set of 21 SNPs from MLST loci provided a comparison between obtained SNP-types with known STs and CCs according to the MLST nomenclature, including the so-called “high-risk international clones”. That correspondence between SNP-typing and MLST data was provided by SQL database and software platform (called SNPTAb, http://snpt.antibiotic.ru:9002/, last accessed on 12 March 2021) [57]. Furthermore, the SNPTAb database was used to store of SNP-typing data with individual isolates data (e.g., source, geographical origin, data of isolation, resistance to carbapenems, and production of carbapenemases). Cluster analysis of obtained SNP profiles and was carried out using the PHYLOViZ 2.0 software [35].
MLST was performed for one isolate using both the University of Oxford and the Institute Pasteur schemes as described previously [14,20]. Obtained MLST sequences were uploaded to PubMLST database (https://pubmlst.org/organisms/acinetobacter-baumannii, last accessed on 12 March 2021) to identify alleles and STs.
4. Conclusions
Nosocomial infections by carbapenem resistant A. baumannii strains emerge sharply in Kazakhstan since 2011. The results of this study indicate a high prevalence of resistance to most antimicrobials, including all carbapenems, aminoglycosides, and fluoroquinolones used to treat infections caused by this pathogen. Colistin was highly active on all A. baumannii isolates. The main mechanism of resistance to carbapenems of A. baumannii was the production of acquired carbapenemases belonging to the OXA-23 group. The spreading of carbapenem resistant A. baumannii strains in Kazakhstan was associated with epidemic “high-risk” clonal groups, predominantly, CG208(92)OXF/CG2PAS (80.8% isolates) and less often CG231(109)OXF/CG1PAS (1.8% isolates). Isolates of these clonal groups were significantly more resistant to aminoglycosides and fluoroquinolones compared to other genetic lines. Furthermore, several isolates were carrying blaOXA-58-like carbapenemase genes and combined into one clonal group CG184OXF/CG218PAS. Given the high probability of A. baumannii strains resistance to the main antibacterial drugs for the treatment of nosocomial infections, the choice of antibiotics for empiric therapy is extremely difficult and requires regular local monitoring of sensitivity in each hospital.
Author Contributions
Conceptualization, A.L. and A.T.; Data curation, I.A.; Formal analysis, E.S. and I.A.; Investigation, A.L. and S.K.; Methodology, A.L. and I.A.; Resources, A.L.; Software, E.S.; Supervision, A.T.; Writing—original draft, A.L. and S.K.; Writing—review & editing, A.L. and A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.H.; Kim, Y.C.; Kim, E.J.; Jung, I.Y.; Jeong, S.J.; Kim, S.Y.; Park, M.S.; Kim, A.; Lee, J.G.; Paik, H.C. Multidrug-Resistant Acinetobacter baumannii Infection in Lung Transplant Recipients: Risk Factors and Prognosis. Infect. Dis. 2019, 51, 493–501. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Available online: https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ (accessed on 30 December 2020).
- Hu, Y.; He, L.; Tao, X.; Meng, F.; Zhang, J. Biofilm May Not Be Necessary for the Epidemic Spread of Acinetobacter baumannii. Sci. Rep. 2016, 6, 32066. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Grisold, A.J.; Luxner, J.; Bedenić, B.; Diab-Elschahawi, M.; Berktold, M.; Wechsler-Fördös, A.; Zarfel, G.E. Diversity of Oxacillinases and Sequence Types in Carbapenem-Resistant Acinetobacter baumannii from Austria. Int. J. Environ. Res. Public Health 2021, 18, 2171. [Google Scholar] [CrossRef]
- Hu, F.P.; Guo, Y.; Zhu, D.M.; Wang, F.; Jiang, X.F.; Xu, Y.C.; Zhang, X.J.; Zhang, C.X.; Ji, P.; Xie, Y.; et al. Resistance Trends among Clinical Isolates in China Reported from CHINET Surveillance of Bacterial Resistance, 2005–2014. Clin. Microbiol. Infect. 2016, 22, S9–S14. [Google Scholar] [CrossRef]
- Shek, E.A.; Sukhorukova, M.V.; Edelstein, M.V.; Skleenova, E.Y.; Ivanchik, N.V.; Shajdullina, E.R.; Kuzmenkov, A.Y.; Dekhnich, A.V.; Kozlov, R.S.; Semyonova, N.V.; et al. Antimicrobial Resistance, Carbapenemase Production, and Genotypes of Nosocomial Acinetobacter spp. Isolates in Russia: Results of Multicenter Epidemiological Study ”MARATHON 2015–2016”. Clin. Microbiol. Antimicrob. Chemother. 2019, 21, 171–180. [Google Scholar] [CrossRef]
