Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coman, A.; Maftei, D.N.; Chereches, R.M.; Zavrotchi, E.; Bria, P.; Dragnea, C.; Mckenzie, P.P.; Valentine, M.A.; Gray, G.C. Avi-an Influenza Surveillance in the Danube Delta Using Sentinel Geese and Ducks. Influ. Res. Treat. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Páll, E.; Niculae, M.; Kiss, T.; Şandru, C.D.; Spînu, M. Human impact on the microbiological water quality of the rivers. J. Med. Microbiol. 2013, 62, 1635–1640. [Google Scholar] [CrossRef]
- Romanescu, G. Alluvial transport processes and the impact of anthropogenic intervention on the Romanian littoral of the Danube Delta. Ocean Coast. Manag. 2013, 73, 31–43. [Google Scholar] [CrossRef]
- Ionita, C.; Bogdan, T.A.; Ipate, I.; Ivana, S.; Ionita, L.; Gogu-Bogdan, M. Strategies for conserving birds biodiversity in Danube del-ta. Ocean Coast. Manag. 2010, 15, 641–644. [Google Scholar]
- Giosan, L.; Donnelly, J.P.; Vespremeanu, E. River delta morphodynamics: Examples from the Danube Delta. River Deltas-Concepts Models Ex. 2005, 83, 393–411. [Google Scholar]
- Danube Delta-UNESCO World Heritage Centre. Available online: https://whc.unesco.org/en/list/588/ (accessed on 12 January 2021).
- Păunescu, C. A study of migration in time of the Sacalin Island, Danube Delta, Romania. Proc. Rom. Acad. Ser. B 2012, 2, 156–160. [Google Scholar]
- Greig, J.; Rajić, A.; Young, I.; Mascarenhas, M.; Waddell, L.; Lejeune, J. A scoping review of the role of wildlife in the transmis-sion of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health 2015, 62, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Imtiaz, M.A.; Sayeed, M.A.; Shaikat, A.H.; Hassan, M.M. Antimicrobial resistance pattern in domestic animal-Wildlife-environmental niche via the food chain to humans with a Bangladesh perspective; A systematic review. BMC Vet. Res. 2020, 16. [Google Scholar] [CrossRef]
- Abulreesh, H.H.; Goulder, R.; Scott, G.W. Wild birds and human pathogens in the context of ringing and migration. Ringing Migr. 2007, 23, 193–200. [Google Scholar] [CrossRef]
- Lord, A.T.K.; Mohandas, K.; Somanath, S.; Ambu, S. Multidrug resistant yeasts in synanthropic wild birds. Ann. Clin. Microbiol. Antimicrob. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Elmberg, J.; Berg, C.; Lerner, H.; Waldenström, J.; Hessel, R. Health perspective. Infect. Ecol. Epidemiol. 2017, 7, 1300450. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leung, P.C.; Qian, P.Y.; Gu, J.D. Antibiotic resistance and plasmid profile of environmental isolates of Vibrio species from Mai Po Nature Reserve, Hong Kong. Ecotoxicology 2006, 15, 371–378. [Google Scholar] [CrossRef]
- Li, B.; Liu, J.; Zhou, S.; Fu, L.; Yao, P.; Chen, L.; Yang, Z.; Wang, X.; Zhang, X.-H. Vertical variation in Vibrio community com-position in Sansha Yongle Blue Hole and its ability to degrade macromolecules. Mar. Life Sci. Technol. 2020, 2, 60–72. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Hunter, A.C.; Murray, J.C. Long-Circulating and Target-Specific Nanoparticles: Theory to Practice. Pharmacol. Rev. 2001, 53, 283–318. [Google Scholar]
- Sarita, G.B. Multiple Antibiotic Resistances of Vibrio Isolates from Coastal and Brackish Water Areas. Am. J. Biochem. Biotechnol. 2005, 1, 201–206. [Google Scholar]
- Chikwendu, C. Multiple Antimicrobial Resistance in Vibrio spp Isolated from River and Aquaculture Water Sources in Imo State, Nigeria. Br. Microbiol. Res. J. 2014, 4, 560–569. [Google Scholar] [CrossRef]
- Brooks, G.F.; Carroll, K.C.; Butel, J.S.; Morse, S.A. Vibrio, Campylobacters, Helicobacter and associated bacteria. Jawetz Melnock Adelberg’s Med. Microbiol. 2007, 27e, 270–279. [Google Scholar]
- Osunla, C.A.; Okoh, A.I. Vibrio Pathogens: A Public Health Concern in Rural Water Resources in Sub-Saharan Africa. Int. J. Environ. Res. Public Health 2017, 14, 1188. [Google Scholar] [CrossRef]
- Adebayo-Tayo, B.C.; Okonko, I.O.; Esen, C.U.; Odu, N.N.; Onoh, C.C.; Igwiloh, N.J.P. Incidence of Potentially Pathogenic Vib-rio Spp. In Fresh Seafood from Itu Creek in Uyo, Akwa Ibom State, Nigeria. World Appl. Sci. J. 2011, 15, 985–991. [Google Scholar]
