Abstract
The aim of the study was to analyze the mechanisms of resistance in extended-spectrum beta-lactamase (ESBL)- and acquired AmpC (qAmpC)-producing Escherichia coli isolates from healthy and sick cats in Portugal. A total of 141 rectal swabs recovered from 98 sick and 43 healthy cats were processed for cefotaxime-resistant (CTXR) E. coli recovery (in MacConkey agar supplemented with 2 µg/mL cefotaxime). The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) method was used for E. coli identification and antimicrobial susceptibility was performed by a disk diffusion test. The presence of resistance/virulence genes was tested by PCR sequencing. The phylogenetic typing and multilocus sequence typing (MLST) were determined by specific PCR sequencing. CTXR E. coli isolates were detected in seven sick and six healthy cats (7.1% and 13.9%, respectively). Based on the synergy tests, 11 of 13 CTXR E. coli isolates (one/sample) were ESBL-producers (ESBL total rate: 7.8%) carrying the following ESBL genes: blaCTX-M-1 (n = 3), blaCTX-M-15 (n = 3), blaCTX-M-55 (n = 2), blaCTX-M-27 (n = 2) and blaCTX-M-9 (n = 1). Six different sequence types were identified among ESBL-producers (sequence type/associated ESBLs): ST847/CTX-M-9, CTX-M-27, CTX-M-1; ST10/CTX-M-15, CTX-M-27; ST6448/CTX-M-15, CTX-M-55; ST429/CTX-M-15; ST101/CTX-M-1 and ST40/CTX-M-1. Three of the CTXR isolates were CMY-2-producers (qAmpC rate: 2.1%); two of them were ESBL-positive and one ESBL-negative. These isolates were typed as ST429 and ST6448 and were obtained in healthy or sick cats. The phylogenetic groups A/B1/D/clade 1 were detected among ESBL- and qAmpC-producing isolates. Cats are carriers of qAmpC (CMY-2)- and ESBL-producing E. coli isolates (mostly of variants of CTX-M group 1) of diverse clonal lineages, which might represent a public health problem due to the proximity of cats with humans regarding a One Health perspective.
1. Introduction
The emergence and fast dissemination of antimicrobial-resistant bacteria (AMR) continue to be a public health concern in both medicine and agriculture [1]. It was estimated that the global consumption of antimicrobial agents in animal food production will increase by 67% between 2010 and 2030 [2]. Accordingly, this global rapid increase of AMR has been mostly attributed to the overuse and misuse of antibiotics in human and veterinary medicine [3,4]. In the veterinary environment, antimicrobial use is discussed as one of the main drivers of antimicrobial resistance development [5,6].
Escherichia coli, a Gram-negative bacterium that normally habits in the gastrointestinal tract of healthy humans and warm-blooded animals, is also an important opportunistic pathogen [6,7]. This commensal microorganism is known to be an important indicator of the antibiotic resistance evolution as well as an eventual reservoir of virulence genes in different ecosystems [8,9].
Urinary tract infections (UTIs) are one of the common health problems among companion animals in European countries. Gram-negative bacteria, mainly E. coli strains, are responsible for 75% of the cases [6,10]. Treatment of UTIs caused by E. coli strains is becoming difficult due to the antibiotic resistance phenomenon. E. coli can act as a reservoir of antimicrobial-resistant genes that can be transmitted to other pathogenic bacteria. Multidrug-resistant (MDR) microorganisms are transmitted among pets, owners and veterinary staff, which leads to their spread within the community [11,12,13].
Extended-spectrum β-lactamase (ESBL), acquired AmpC β-lactamase (qAmpC) and carbapenemase production in Enterobacteriaceae are three of the major concerns due to their rapid emergence in humans and animals during the last years [9,13,14,15].
Escherichia coli isolates harbor intrinsic and low-expressed chromosomic AmpC β-lactamases (cAmpC) that do not confer resistance to β-lactams; nevertheless, when overexpressed (generally by mutations at either the promoter or the attenuator of the structural gene), they produce a resistance to amoxicillin-clavulanic acid, cefoxitin and third generation cephalosporins [16]. E. coli isolates can also acquire different AmpC β-lactamases (qAmpC), also conferring an AmpC phenotype. During the last years, different qAmpC types have been reported (as CMY, DHA, MOX, FOX and ACC) or the qAmpC β-lactamases family (EBC), being genes that are mostly plasmid located. The first plasmid-mediated qAmpC, named CMY-1, was described in South Korea (1989). Recent studies have described an increase in specific qAmpC enzymes among hospital or community infections caused by Enterobacteriaceae [17,18]. The DHA-1 and CMY-2 variants are the most common qAmpC types among Enterobacteriaceae implicated in human infections in Portugal [18]. During the last years, the CMY-2 enzyme has been increasingly reported in humans and animals, frequently associated with lineages ST429, ST354 or ST57, among others [17,19,20,21]. According to recent studies in Portugal, qAmpC-producing E. coli was reported (CMY-2) among sick pets, mostly associated with ST648 [6], and among healthy humans, associated with lineages ST4953 and ST665 [22]. Moreover, CMY-2 was also reported among Portuguese clinical E. coli isolates associated with a diversity of lineages [6,18].