- Khurshid, M.; Rasool, M.H.; Ashfaq, U.A.; Aslam, B.; Waseem, M.; Xu, Q.; Zhang, X.; Guo, Q.; Wang, M. Dissemination of blaOXA-23-Harbouring Carbapenem-Resistant Acinetobacter baumannii Clones in Pakistan. J. Glob. Antimicrob. Resist. 2020, 21, 357–362. [Google Scholar] [CrossRef]
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H.; Garnacho-Montero, J.; et al. High Incidence of Pandrug-Resistant Acinetobacter baumannii Isolates Collected from Patients with Ventilator-Associated Pneumonia in Greece, Italy and Spain as Part of the MagicBullet Clinical Trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-Resistant Acinetobacter baumannii: Beyond Carbapenem Resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef]
- Tavares, L.C.B.; De Vasconcellos, F.M.; De Sousa, W.V.; Rocchetti, T.T.; Mondelli, A.L.; Ferreira, A.M.; Montelli, A.C.; Sadatsune, T.; Tiba-Casas, M.R.; Camargo, C.H. Emergence and Persistence of High-Risk Clones among MDR and XDR A. baumannii at a Brazilian Teaching Hospital. Front. Microbiol. 2019, 10, 2898. [Google Scholar] [CrossRef]
- Giannouli, M.; Tomasone, F.; Agodi, A.; Vahaboglu, H.; Daoud, Z.; Triassi, M.; Tsakris, A.; Zarrilli, R. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Strains in Intensive Care Units of Multiple Mediterranean Hospitals. J. Antimicrob. Chemother. 2009, 63, 828–830. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The Population Structure of Acinetobacter baumannii: Expanding Multiresistant Clones from an Ancestral Susceptible Genetic Pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Nemec, A.; Dijkshoorn, L.; Van Der Reijden, T.J.K. Long-Term Predominance of Two Pan-European Clones among Multi-Resistant Acinetobacter baumannii Strains in the Czech Republic. J. Med. Microbiol. 2004, 53, 147–153. [Google Scholar] [CrossRef]
- Karah, N.; Sundsfjord, A.; Towner, K.; Samuelsen, Ø. Insights into the Global Molecular Epidemiology of Carbapenem Non-Susceptible Clones of Acinetobacter baumannii. Drug Resist. Updat. 2012, 15, 237–247. [Google Scholar] [CrossRef]
- Hamidian, M.; Nigro, S.J. Emergence, Molecular Mechanisms and Global Spread of Carbapenem-Resistant Acinetobacter baumannii. Microb. Genom. 2019, 5. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C.J. Using MLST to Study Bacterial Variation: Prospects in the Genomic Era. Future Microbiol. 2014, 9, 623–630. [Google Scholar] [CrossRef]
- Kim, M.H.; Jeong, H.; Sim, Y.M.; Lee, S.; Yong, D.; Ryu, C.M.; Choi, J.Y. Using Comparative Genomics to Understand Molecular Features of Carbapenem-Resistant Acinetobacter baumannii from South Korea Causing Invasive Infections and Their Clinical Implications. PLoS ONE 2020, 15. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a Multilocus Sequence Typing Scheme for Characterization of Clinical Isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Gaiarsa, S.; Batisti Biffignandi, G.; Esposito, E.P.; Castelli, M.; Jolley, K.A.; Brisse, S.; Sassera, D.; Zarrilli, R. Comparative Analysis of the Two Acinetobacter baumannii Multilocus Sequence Typing (MLST) Schemes. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Castillo-Ramírez, S.; Graña-Miraglia, L. Inaccurate Multilocus Sequence Typing of Acinetobacter baumannii. Emerg. Infect. Dis. 2019, 25, 186–187. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Tassios, P.T.; Dijkshoorn, L.; Haeggman, S.; Cookson, B.; Fry, N.K.; Fussing, V.; Green, J.; Feil, E.; Gerner-smidt, P.; et al. Guidelines for the Validation and Application of Typing Methods for Use in Bacterial Epidemiology. Clin. Microbiol. Infect. 2007, 13, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Pournaras, S.; Gogou, V.; Giannouli, M.; Dimitroulia, E.; Dafopoulou, K.; Tsakris, A.; Zarrilli, R. Single-Locus-Sequence-Based Typing of blaOXA-51-like Genes for Rapid Assignment of Acinetobacter baumannii Clinical Isolates to International Clonal Lineages. J. Clin. Microbiol. 2014, 52, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Gundi, V.A.K.B.; Dijkshoorn, L.; Burignat, S.; Raoult, D.; La Scola, B. Validation of Partial rpoB Gene Sequence Analysis for the Identification of Clinically Important and Emerging Acinetobacter Species. Microbiology 2009, 155, 2333–2341. [Google Scholar] [CrossRef]