- Adeleye, I.A.; Daniels, F.V.; Enyinnia, V.A. Characterization and pathogenicity of Vibrio spp. contaminating seafoods in La-gos, Nigeria. Internet J. Food Saf. 2010, 12, 1–9. [Google Scholar]
- Ballal, M.; Shetty, V.; Bangera, S.R.; Prabhu, M.; Umakanth, S. Vibrio furnissii, an emerging pathogen causing acute gastroen-teritis: A Case Report. JMM Case Rep. 2017, 4. [Google Scholar] [CrossRef]
- Willey, J.M.; Sherwood, L.M.; Woolverton, C.J. Presscott, Harley, and Klein’s Microbiology; McGraw-Hill: New York, NY, USA, 2008; ISBN 0073302082. [Google Scholar]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Prim. 2018, 4, 8. [Google Scholar] [CrossRef]
- Montánchez, I.; Ogayar, E.; Plágaro, A.H.; Esteve-Codina, A.; Gómez-Garrido, J.; Orruño, M.; Arana, I.; Kaberdin, V.R. Analysis of Vibrio harveyi adaptation in sea water microcosms at elevated temperature provides insights into the putative mechanisms of its persistence and spread in the time of global warming. Sci. Rep. 2019, 9, 289. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.K.; Garg, S.; Mitra, R.; Basu, A.; Rajendran, K.; Dutta, D.; Bhattacharya, S.K.; Shimada, T.; Takeda, T.; Takeda, Y.; et al. Temporal Shifts in Traits of Vibrio cholerae Strains Isolated from Hospitalized Patients in Calcutta: A 3-Year (1993 to 1995) Analysis. J. Clin. Microbiol. 1996, 34, 2537–2543. [Google Scholar] [CrossRef]
- Okoh, A.I.; Igbinosa, E.O. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010, 10, 143. [Google Scholar] [CrossRef]
- Das, B.; Verma, J.; Kumar, P.; Ghosh, A.; Ramamurthy, T. Antibiotic resistance in Vibrio cholerae: Understanding the ecology of resistance genes and mechanisms. Vaccine 2020, 38, A83–A92. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yu, D.; Yue, J.; Kan, B. Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci. Rep. 2016, 6, 22733. [Google Scholar] [CrossRef] [PubMed]
- Hochhut, B.; Lotfi, Y.; Mazel, D.; Faruque, S.M.; Woodgate, R.; Waldor, M.K. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 2001, 45, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Morita, D.; Ghosh, A.; Chowdhury, G.; Mukhopadhyay, A.K.; Okamoto, K.; Ramamurthy, T. Altered Integrative and Conjugative Elements (ICEs) in Recent Vibrio cholerae O1 Isolated From Cholera Cases, Kolkata, India. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Waldor, M.K.; Tscha¨pe, H.; Tscha¨pe, T.; Mekalanos, J.J. A New Type of Conjugative Transposon Encodes Resistance to Sul-famethoxazole, Trimethoprim, and Streptomycin in Vibrio cholerae O139. J. Bacteriol. 1996, 178, 4157–4165. [Google Scholar] [CrossRef]
- Kitaoka, M.; Miyata, S.T.; Unterweger, D.; Pukatzki, S. Antibiotic resistance mechanisms of Vibrio cholerae. J. Med. Microbiol. 2011, 60, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Dengo-Baloi, L.C.; Semá-Baltazar, C.A.; Manhique, L.V.; Chitio, J.E.; Inguane, D.L.; Langa, J.P. Antibiotics resistance in El Tor Vibrio cholerae 01 isolated during cholera outbreaks in Mozambique from 2012 to 2015. PLoS ONE 2017, 12, e0181496. [Google Scholar] [CrossRef]
- Falbo, V.; Carattoli, A.; Tosini, F.; Pezzella, C.; Dionisi, A.M.; Luzzi, I. Antibiotic Resistance Conferred by a Conjugative Plas-mid and a Class I Integron in Vibrio cholerae O1 El Tor Strains Isolated in Albania and Italy. Am. Soc. Microbiol. 1999, 43, 693–696. [Google Scholar]
- Amita, M.; Chowdhury, S.R.; Thungapathra, M.; Ramamurthy, T.; Nair, G.B.; Ghosh, A. Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae 0139 outbreak, Calcutta, India. Emerg. Infect. Dis. 2003, 9, 500–502. [Google Scholar] [CrossRef]
- Mohapatra, H.; Mohapatra, S.S.; Mantri, C.K.; Colwell, R.R.; Singh, D. V Vibrio cholerae non-O1, non-O139 strains isolated before 1992 from Varanasi, India are multiple drug resistant, contain intSXT, dfr18 and aadA5 genes. Environ. Microbiol. 2008, 10, 866–873. [Google Scholar] [CrossRef]
- Bhanumathi, R.; Sabeena, F.; Isac, S.R.; Shukla, B.N.; Singh, D. V Molecular Characterization of Vibrio cholerae O139 Bengal Isolated from Water and the Aquatic Plant Eichhornia crassipes in the River Ganga, Varanasi, India. Appl. Environ. Microbiol. 2003, 69, 2389–2394. [Google Scholar] [CrossRef]