The number of reports concerning ESBLs from E. coli isolates in companion animals has also increased [23,24]. Moreover, CTX-M type enzymes have formed a rapidly growing family of ESBLs in bacteria from human infections. According to a review done by Dandachi et al. [25], the CTX-M group 1 seems to prevail in animals in the Mediterranean Basin followed by SHV-12 and CTX-M group 9. The pandemic-specific ST131 clone with multiresistance and a high virulence potential is largely associated with the global increase of these ESBL-producers.
In addition, shiga toxin (stx)-producing Escherichia coli (STEC) strains have emerged as an important cause of serious human gastrointestinal disease during the last years. Pets can also be reservoirs of STEC and enteropathogenic E. coli (EPEC) strains. Recent studies have reported its presence among E. coli isolated from wildlife animals [14,26].
Companion animals (including cats) may play an important role in the spread of resistant bacteria due to their frequent and close contact with humans. Therefore, the risk of animal to human transference of such bacteria is a significant concern [3]. Regarding the global situation of antimicrobial resistance, many studies have been performed and current data are available particularly in human settings [6,9]. Likewise, other authors either from Europe [27,28,29] or from other continents [30,31,32,33] have focused on the study of E. coli isolates from pets. In Portugal, studies about AMR with a detection of ESBL and/or qAmpC in E. coli isolates recovered from humans [18,34,35] and healthy or sick pets [36,37,38] were communicated. Despite the scarcity of data about healthy and sick cats, all cited surveys showing a worrying picture.
To our knowledge, this study is the first report that combines AMR among E. coli isolates from both healthy and sick cats in Portugal. It aimed to analyze the occurrence and molecular epidemiology of qAmpC- and ESBL-producing E. coli isolates originating from healthy cats (kennel and house cats) and sick cats (of seven different hospitals) in the Portuguese territory.
2. Results
2.1. Cefotaxime-Resistant (CTXR) E. coli Isolates and Antimicrobial Susceptibility Testing
Of the 141 fecal samples analyzed (98 from sick cats and 43 from healthy cats), CTXR E. coli isolates were detected in 13 (seven from sick and six from healthy cats; rates of 7.1% and 13.9%, respectively) and one isolate per positive sample was further characterized (corresponding to 12 mixed breed cats and one Siamês). Regarding the healthy cats, five of the positive samples came from healthy cats from their owners and the other sample from a cattery located in Vila Real (Table 1). Regarding the sick animals, CTXR E. coli isolates were recovered from samples of five different veterinary clinics/hospitals (Figure 1) distributed as follows: Vila Real (two isolates from Veterinary Clinic Quinchosos and one isolate from Transmonvete), Bragança (two isolates from Veterinary Clinic Macedo de Cavaleiros) and Leiria (two isolates from Veterinary Clinic Guia) (Table 1).

Table 1.
Phenotypic and molecular features of the CTXR E. coli isolates recovered from healthy and sick cats in Portugal.
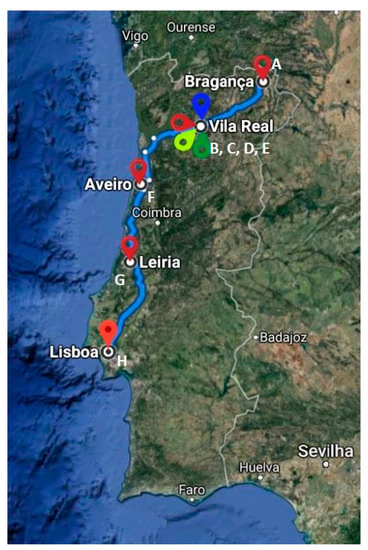
Figure 1.
Geographic location of the different areas where the fecal samples from cats were collected in Portugal. A—Veterinary clinic from Macedo de Cavaleiros (Bragança); B—Kennel (Vila Real); C—Veterinary clinic from Quinchosos (Vila Real); D—Transmonvete (Vila Real), E—Veterinary hospital from Trás os Montes HVTM (Vila Real); F—Veterinary clinic from Vouga (Aveiro); G—Veterinary clinic from Guia (Leiria); H—Veterinary hospital from São Bento (Lisboa).
The 13 CTXR E. coli isolates showed high rates of resistance for cefoxitin (61.5%), amoxicillin + clavulanic acid (76.9%), tetracycline (53.8%), ciprofloxacin (38.5%) and trimethoprim + sulfamethoxazole (30.8%) (Table 1). Resistance to chloramphenicol, gentamicin and streptomycin were also observed in three isolates in each one (23%). Consequently, most of the isolates showed a multidrug-resistance (MDR) phenotype (92.3%) including resistance to at least three classes of antimicrobial agents.