- Higgins, P.G.; Lehmann, M.; Wisplinghoff, H.; Seifert, H. gyrB Multiplex PCR to Differentiate between Acinetobacter calcoaceticus and Acinetobacter Genomic Species 3. J. Clin. Microbiol. 2010, 48, 4592–4594. [Google Scholar] [CrossRef]
- Turton, J.F.; Gabriel, S.N.; Valderrey, C.; Kaufmann, M.E.; Pitt, T.L. Use of Sequence-Based Typing and Multiplex PCR to Identify Clonal Lineages of Outbreak Strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007, 13, 807–815. [Google Scholar] [CrossRef]
- Suzuki, M.; Hosoba, E.; Matsui, M.; Arakawa, Y. New PCR-Based Open Reading Frame Typing Method for Easy, Rapid, and Reliable Identification of Acinetobacter baumannii International Epidemic Clones without Performing Multilocus Sequence Typing. J. Clin. Microbiol. 2014, 52, 2925–2932. [Google Scholar] [CrossRef]
- Sousa, C.; Silva, L.; Grosso, F.; Lopes, J.; Peixe, L. Development of a FTIR-ATR Based Model for Typing Clinically Relevant Acinetobacter baumannii Clones Belonging to ST98, ST103, ST208 and ST218. J. Photochem. Photobiol. B Biol. 2014, 133, 108–114. [Google Scholar] [CrossRef]
- Sheck, E.; Fedintsev, A.; Skleenova, E.; Martinovich, A.; Edelstein, M. Development of a High-Throughput Single Nucleotide Polymorphism Typing Method for Acinetobacter baumannii (SNPTAb). In Proceedings of the 25th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Copenhagen, Denmark, 25–28 April 2015. [Google Scholar]
- Jean, S.S.; Hsueh, P.R. High Burden of Antimicrobial Resistance in Asia. Int. J. Antimicrob. Agents 2011, 37, 291–295. [Google Scholar] [CrossRef]
- Karaiskos, I.; Giamarellou, H. Multidrug-Resistant and Extensively Drug-Resistant Gram-Negative Pathogens: Current and Emerging Therapeutic Approaches. Expert Opin. Pharmacother. 2014, 15, 1351–1370. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical Breakpoints–Breakpoints and Guidance. Ver. 11.0. 2021. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 12 March 2021).
- Zarrilli, R.; Pournaras, S.; Giannouli, M.; Tsakris, A. Global Evolution of Multidrug-Resistant Acinetobacter baumannii Clonal Lineages. Int. J. Antimicrob. Agents 2013, 41, 11–19. [Google Scholar] [CrossRef]
- Nascimento, M.; Sousa, A.; Ramirez, M.; Francisco, A.P.; Carriço, J.A.; Vaz, C. PHYLOViZ 2.0: Providing Scalable Data Integration and Visualization for Multiple Phylogenetic Inference Methods. Bioinformatics 2017, 33, 128–129. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Genetic Structures at the Origin of Acquisition and Expression of the Carbapenem-Hydrolyzing Oxacillinase Gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 1442–1448. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Poirel, L.; Marqué, S.; Héritier, C.; Segonds, C.; Chabanon, G.; Nordmann, P. OXA-58, a Novel Class D β-Lactamase Involved in Resistance to Carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 202–208. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P. Carbapenem Resistance in Acinetobacter baumannii: Mechanisms and Epidemiology. Clin. Microbiol. Infect. 2006, 12, 826–836. [Google Scholar] [CrossRef]
- Wybo, I.; Blommaert, L.; De Beer, T.; Soetens, O.; De Regt, J.; Lacor, P.; Piérard, D.; Lauwers, S. Outbreak of Multidrug-Resistant Acinetobacter baumannii in a Belgian University Hospital after Transfer of Patients from Greece. J. Hosp. Infect. 2007, 67, 374–380. [Google Scholar] [CrossRef]
- Mendes, R.E.; Spanu, T.; Deshpande, L.; Castanheira, M.; Jones, R.N.; Fadda, G. Clonal Dissemination of Two Clusters of Acinetobacter baumannii Producing OXA-23 or OXA-58 in Rome, Italy. Clin. Microbiol. Infect. 2009, 15, 588–592. [Google Scholar] [CrossRef]
- Karampatakis, T.; Antachopoulos, C.; Tsakris, A.; Roilides, E. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii in Greece: An Extended Review (2000–2015). Future Microbiol. 2017, 12, 801–815. [Google Scholar] [CrossRef]