- Seman, M.; Prokšová, M.; Rosinský, J.; Ferianc, P. Isolation, identification, and characterization of Vibrio cholerae from the Danube River in Slovakia. Folia Microbiol. 2012, 57, 191–197. [Google Scholar] [CrossRef]
- Oprea, M.; Njamkepo, E.; Cristea, D.; Zhukova, A.; Clark, C.G.; Kravetz, A.N.; Monakhova, E.; Ciontea, A.S.; Cojocaru, R.; Rauzier, J.; et al. The seventh pandemic of cholera in Europe revisited by microbial genomics. Nat. Commun. 2020, 11, 5347. [Google Scholar] [CrossRef]
- Nandi, B.; Nandy, R.K.; Mukhopadhyay, S.; Nair, G.B.; Shimada, T.; Ghose, A.C. Rapid Method for Species-Specific Identifi-cation of Vibrio cholerae Using Primers Targeted to the Gene of Outer Membrane Protein OmpW. J. Clin. Microbiol. 2000, 38, 4145–4151. [Google Scholar] [CrossRef]
- Lembke, M.; Höfler, T.; Walter, A.N.; Tutz, S.; Fengler, V.; Schild, S.; Reidl, J. Host stimuli and operator binding sites control-ling protein interactions between virulence master regulator ToxR and ToxS in Vibrio cholerae. Mol. Microbiol. 2020, 114, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple Antibiotic Resistance Indexing of Escherichia coli to Identify High-Risk Sources of Fecal Con-tamination of Foodst. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Tambekar, D.H.; Dhanorkar, D.V.; Gulhane, S.R.; Khandelwal, V.K.; Dudhane, M.N. Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. Afr. J. Biotechnol. 2006, 5, 1562–1565. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Kirs, M.; Depaola, A.; Fyfe, R.; Jones, J.L.; Krantz, J.; Van Laanen, A.; Cotton, D.; Castle, M. A survey of oysters (Crassostrea gigas) in New Zealand for Vibrio parahae-molyticus and Vibrio vulnificus. Int. J. Food Microbiol. 2010, 147, 149–153. [Google Scholar] [CrossRef]
- Johnson, C.N.; Bowers, J.C.; Griffitt, K.J.; Molina, V.; Clostio, R.W.; Pei, S.; Laws, E.; Paranjpye, R.N.; Strom, M.S.; Chen, A.; et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the Coastal and Estuarine Waters of Louisiana, Maryland, Mississippi, and Washington (United States). Am. Soc. Microbiol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Kokashvili, T.; Whitehouse, C.A.; Tskhvediani, A.; Grim, C.J.; Elbakidze, T.; Mitaishvili, N.; Janelidze, N.; Jaiani, E.; Haley, B.J.; Lashkhi, N.; et al. Occurrence and Diversity of Clinically Important Vibrio Species in the Aquatic Environment of Geor-gia. Front. Public Health 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-H.; Hwang, C.-K.; Chin, C.; Lin, H.-H.; Wong, W.-W.; Liu, C.-Y. Severe watery diarrhoea and bacteraemia caused by Vibrio fluvialis. J. Infect. 2006, 52, 95–98. [Google Scholar] [CrossRef]
- Horseman, M.A.; Surani, S. A comprehensive review of Vibrio vulnificus: An important cause of severe sepsis and skin and soft-tissue infection. Int. J. Infect. Dis. 2011, 15, 157–166. [Google Scholar] [CrossRef]
- Chase, E.; Harwood, V.J. Comparison of the Effects of Environmental Parameters on Growth Rates of Vibrio vulnificus Bio-types I, II, and III by Culture and Quantitative PCR Analysis. Appl. Environ. Microbiol. 2011, 77, 4200–4207. [Google Scholar] [CrossRef]
- Heng, S.P.; Letchumanan, V.; Deng, C.Y.; Mutalib, N.S.A.; Khan, T.M.; Chuah, L.H.; Chan, K.G.; Goh, B.H.; Pusparajah, P.; Lee, L.H. Vibrio vulnificus: An environmental and clinical burden. Front. Microbiol. 2017, 8, 997. [Google Scholar] [CrossRef]
- Faruque, S.M.; Albert, M.J.; Mekalanos, J.J. Epidemiology, Genetics, and Ecology of Toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 1998, 62, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Faruque, S.M.; Islam, M.J.; Ahmad, Q.S.; Biswas, K.; Faruque AS, G.; Nair, G.B.; Sack, R.B.; Sack, D.A.; Mekalanos, J.J. An Improved Technique for Isolation of Environmental Vibrio cholerae with Epi-demic Potential: Monitoring the Emergence of a Multiple-Antibiotic–Resistant Epidemic. J. Infect. Dis. 2006, 193, 1029–1036. [Google Scholar] [CrossRef]
- Rodriguez Ojeda, J.A.; Chadi, I.K. Vibrio Cholerae; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK526099/ (accessed on 18 March 2020).