2.2. Genes Encoding ESBLs and qAmpC Enzymes as Well as Other Resistance/Virulence Genes
Based on the synergy tests, 11 E. coli isolates were found to be ESBL-producers (7.8% of total samples; 11.6% of healthy cats and 8.1% of sick cats). Five different blaCTX-M variants were detected among our ESBL-producing E. coli (number of isolates): blaCTX-M-1 (3), blaCTX-M-15 (3), blaCTX-M-55 (2), blaCTX-M-27 (2) and blaCTX-M-9 (1) (Table 1). Furthermore, the blaTEM gene was detected in eight of the 11 ESBL-positive E. coli isolates (72.7%) in association with the blaCTX-M gene. One isolate carried the blaSHV-28 gene together with blaCTX-M-27 and blaTEM genes.
Eight of the 13 CTXR isolates showed a resistance to cefoxitin and amoxicillin clavulanic acid, a phenotype compatible with a qAmpC. After testing the presence of qAmpC genes by PCR, three isolates gave a positive result for the blaCMY-2 gene (two ESBL-positive and one ESBL-negative); these isolates were obtained from two healthy cats and one sick cat. Taken together, qAmpC-carrying isolates were detected in three of the 141 animals tested (2.1%) and in two cases associated with ESBLs.
Considering all of the 13 CTXR isolates, the tetA gene was detected among seven tetracycline-resistant isolates and the combination of both tetA and tetB genes was detected in one additional isolate (Table 1). The integrase of class 1 integrons (int1) was revealed among four SXT-resistant isolates and they carried the sul1 and/or sul2 genes. Three chloramphenicol-resistant isolates were detected but they were negative for cmlA, catA and floR genes. Two ciprofloxacin-resistant isolates carried the aac(6’)-Ib-cr gene and one amikacin-resistant isolate carried the armA gene. None of the CTXR isolates contained the mcr1,4 resistance genes nor the sxt-1, sxt-2 or eae virulence genes.
2.3. Phylogenetic Groups and MLST Typing
The 13 CTXR E. coli isolates were ascribed to phylogroups B1 (n = 4), A (n = 3), D (n = 2) and clade 1 (n = 1); three additional isolates were not conclusive regarding the phylogroup guidelines according with Clermont et al. [39]. MLST was performed in the twelve ESBL- and qAmpC-producing E. coli isolates and the following sequence types were obtained (sequence type/associated ESBL types (phylogroups)): ST847/CTX-M-9 or CTX-M-27 or CTX-M-1 (B1, one isolate); ST10/CTX-M-15 or CTX-M-27 (A); ST6448/CTX-M-15 or CTX-M-55 (B1); ST429/CTX-M-15 (clade 1); ST101/CTX-M-1 (D); ST40/CTX-M-1 (B1) and ST429/CMY-2 (D) (Table 1). A diversity of CTX-M variants was detected among healthy (CTX-M-1, -9, -15 and-27) and sick cats (CTX-M-1, -15 and -55).
3. Discussion
Worldwide, it is known that antimicrobial-resistant bacteria have hugely disseminated in both clinical and non-clinical settings. In our study, 141 fecal samples were studied from healthy and sick cats in Portugal to estimate the occurrence of ESBL- and qAmpC-producing E. coli isolates. Almost 8% of the total fecal samples analyzed contained ESBL-producers and 2.1% of samples were confirmed as qAmpC-producers (carrying the blaCMY-2 gene). Two isolates co-produced ESBLs and qAmpC enzymes. We could not detect the mechanism of β-lactam resistance in one CTXR ESBL-negative E. coli isolate with an AmpC phenotype; however, it cannot be discarded the presence of other non-tested acquired ampC genes or the hyperproduction of the chromosomal ampC gene. One limitation of this study is the selection of only one CTXR isolate/positive sample; with more variability of ESBL or qAmpC-producers we could have detected if more CTXR isolates were selected in each positive sample.
The detection of CMY-2-producing E. coli isolates was previously reported in healthy and sick dogs in Italy [29,40], healthy dogs in Mexico [21] and sick dogs and cats in Portugal [6] and the United States [41] among others. Moreover, the CMY-2 enzyme was recently reported among healthy humans in Portugal (0.48%) [22] as well as in human clinical settings (1.04%) [6] with a similar prevalence as the one detected in our study (2.1%). The One Health perspective can be considered because of possible horizontal transmission among pets and humans. Freitas et al. [34] detected the CMY-2 encoding gene among 6% (3/50) of qAmpC-producers in different Portuguese hospitals.