- Merkier, A.K.; Catalano, M.; Ramírez, M.S.; Quiroga, C.; Orman, B.; Ratier, L.; Famiglietti, A.; Vay, C.; Di Martino, A.; Kaufman, S.; et al. Polyclonal Spread of bla(OXA-23) and bla(OXA-58) in Acinetobacter baumannii Isolates from Argentina. J. Infect. Dev. Ctries. 2008, 2, 235–240. [Google Scholar] [CrossRef][Green Version]
- Huang, X.Z.; Chahine, M.A.; Frye, J.G.; Cash, D.M.; Lesho, E.P.; Craft, D.W.; Lindler, L.E.; Nikolich, M.P. Molecular Analysis of Imipenem-Resistant Acinetobacter baumannii Isolated from US Service Members Wounded in Iraq, 2003–2008. Epidemiol. Infect. 2012, 140, 2302–2307. [Google Scholar] [CrossRef]
- Lopes, B.S.; Al-Hassan, L.; Amyes, S.G.B. ISAba825 Controls the Expression of the Chromosomal blaOXA-51-like and the Plasmid Borne blaOXA-58 Gene in Clinical Isolates of Acinetobacter baumannii Isolated from the USA. Clin. Microbiol. Infect. 2012, 18, E446–E451. [Google Scholar] [CrossRef]
- Lee, Y.-T.; Huang, L.-Y.; Chiang, D.-H.; Chen, C.-P.; Chen, T.-L.; Wang, F.-D.; Fung, C.-P.; Siu, L.-K.; Cho, W.-L. Differences in Phenotypic and Genotypic Characteristics among Imipenem-Non-Susceptible Acinetobacter Isolates Belonging to Different Genomic Species in Taiwan. Int. J. Antimicrob. Agents 2009, 34, 580–584. [Google Scholar] [CrossRef]
- Koh, T.H.; Tan, T.T.; Khoo, C.T.; Ng, S.Y.; Tan, T.Y.; Hsu, L.Y.; Ooi, E.E.; Van Der Reijden, T.J.K.; Dijkshoorn, L. Acinetobacter calcoaceticus-Acinetobacter baumannii Complex Species in Clinical Specimens in Singapore. Epidemiol. Infect. 2012, 140, 535–538. [Google Scholar] [CrossRef]
- Chen, L.; Tan, P.; Zeng, J.; Yu, X.; Cai, Y.; Liao, K.; Guo, P.; Chen, Y.; Wu, Z.; Qu, P.; et al. Impact of an Intervention to Control Imipenem-Resistant Acinetobacter baumannii and Its Resistance Mechanisms: An 8-Year Survey. Front. Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Kuzmenkov, A.Y.; Trushin, I.V.; Vinogradova, A.G.; Avramenko, A.A.; Sukhorukova, M.V.; Malhotra-Kumar, S.; Dekhnich, A.V.; Edelstein, M.V.; Kozlov, R.S. AMRmap: An Interactive Web Platform for Analysis of Antimicrobial Resistance Surveillance Data in Russia. Front. Microbiol. 2021, 12, 377. [Google Scholar] [CrossRef]
- Tafreshi, N.; Babaeekhou, L.; Ghane, M. Antibiotic Resistance Pattern of Acinetobacter baumannii from Burns Patients: Increase in Prevalence of blaOXA-24-like and blaOXA-58-like Genes. Iran. J. Microbiol. 2019, 11, 502–509. [Google Scholar] [CrossRef]
- Li, S.; Duan, X.; Peng, Y.; Rui, Y. Molecular Characteristics of Carbapenem-Resistant Acinetobacter Spp. From Clinical Infection Samples and Fecal Survey Samples in Southern China. BMC Infect. Dis. 2019, 19. [Google Scholar] [CrossRef]
- Zarrilli, R.; Bagattini, M.; Migliaccio, A.; Esposito, E.P.; Triassi, M. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii in Italy. Ann. Di Ig Med. Prev. E Di Comunità 2020. [Google Scholar] [CrossRef]
- Silva, L.; Grosso, F.; Rodrigues, C.; Ksiezarek, M.; Ramos, H.; Peixe, L. The Success of Particular Acinetobacter baumannii Clones: Accumulating Resistance and Virulence inside a Sugary Shield. J. Antimicrob. Chemother. 2021, 76, 305–311. [Google Scholar] [CrossRef]
- Nakamura, R.K.; Tompkins, E. Nosocomial Infections. Compend. Contin. Educ. Vet. 2012, 34, E1–E10. [Google Scholar] [PubMed]
- Hughes, J.M. Nosocomial Infection Surveillance in the United States: Historical Perspective. Infect. Control. 1987, 8, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Myakishev, M.V.; Khripin, Y.; Hu, S.; Hamer, D.H. High-Throughput SNP Genotyping by Allele-Specific PCR with Universal Energy-Transfer-Labeled Primers. Genome Res. 2001, 11, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Fedintsev, A.; Sheck, E.; Trushin, I.; Avramenko, A.; Edelstein, M. Development of an Online Database and Web Application for Analysis of SNP Typing Data of Acinetobacter baumannii. Proccedings of 26th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Amsterdam, The Netherlands, 9–12 April 2016. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).