- Benskin, C.M.W.H.; Wilson, K.; Jones, K.; Hartley, I.R. Bacterial pathogens in wild birds: A review of the frequency and effects of infection. Biol. Rev. 2009, 84, 349–373. [Google Scholar] [CrossRef] [PubMed]
- Laviad-Shitrit, S.; Izhaki, I.; Halpern, M. Accumulating evidence suggests that some waterbird species are potential vectors of Vibrio cholerae. PLoS Pathog. 2019, 15, e1007814. [Google Scholar] [CrossRef] [PubMed]
- Lutz, C.; Erken, M.; Noorian, P.; Sun, S.; McDougald, D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef]
- Miyasaka, J.; Yahiro, S.; Arahira, Y.; Tokunaga, H.; Katsuki, K.; Hara-Kudo, Y. Isolation of Vibrio parahaemolyticus and Vibrio vulnificus from wild aquatic birds in Japan. Epidemiol. Infect. 2006, 134, 780–785. [Google Scholar] [CrossRef]
- Ogg, J.E.; Ryder, R.A.; Smith, H.L. Isolation of Vibrio cholerae from Aquatic Birds in Colorado and Utah. Appl. Environ. Microbiol. 1989, 55, 95–99. [Google Scholar] [CrossRef]
- Liang, W.; Wang, L.; Liang, P.; Zheng, X.; Zhou, H.; Zhang, J.; Zhang, L.; Kan, B. Sequence polymorphisms of rfbT among the Vibrio cholerae O1 strains in the Ogawa and Inaba serotype shifts. BMC Microbiol. 2013, 13, 173. [Google Scholar] [CrossRef]
- Krukonis, E.S.; Yu, R.R.; DiRita, V.J. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: Distinct roles for two mem-brane-localized transcriptional activators on a single promoter. Mol. Microbiol. 2000, 38, 67–84. [Google Scholar] [CrossRef]
- Kunkle, D.E.; Bina, T.F.; Bina, X.R.; Bina, J.E. Vibrio cholerae OmpR Represses the ToxR Regulon in Response to Membrane Intercalating Agents That Are Prevalent in the Human Gastrointestinal Tract. Am. Soc. Microbiol. 2020. [Google Scholar] [CrossRef]
- Li, J.; Yie, J.; Foo, W.T.; Ling Julia, M.L.; Huaishu, X.; Norman, Y.S.W. Antibiotic resistance and plasmid profiles of Vibrio isolates from cultured silver sea bream, Sparus sarba. Mar. Poll. Bull. 1999, 39, 245–249. [Google Scholar] [CrossRef]
- You, K.G.; Bong, C.W.; Lee, C.W. Antibiotic resistance and plasmid profiling of Vibrio spp. in tropical waters of Peninsular Malaysia. Environ. Monit. Assess. 2016, 188, 171. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xu, L.; Chen, H.; Liu, S.; Guo, Z.; Cheng, C.; Ma, H.; Feng, J. Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in South China. Sci. Rep. 2020, 10, 14329. [Google Scholar] [CrossRef]