Our findings confirm that ESBLs in companion animals have increased rapidly as mentioned by other authors [24,42,43]. In Brazil, a rate of 24.8% was found in healthy stray cats [32]. However, no ESBL-positive isolates were identified in other studies done in healthy cats from Portugal [38] or in sick cats in Turkey [10]. Furthermore, no CTXR was found in E. coli isolates from cats in Oporto (Portugal) [44]. In the Netherlands, 25% of cats with diarrhea carried ESBL-producing isolates and none of the healthy cats’ isolates (0%) was positive for ESBLs [43]. Furthermore, Piccolo et al. [40] recently found 26.2% of ESBL-producing E. coli among diseased cats in Italy. Moreover, a high rate of ESBL-producing E. coli isolates was reported among clinical samples from pets in Singapore (40%) [31]. Curiously, in our study, the ESBL prevalence was higher in healthy cats (11.6.%) than in sick animals (8.1%); although the reason for this difference is unknown, we cannot discard the influence of the different number of animals included in each group. In this respect, Hordijk et al. reported that 25% of sick cats were ESBL-producers but no ESBL production was detected among healthy cats in the Netherlands [45]. Different studies have been performed in Portugal regarding ESBL-producing E. coli isolates recovered from sick and healthy dogs and cats [6,38]. Nevertheless, to our knowledge, this is the first report of the characterization of qAmpC- and ESBL-producing E. coli isolates from healthy and sick cats simultaneously in the same work and country. Moreover, our results about the MDR phenotype join information with other studies already performed in isolates of companion animals from different origins and countries [24,30,40].
In the present report, the high abundance of the blaCTX−M−1 and blaCTX−M−15 variants among the ESBL-positive isolates (6/11) (Table 1) was consistent with studies based on isolates from humans and farm animals in which blaCTX−M−1 was either the most or second most predominant ESBL gene. It is important to note that CTX-M-15 has emerged as the predominant ESBL type in E. coli isolates in healthy and sick humans [9,46]. Our results are also in accordance with the data reported by Day et al. [15] in which the prevalence of blaCTX-M-1 and blaCTX-M-15 in human isolates from the Netherlands and Germany was high. Furthermore, Hordijk et al. [43] indicated that diarrheic cats were positive for blaCTX−M−1 and blaCTX−M−15. The first report of CTX-M-1 in Portugal was performed in one healthy dog [38]. Carattoli et al. [29] also reported the blaCX-M-1 gene in two healthy cats in Italy. Furthermore, Marques et al. [6] observed that five out of 220 cats with a UTI carried E. coli isolates with CTX-M-15, CTX-M-1, CTX-M-32 or CTX-M-9 variants.
The phylogenetic group B1 represented the most prevalent group followed by A (n = 3) and D (n = 2) among the CTXR isolates of this study. The phylogenetic group B2 was nevertheless the most frequently detected among E. coli isolates implicated in urinary tract infections of cats and dogs in a previous study [10].
In the last decade, E. coli of clonal group ST131 has emerged as an important clinical health concern causing multidrug-resistant (MDR) infections worldwide in animals and humans [9,47]. However, this epidemic clone was not found among these cats’ fecal samples. Concerning the sequence types found in this study, ST847 (n = 3), ST6448 (n = 3), ST10 (n = 2), ST101, ST429 and ST40 were detected in blaCTX-M-1,9,27, blaCTX-M-15,55/blaCMY-2, blaCTX-M-15,27, blaCTX-M-1, blaCTX-M-15/blaCMY-2 and blaCTX-M-1-containing strains, respectively (Table 1). A previous report has indicated the detection of ST847 among avian ESBL-E. coli isolates in Germany [48]. The same authors verified that the most common multilocus sequence types (STs) in ESBL-producers, for which whole genome sequencing was performed, were ST131 (25.6%) followed by ST10. Day et al. [15] reported also the presence of ST10 from human isolates in Germany and the Netherlands. To our knowledge, the ST101 is an international sequence type frequently detected in pigs [49] and broilers [50] as well as in clinical settings; this clone can be considered to be a candidate for the zoonotic transmission to the human population.
In order to compare the data of healthy and sick cats, the healthy cats were frequently colonized by ESBL-producing E. coli (different variants of CTX-M enzymes; namely, CTX-M-15,27,9) and qAmpC production (associated with CMY-2) with a predominance of ST429 and ST6448. On the other hand, sick cats were associated with CTX-M-1,15,55 and with a predominance of ST6448. In all cases, the heterogenicity of phylogroups was observed.
The ST6448 lineage was detected in three of our isolates carrying the genes encoding CTX-M-15 or CTX-M-55 (associated with CMY-2 in one isolate); this lineage has also been found in previous studies of our team in two CTX-M-15-producing isolates of sick dogs in Portugal [51] and in one CTX-M-55-producing isolate of a vulture from the Canary Islands [52]. To our knowledge, there is only one previous report related to the detection of this clone in humans, performed in healthy children in Sweden [53]. Furthermore, this clone was also detected in E. coli isolates from broilers in Algeria, associated with the B1 phylogroup [54].
Regarding the ST40 clone, a relation between diarrheal disease and the presence of this lineage (containing specific virulence genes) was recently found among clinical isolates from patients in the United Kingdom [55]. Furthermore, the ST429 lineage was found in our study in CMY-2- and CTX-M-15-producing isolates of healthy cats. The ST429 lineage was previously reported among CMY-2-producing isolates of a poultry origin in Spain [50], Germany [20] and, more recently, in the Czech Republic [56], mostly implicated in avian diseases [56]. This lineage was not detected among the CMY-2-producing E. coli isolates detected in humans in the north of Portugal [18,22,34]. However, CMY-2 was identified among Enterobacteriaceae in a Portuguese hospital [57].
4. Materials and Methods
4.1. Animals and Sampling
A total of 141 fecal samples were collected using standardized procedures from 98 hospitalized cats (69.5%) recovered from seven different veterinary clinic/hospitals and from 43 healthy cats (30.5%), from three local kennels and 40 local houses between January and April 2017 (one sample per animal). Veterinary clinics/hospitals were located in the following Portuguese territories: Braganza (A), Vila Real (B, C, D, E), Aveiro (F), Leiria (G) and Lisbon (H) (Figure 1). The samples of sick cats were collected from those animals that attended the hospital and stayed hospitalized; in the case of healthy cats, the fecal samples were collected after obtaining the owner’s permission or in collaboration with a local kennel. The samples were then dispatched immediately to the Microbiology Laboratory of the University of Trás-os-Montes and Alto-Douro (UTAD) located in Vila Real (Portugal) for processing.
4.2. Escherichia Coli Isolation
From each fecal sample, a small portion of 2 g was taken and diluted in Brain Heart Infusion (BHI) broth and incubated in an aerobic condition for 24 h at 37 °C. They were then inoculated with swabs onto MacConkey agar plates supplemented with 2 μg/mL of cefotaxime (MC + CTX). The plates were then incubated at 37 °C during 24 h and one colony per sample with the morphological aspect of E. coli was selected and identified by conventional biochemical tests (IMViC: Indol, Methyl-red, Voges–Proskauer and Citrate).
The matrix-assisted laser desorption/ionization time-of-flight mass spectrometry method (MALDI-TOF MS, Bruker) was applied in this study to confirm the bacterial species identification and E. coli isolates were kept at −80 °C for further analysis.
4.3. Susceptibility Testing
Antimicrobial susceptibility testing was performed using the Kirby–Bauer disk diffusion method on Mueller–Hinton agar according to the Clinical Laboratory Standards Institute guidelines [58]. The susceptibility of isolates was tested for the following antibiotics (μg/disk): ampicillin (10), amoxicillin + clavulanic acid (20/10), cefoxitin (30), cefotaxime (30), ceftazidime (30), aztreonam (30), imipenem (10), gentamicin (10), amikacin (30), streptomycin (10), nalidixic acid (30), ciprofloxacin (5), tetracycline (30), trimethoprim-sulfamethoxazole (1.25 ± 23.75) and chloramphenicol (30). The plates were incubated aerobically for 24 h at 37 °C.
In addition, the screening of phenotypic ESBL production was carried out in the CTXR isolates by the double disk synergy test using cefotaxime, ceftazidime and amoxicillin/clavulanic acid discs [58]. A presumptive qAmpC phenotype was assigned to the CTXR E. coli isolates that showed resistance to cefoxitin (FOXR) and intermediate susceptibility or resistance to amoxicillin-clavulanic acid (AMCI/R) [34,59]; the presence of qAmpC resistance genes was tested in those isolates.
4.4. Antibiotic Resistance Genes
Genomic DNA was obtained from E. coli isolates using the boiling lysis method [60]. The presence of antibiotic resistance genes was investigated by PCR and subsequent sequencing of the obtained amplicons (when required). The presence of β-lactamase genes was analyzed in the CTXR isolates (blaTEM, blaSHV, blaOXA and blaCTX-M of different groups) and the genes blaCMY, blaDHA, blaFOX, blaMOX and blaACC were also tested in the isolates with a presumptive qAmpC phenotype; all of these isolates were also tested for the mcr-1 and mcr-4 genes associated with colistin resistance [61,62,63,64]. Resistance phenotypes and isolates were also checked (by PCR sequencing when required) for gene encoding resistance to tetracycline (tet(A) and tet(B)), sulfonamides (sul1 and sul2), chloramphenicol (catA, cmlA and florR), ciprofloxacin (aac(6´)-Ib-cr) and amikacin (aph(3´)-VI and armA) [62,63,64,65,66]. The analysis of DNA sequences was performed with the BLAST program available at the National Center for Biotechnology Information.
Moreover, the presence of the int1 gene, encoding the integrase of class 1 integrons, was also analyzed [67] as well as the presence of eae and sxt1,2 genes (encoding the intimin and the shiga toxin, respectively) [19]. Positive and negative controls of the collection of the University of La Rioja (Spain) were included in all assays.
4.5. Molecular Typing of E. coli Isolates
The multilocus sequence typing (MLST) with seven housekeeping genes (fumC, adk, purA, icd, recA, mdh and gyrB) was carried out according to the protocol on the PubMLST (Public databases for molecular typing and microbial genome diversity) website (https://pubmlst.org/escherichia/) (access date on 1 January 2021). The allele combination was determined after sequencing the seven genes amplified by PCR and the sequence type (ST) was identified. A phylogenetic classification of the isolates was also performed according to the existence of arpA, chuA, yjaA and TSPE4.C2 genes as previously reported by Clermont et al. combined with the presence of the E- or C-specific primers [39]. In this line, the E. coli isolates were assigned to one of the eight phylogenetic groups (A, B1, B2, C, D, E, F and clade 1) by multiplex PCR.