- Matyar, F.; Kaya, A.; Dinçer, S. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from sea-water, shrimp and sediment in Iskenderun Bay, Turkey. Sci. Total Environ. 2008, 407, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Karunasagar, I.; Shivakumaraswamy, S.K.; Deekshit, V.K.; Vittal, R.; Akhila, D.S.; Mundanda, D.M.; Raj, J.R.M.; Chakraborty, A. Phenotypic & genotypic study of antimicrobial profile of bacteria isolates from environmental samples. Indian J. Med. Res. 2019, 149, 232–239. [Google Scholar]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Fallacara, D.M.; Monahan, C.M.; Morishita, T.Y.; Diseases, R.F.W. Fecal shedding and antimicrobial susceptibility of selected bacterial pathogens and a survey of intestinal parasites in free-living waterfowl. Avian Dis. 2001, 45, 128–135. [Google Scholar] [CrossRef]
- Dolejska, M.; Cizek, A.; Literak, I. High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from Black-headed Gulls in the Czech Republic. J. Appl. Microbiol. 2007, 103, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Järhult, J.D. Antibiotic resistance in wild birds. Upsala J. Med. Sci. 2014, 119, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment, and human health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef]
- Suárez-Pérez, A.; Corbera, J.A.; González-Martín, M.; Donázar, J.A.; Rosales, R.S.; Morales, M.; Tejedor-Junco, M.T. Micro-organisms Resistant to Antimicrobials in Wild Canarian Egyptian Vultures (Neophron percnopterus majorensis). Animals 2020, 10, 970. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.H.; Khor, W.C.; Quek, J.Y.; Low, Z.X.; Arivalan, S.; Humaidi, M.; Chua, C.; Seow, K.L.G.; Guo, S.; Tay, M.Y.F.; et al. Oc-currence and Antimicrobial Resistance Traits of Escherichia coli from Wild Birds and Rodents in Singapore. Int. J. Environ. Res. Public Health 2020, 17, 5606. [Google Scholar] [CrossRef]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.D.; Wieler, L.H.; Ewers, C. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet. Microbiol. 2010, 144, 219–225. [Google Scholar] [CrossRef]
- Gandra, S.; Alvarez-Uria, G.; Turner, P.; Joshi, J.; Limmathurotsakul, D.; van Doorn, H.R. Antimicrobial Resistance Surveil-lance in Low- and Middle-Income Countries: Progress and Challenges in Eight South Asian and Southeast Asian Countries. Clin. Microbiol. Rev. 2020, 33, e00048-19. [Google Scholar] [CrossRef] [PubMed]
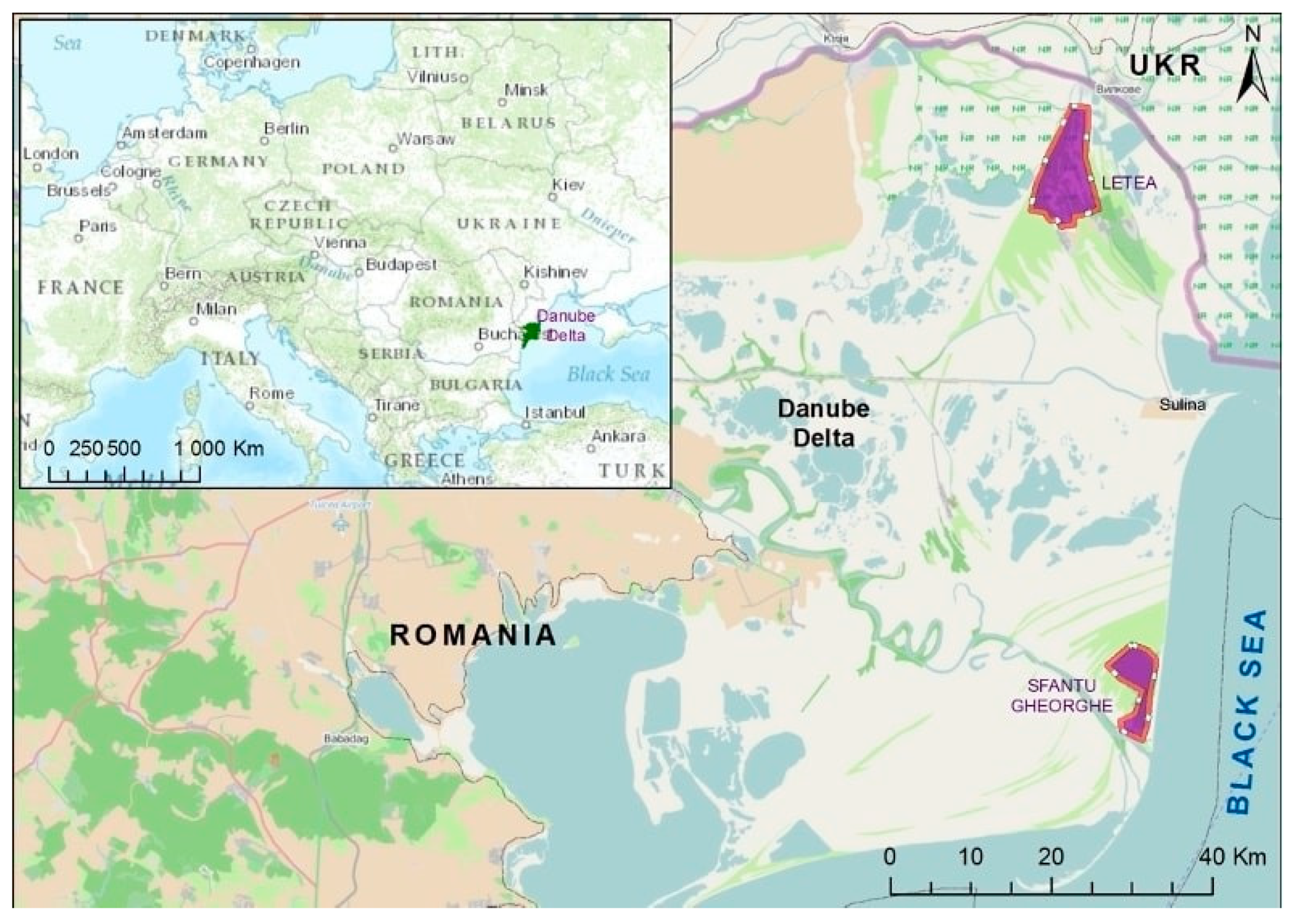
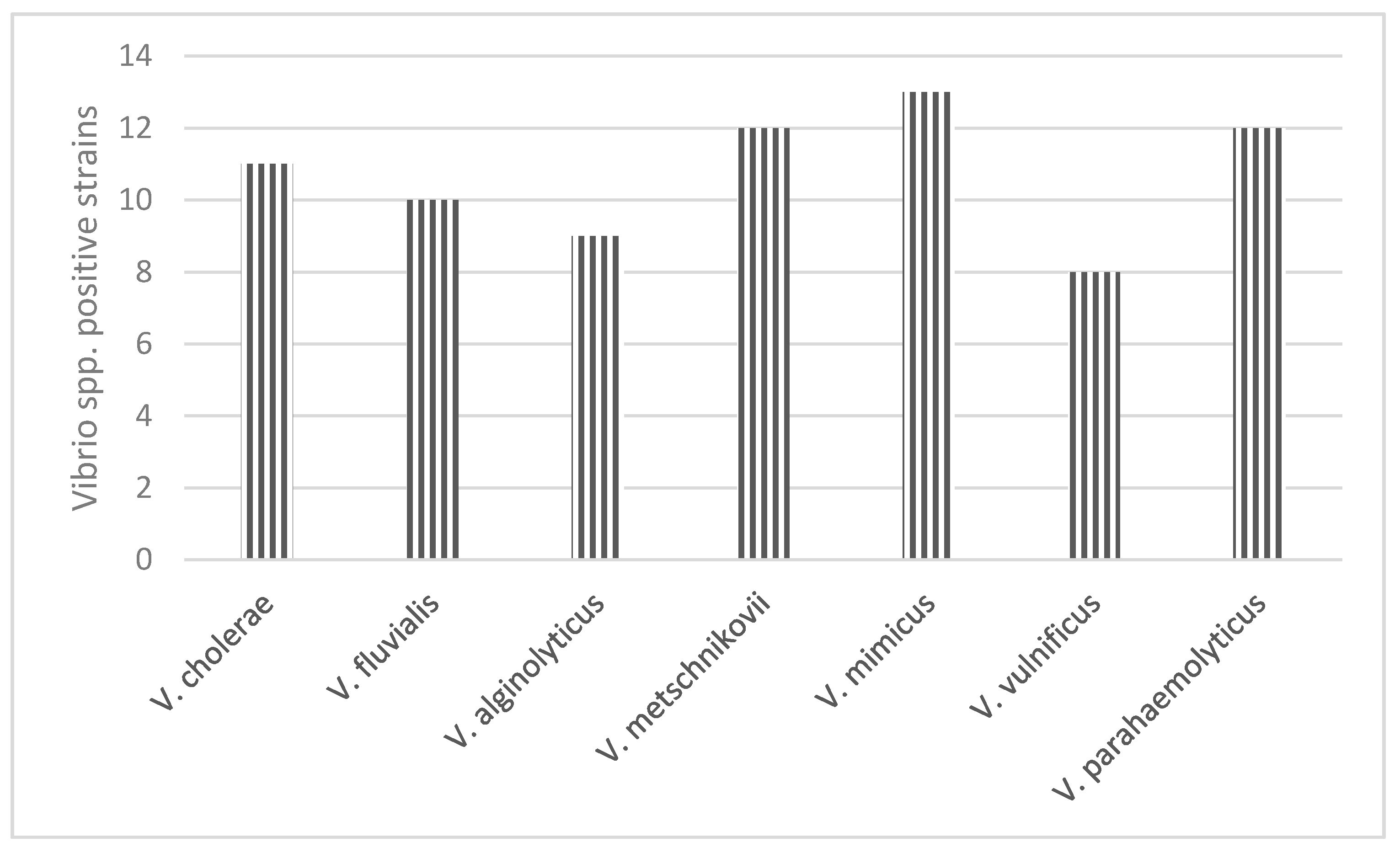
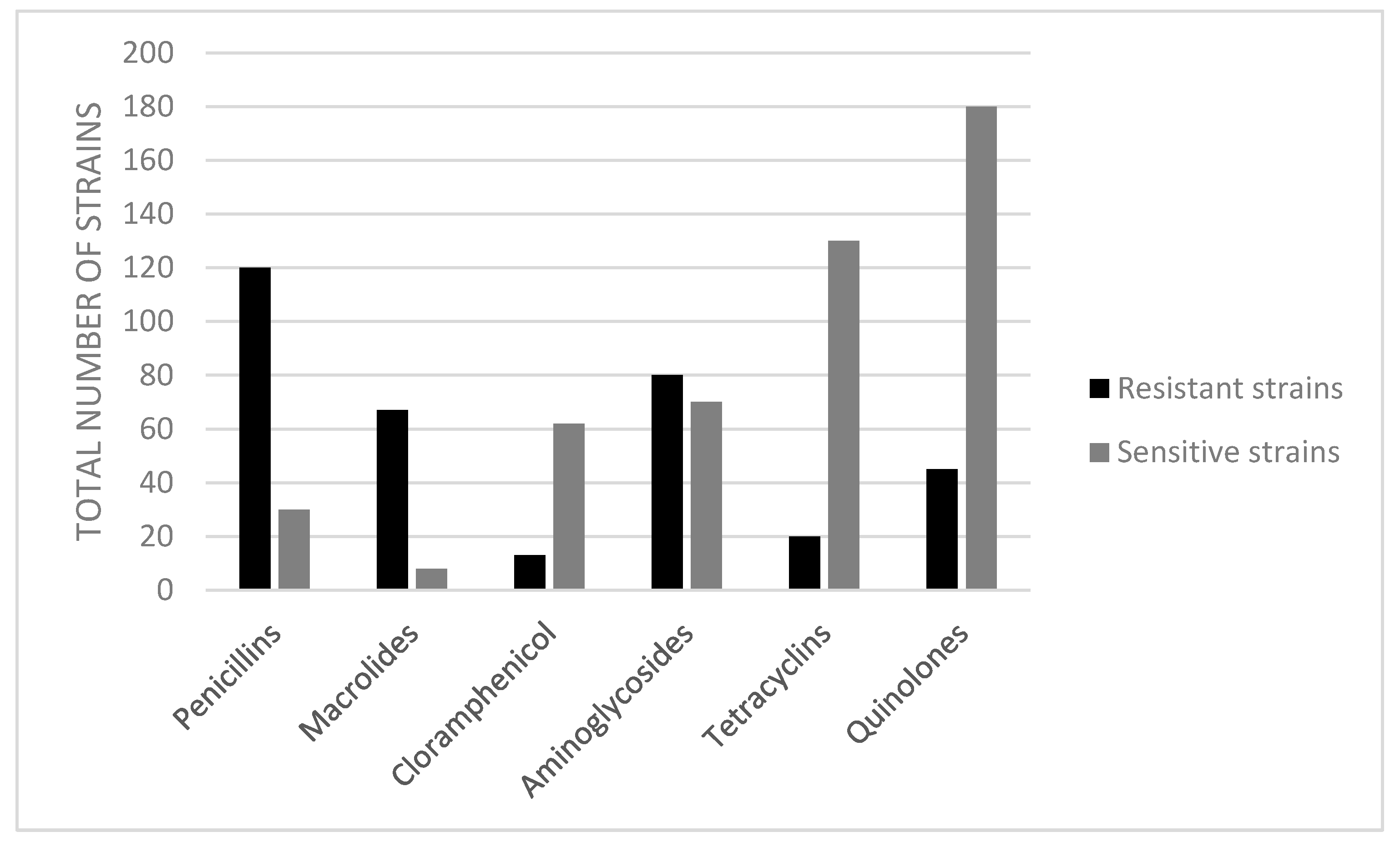
| Species | Habitat Type | No. Captured Birds | Positive Samples for Vibrio spp. | Bacterial Isolate | PCR | |
|---|---|---|---|---|---|---|
| ompW | toxR | |||||
| Squacco heron (Ardeola ralloides) | Migratory | 2 | 2 | V. cholerae | + | + |
| V. alginolyticus | - | + | ||||
| V. parahaemolyticus | - | - | ||||
| Common greenshank (Tringa nebularia) | Migratory | 2 | 2 | V. fluvialis | - | + |
| V. alginolyticus | - | - | ||||
| Reed bunting (Emberiza schoeniclus) | Migratory | 2 | 1 | V. metschnikovii | - | - |
| V. fluvialis | - | - | ||||
| V. parahaemolyticus | - | - | ||||
| Song thrush (Turdus philomelos) | Migratory | 2 | 1 | V. cholerae | + | - |
| V. alginolyticus | - | - | ||||
| V. mimicus | - | - | ||||
| Eurasian hobby (Falco subbuteo) | Migratory | 2 | 2 | V. cholerae | + | + |
| V. fluvialis | - | - | ||||
| V. alginolyticus | - | - | ||||
| V. mimicus | - | - | ||||
| Wood sandpiper (Tringa glareola) | Migratory | 2 | 2 | V. mimicus | - | - |
| V. vulnificus | - | - | ||||
| Black-crowned night heron (Nycticorax nycticorax) | Migratory | 2 | 2 | V. vulnificus | - | - |
| V. parahaemolyticus | - | - | ||||
| Common snipe (Gallinago gallinago) | Migratory | 3 | 2 | V. cholerae | + | + |
| V. mimicus | - | - | ||||
| Eurasian sparrowhawk (Accipiter nisus) | Migratory | 2 | 1 | V. cholerae | + | - |
| V. parahaemolyticus | - | - | ||||
| Common chaffinch (Fringilla coelebs) | Migratory | 2 | 2 | V. mimicus | - | - |
| Eurasian teal (Anas crecca) | Migratory | 2 | 1 | V. mimicus | - | - |
| V. vulnificus | - | - | ||||
| - | - | |||||
| Eurasian tree sparrow (Passer montanus) | Sedentary | 2 | 2 | V. cholerae | + | - |
| V. fluvialis | - | - | ||||
| Lesser whitethroat (Sylvia curucca) | Migratory | 1 | 1 | V. metschnikovii | - | - |
| V. mimicus | - | - | ||||
| V. vulnificus | - | - | ||||
| Red-backed shrike (Lanius collurio) | Migratory | 1 | 1 | V. fluvialis | - | - |