5. Conclusions
The present survey showed an increased risk of AMR in fecal E. coli strains from cats, which is consistent with the results of several previous studies in different animal species.
Although the prevalence of ESBL-producing E. coli in cats was higher than the corresponding qAmpC production (7.8% versus 2.1%), cats are frequent carriers of acquired mechanisms associated with CTXR in E. coli isolates, harboring mostly the genes encoding CTX-M-1, CTX-M-15 and CTX-M-55 enzymes as well as the CMY-2 gene. Regarding the One Health approach, these resistance genes can be transferred among cats to humans by the transference of resistant bacteria or resistance genes. It is important to note that a high diversity of genetic lineages/sequence types was found in this research. To our knowledge, this is the first time that ESBL-encoding genes have been studied simultaneously in healthy and sick cats in Portugal as well as one of the few providing qAmpC-producing Enterobacteriaceae occurrence in healthy pets in this country. More studies should be carried out in the future to track the evolution of this type of β-lactamase in different environments.
Author Contributions
I.C.: conceptualization, methodology, investigation, resources, data curation, writing and rewriting; N.S.C.: investigation, data curation, writing—review and editing; R.C.: sampling, methodology; C.M.: sampling, methodology; P.P. (Paulo Pimenta): sampling, methodology; A.R.P.: sampling, methodology; S.M.-Á.: investigation, data curation; S.R.: sampling, methodology; V.S.: writing—review and editing; G.I.: conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition; C.T.: conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition; P.P. (Patrícia Poeta): conceptualization, methodology, validation, resources, data curation, writing—review and editing, visualization, supervision, project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
I.C. gratefully acknowledges the financial support of “Fundação para a Ciência e Tecnologia” (FCT—Portugal) related to PhD grant, through the reference SFRH/BD/133266/2017 (Medicina Clínica e Ciências da Saúde). Part of this study was financed by the project SAF2016-76571-R from the Agencia Estatal de Investigation (AEI) of Spain and FEDER of EU. N.S.C. was awarded a grant for the year 2018, from the Algerian Ministry of Higher Education and Scientific Research (The PNE Program), under the direction of Prof. Carmen Torres. V.S. is grateful to FCT (Fundação para a Ciência e a Tecnologia) for her PhD Grant (SFRH/BD/137947/2018).
Acknowledgments
The authors would like to thank Carla Oliveira from Transmonvete Veterinary Clinic and also Anabela Pais from Quinchosos Veterinary center who helped us with the collection of fecal samples of pets in Vila Real (Portugal). In the same line, we also would like to thank for the collaboration with the Local Kennel located with Vila Real.
Conflicts of Interest
The authors confirm that there are no known conflicts of interest associated with this publication. Preliminary data were presented as a poster entitled “Detection of blaCTX M genes and blaTEM genes in ESBL-producing Escherichia coli isolated from cats in Portugal” in BMAT 2018 Congress (Tunisia) and as part of an awarded oral presentation named “High frequency of ESBL-E. coli-producers in pets in Portugal with detection of ST131 clone carrying different variants of CTX-M genes” in IC2AR 2019 (Portugal).
References
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Carvalho, I.; Silva, N.; Carrola, J.; Silva, V.; Currie, C.; Igrejas, G.; Poeta, P. Antibiotic Resistance. In Antibiotic Drug Resistance; Capelo-Martínez, J.L., Igrejas, G., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 239–259. [Google Scholar] [CrossRef]
- Alonso, C.A.; Michael, G.B.; Li, J.; Somalo, S.; Simon, C.; Wang, Y.; Kaspar, H.; Kadlec, K.; Torres, C.; Schwarz, S. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 2017, 72, 1589–1596. [Google Scholar] [CrossRef]
- Schmitt, K.; Lehner, C.; Schuller, S.; Schüpbach-Regula, G.; Mevissen, M.; Peter, R.; Müntener, C.R.; Naegeli, H.; Willi, B. Antimicrobial use for selected diseases in cats in Switzerland. BMC Vet. Res. 2019, 15, 94. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Poeta, P.; Costa, D.; Sáenz, Y.; Klibi, N.; Ruiz-Larrea, F.; Rodrigues, J.; Torres, C. Characterization of Antibiotic Resistance Genes and Virulence Factors in Faecal enterococci of Wild Animals in Portugal. J. Vet. Med. 2005, 52, 396–402. [Google Scholar] [CrossRef]
- Abdallah, H.M.; Reuland, E.A.; Wintermans, B.B.; Al Naiemi, N.; Koek, A.; Abdelwahab, A.M.; Ammar, A.M.; Mohamed, A.A.; Vandenbroucke-Grauls, C.M.J.E. Extended-spectrum β-lactamases and/or carbapenemases-producing Enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS ONE 2015, 10, e0136052. [Google Scholar] [CrossRef]
- Cortes-Cortes, G.; Lozano-Zarain, P.; Torres, C.; Alonso, C.A.; Rios-Torres, A.M.; Castaneda, M.; Lopez-Pliego, L.; Navarro, A.; Del Carmen Rocha-Gracia, R. Extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans in Mexico, including subclone ST131-B2-O25:H4-H30-Rx. J. Glob. Antimicrob. Resist. 2017, 9, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Bagcigil, F.; Dokuzeylül, B.; Gocmen, H.; Yalcin, E.; İkiz, S.; Özgür, N.Y.; Ak, S. The Characterization of Escherichia coli Strains Isolated from Urinary Tract Infections in Cats and Dogs. JAES 2018, 3, 131–136. [Google Scholar]
- Zhang, P.L.C.; Shen, X.; Chalmers, G.; Reid-Smith, R.J.; Slavic, D.; Dick, H.; Boerlin, P. Prevalence and mechanisms of extended-spectrum cephalosporin resistance in clinical and fecal Enterobacteriaceae isolates from dogs in Ontario, Canada. Vet. Microbiol. 2018, 213, 82–88. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect.CMI 2018, 24, 1241–1250. [Google Scholar] [CrossRef]
- Alcala, L.; Alonso, C.A.; Simon, C.; Gonzalez-Esteban, C.; Oros, J.; Rezusta, A.; Ortega, C.; Torres, C. Wild Birds, Frequent Carriers of Extended-Spectrum β-Lactamase (ESBL) Producing Escherichia coli of CTX-M and SHV-12 Types. Microb. Ecol. 2016, 72, 861–869. [Google Scholar] [CrossRef]
- Alonso, C.A.; Mora, A.; Diaz, D.; Blanco, M.; Gonzalez-Barrio, D.; Ruiz-Fons, F.; Simon, C.; Blanco, J.; Torres, C. Occurrence and characterization of stx and/or eae-positive Escherichia coli isolated from wildlife, including a typical EPEC strain from a wild boar. Vet. Microbiol. 2017, 207, 69–73. [Google Scholar] [CrossRef]
- Day, M.J.; Rodriguez, I.; van Essen-Zandbergen, A.; Dierikx, C.; Kadlec, K.; Schink, A.K.; Wu, G.; Chattaway, M.A.; DoNascimento, V.; Wain, J.; et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016, 71, 1178–1182. [Google Scholar] [CrossRef]
- Brinas, L.; Zarazaga, M.; Sáenz, Y.; Ruiz-Larrea, F.; Torres, C. Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 2002, 46, 3156–3163. [Google Scholar] [CrossRef]
- Koga, V.L.; Maluta, R.P.; da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of South Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.G.; Novais, Â.; Rodrigues, C.; Nascimento, R.; Freitas, F.; Machado, E.; Peixe, L. Dynamics of clonal and plasmid backgrounds of Enterobacteriaceae producing acquired AmpC in Portuguese clinical settings over time. Int. J. Antimicrob. Agents 2019, 53, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Oikarainen, P.E.; Pohjola, L.K.; Pietola, E.S.; Heikinheimo, A. Direct vertical transmission of ESBL/pAmpC-producing Escherichia coli limited in poultry production pyramid. Vet. Microbiol. 2019, 231, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Kasbohrer, A.; Schwarz, S.; Rosler, U.; Kreienbrock, L.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Gracia, R.C.; Cortes-Cortes, G.; Lozano-Zarain, P.; Bello, F.; Martinez-Laguna, Y.; Torres, C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 beta-lactamases. Vet. Microbiol. 2015, 203, 315–319. [Google Scholar] [CrossRef]
- Gonçalves Ribeiro, T.; Novais, Â.; Machado, E.; Peixe, L. Acquired AmpC β-Lactamases among Enterobacteriaceae from Healthy Humans and Animals, Food, Aquatic and Trout Aquaculture Environments in Portugal. Pathogens 2020, 9, 273. [Google Scholar] [CrossRef]
- Maeyama, Y.; Taniguchi, Y.; Hayashi, W.; Ohsaki, Y.; Osaka, S.; Koide, S.; Tamai, K.; Nagano, Y.; Arakawa, Y.; Nagano, N. Prevalence of ESBL/AmpC genes and specific clones among the third-generation cephalosporin-resistant Enterobacteriaceae from canine and feline clinical specimens in Japan. Vet. Microbiol. 2018, 216, 183–189. [Google Scholar] [CrossRef]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyorala, S.; Ruzauskas, M.; et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef]
- Dandachi, I.; Chabou, S.; Daoud, Z.; Rolain, J.M. Prevalence and Emergence of Extended-Spectrum Cephalosporin-, Carbapenem- and Colistin-Resistant Gram Negative Bacteria of Animal Origin in the Mediterranean Basin. Front. Microbiol. 2018, 9, 2299. [Google Scholar] [CrossRef] [PubMed]