| V. alginolyticus | - | - | ||||
| V. metschnikovii | - | - | ||||
| V. parahaemolyticus | - | - | ||||
| Icterine warbler (Hippolais icterina) | Migratory | 3 | 2 | V. parahaemolyticus | - | - |
| Red-footed falcon (Falco vespertinus) | Migratory | 2 | 2 | V. cholerae | + | - |
| V. fluvialis | - | - | ||||
| V. alginolyticus | - | - | ||||
| V. metschnikovii | - | - | ||||
|
Garden warbler (Sylvia borin) | Migratory | 4 | 3 | V. vulnificus | - | - |
| V. parahaemolyticus | - | - | ||||
|
Common whitethroat (Sylvia communis) | Migratory | 5 | 4 | V. alginolyticus | - | - |
| V. metschnikovii | - | - | ||||
|
Common kingfisher (Alcedo atthis) | Migratory | 1 | 1 | V. cholerae | - | - |
| V. fluvialis | - | - | ||||
| V. metschnikovii | - | - | ||||
| V. mimicus | - | - | ||||
|
Eurasian blackcap (Sylvia atricapilla) | Migratory | 1 | 1 | V. parahaemolyticus | - | - |
|
Barred warbler (Sylvia nisoria) | Migratory | 3 | 1 | V. metschnikovii | - | - |
| V. vulnificus | - | - | ||||
| Black-and-white magpie (Pica pica) | Sedentary | 5 | 2 | V. cholerae | + | + |
| V. metschnikovii | - | |||||
| V. parahaemolyticus | - | - | ||||
| Western jackdaw (Corvus monedula) | Sedentary | 4 | 2 | V. cholerae | - | - |
| V. fluvialis | - | - | ||||
| Hooded crow (Corvus corone cornix) | Sedentary | 7 | 3 | V. metschnikovii | - | - |
| V. mimicus | - | - | ||||
| V. vulnificus | - | - | ||||
| V. parahaemolyticus | - | - | ||||
| Eurasian tree sparrow (Passer montanus) | Sedentary | 4 | 2 | V. cholerae | + | - |
| V. fluvialis | - | - | ||||
| Eurasian blue tit (Parus caeruleus) | Sedentary | 2 | 2 | V. metschnikovii | - | - |
| V. mimicus | - | - | ||||
| V. vulnificus | - | - | ||||
| V. parahaemolyticus | - | - | ||||
| Long tailed tit (Aegithalos caudatus) | Sedentary | 2 | 0 | - | - | - |
| Bearded reedling (Panurus biarmicus) | Sedentary | 2 | 1 | V. alginolyticus | - | - |
| V. metschnikovii | - | - | ||||
| V. mimicus | - | - | ||||
| Common chaffinch (Fringilla coelebs) | Sedentary | 3 | 2 | V. cholerae | - | - |
| V. mimicus | - | - | ||||
| Hawfinch (Coccothraustes coccothraustes) | Sedentary | 8 | 5 | V. alginolyticus | - | - |
| V. metschnikovii | - | - | ||||
| V. mimicus | - | - | ||||
| Great tit (Parus major) | Sedentary | 2 | 2 | V. fluvialis | - | - |
| V. mimicus | - | - | ||||
| Antimicrobial Class | Drug | Total Number of Resistant Strains Isolated from Migratory Birds, by Antimicrobial | Total Number of Resistant Strains Isolated from Sedentary Birds, by Antimicrobial | MAR Index for the Tested Antibiotics |
|---|---|---|---|---|
| Penicillins | Penicillin | 48 | 18 | 0.8 |
| Ampicillin | 37 | 17 | 0.6 | |
| Macrolides | Erythromycin | 47 | 20 | 0.8 |
| Cloramphenicol | Chloramphenicol | 12 | 1 | 0.1 |
| Aminoglycosides | Amikacin | 45 | 17 | 0.7 |
| Kanamycin | 11 | 7 | 0.2 | |
| Tetracyclines | Oxytetracycline | 10 | 0 | 0.1 |
| Tetracycline | 8 | 2 | 0.1 | |
| Quinolones | Enrofloxacin | 11 | 4 | 0.1 |
| Marbofloxacin | 10 | 5 | 0.1 | |
| Ciprofloxacin | 9 | 6 | 0.1 |
| Number of Antibiotic Classes to Which Isolated Strains Showed Resistance | Number/% of Resistant Strains | Resistance Type |
|---|---|---|
| 1 | 1 (1.32) | - |
| 2 | 5 (6.58) | MAR |
| 3 | 35 (46.05) | MDR |
| 4 | 21 (27.63) | XDR |
| 5 | 10 (13.16) | XDR |
| 6 | 4 (5.26) | XDR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Páll, E.; Niculae, M.; Brudașcă, G.F.; Ravilov, R.K.; Șandru, C.D.; Cerbu, C.; Olah, D.; Zăblău, S.; Potârniche, A.V.; Spinu, M.; et al. Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania. Antibiotics 2021, 10, 333. https://doi.org/10.3390/antibiotics10030333
Páll E, Niculae M, Brudașcă GF, Ravilov RK, Șandru CD, Cerbu C, Olah D, Zăblău S, Potârniche AV, Spinu M, et al. Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania. Antibiotics. 2021; 10(3):333. https://doi.org/10.3390/antibiotics10030333
Chicago/Turabian StylePáll, Emöke, Mihaela Niculae, Gheorghe F. Brudașcă, Rustam Kh. Ravilov, Carmen Dana Șandru, Constantin Cerbu, Diana Olah, Sergiu Zăblău, Adrian Valentin Potârniche, Marina Spinu, and et al. 2021. "Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania" Antibiotics 10, no. 3: 333. https://doi.org/10.3390/antibiotics10030333
APA StylePáll, E., Niculae, M., Brudașcă, G. F., Ravilov, R. K., Șandru, C. D., Cerbu, C., Olah, D., Zăblău, S., Potârniche, A. V., Spinu, M., Duca, G., Rusu, M., Rzewuska, M., & Vasiu, A. (2021). Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania. Antibiotics, 10(3), 333. https://doi.org/10.3390/antibiotics10030333