- Obodoechi, L.O.; Carvalho, I.; Chenouf, N.S.; Martínez-Álvarez, S.; Sadi, M.; Nwanta, J.A.; Chah, K.F.; Torres, C. Antimicrobial resistance in Escherichia coli isolates from frugivorous (Eidolon helvum) and insectivorous (Nycteris hispida) bats in Southeast Nigeria, with detection of CTX-M-15 producing isolates. CIMID 2021, 75, 101613. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Stamm, I.; Grobbel, M.; Kopp, P.A.; Guerra, B.; Stubbe, M.; Doi, Y.; Zong, Z.; Kola, A.; et al. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: Another pandemic clone combining multiresistance and extraintestinal virulence? J. Antimicrob. Chemother. 2014, 69, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Saenz, Y.; Zarazaga, M.; Brinas, L.; Lantero, M.; Ruiz-Larrea, F.; Torres, C. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain. Int. J. Antimicrob. Agents 2001, 18, 353–358. [Google Scholar] [CrossRef]
- Carattoli, A.; Lovari, S.; Franco, A.; Cordaro, G.; Di Matteo, P.; Battisti, A. Extended-Spectrum β-Lactamases in Escherichia coli Isolated from Dogs and Cats in Rome, Italy, from 2001 to 2003. Antimicrob. Agents Chemother. 2005, 49, 833–835. [Google Scholar] [CrossRef]
- Chung, Y.S.; Hu, Y.S.; Shin, S.; Lim, S.K.; Yang, S.J.; Park, Y.H.; Park, K.T. Mechanisms of quinolone resistance in Escherichia coli isolated from companion animals, pet-owners, and non-pet-owners. Int. J. Vet. Sci. 2017, 18, 449–456. [Google Scholar] [CrossRef]
- Hartantyo, S.H.P.; Chau, M.L.; Fillon, L.; Ariff, A.Z.B.M.; Kang, J.S.L.; Aung, K.T.; Gutiérrez, R.A. Sick pets as potential reservoirs of antibiotic-resistant bacteria in Singapore. Antimicrob. Resist. Infect. Control. 2018, 7, 106. [Google Scholar] [CrossRef]
- Melo, L.C.; Oresco, C.; Leigue, L.; Netto, H.M.; Melville, P.A.; Benites, N.R.; Saras, E.; Haenni, M.; Lincopan, N.; Madec, J.Y. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 2018, 221, 59–66. [Google Scholar] [CrossRef]
- Sallem, R.B.; Gharsa, H.; Slama, K.B.; Rojo-Bezares, B.; Estepa, V.; Porres-Osante, N.; Jouini, A.; Klibi, N.; Saenz, Y.; Boudabous, A.; et al. First detection of CTX-M-1, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector Borne Zoonotic Dis. 2013, 13, 98–102. [Google Scholar] [CrossRef]
- Freitas, F.; Machado, E.; Ribeiro, T.G.; Novais, Â.; Peixe, L. Long-term dissemination of acquired AmpC β-lactamases among Klebsiella spp. and Escherichia coli in Portuguese clinical settings. Eur J. Clin. Microbiol. Infect. Dis. 2014, 33, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Amador, P.; Prudêncio, C.; Tomaz, C.T.; Tavares-Ratado, P.; Fernandes, R. ESBL and AmpC β-Lactamases in Clinical Strains of Escherichia coli from Serra da Estrela, Portugal. Medicina 2019, 55, 272. [Google Scholar] [CrossRef] [PubMed]
- Feria, C.; Ferreira, E.; Correia, J.D.; Goncalves, J.; Canica, M. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 2002, 49, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Feria, C.P.; Correia, J.D.; Goncalves, J.; Machado, J. Detection of virulence factors in uropathogenic Escherichia coli isolated from humans, dogs and cats in Portugal. Adv. Exp. Med. Biol. 2000, 485, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Poeta, P.; Briñas, L.; Sáenz, Y.; Rodrigues, J.; Torres, C. Detection of CTX-M-1 and TEM-52 β-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 2004, 54, 960–961. [Google Scholar] [CrossRef][Green Version]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, F.L.; Belas, A.; Foti, M.; Fisichella, V.; Marques, C.; Pomba, C. Detection of multidrug resistance and extended-spectrum/plasmid-mediated AmpC beta-lactamase genes in Enterobacteriaceae isolates from diseased cats in Italy. Med. Surg. 2020, 22, 613–622. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, A.; Hutton, T.A.; Schifferli, D.M.; Rankin, S.C. First Detection of CTX-M and SHV Extended-Spectrum β-Lactamases in Escherichia coli Urinary Tract Isolates from Dogs and Cats in the United States. Antimicrob. Agents Chemother. 2010, 54, 3489–3492. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Hordijk, J.; Schoormans, A.; Kwakernaak, M.; Duim, B.; Broens, E.; Dierikx, C.; Mevius, D.; Wagenaar, J.A. High prevalence of fecal carriage of extended spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013, 4, 242. [Google Scholar] [CrossRef]
- Leite-Martins, L.R.; Mahu, M.I.; Costa, A.L.; Mendes, A.; Lopes, E.; Mendonca, D.M.; Niza-Ribeiro, J.J.; de Matos, A.J.; da Costa, P.M. Prevalence of antimicrobial resistance in enteric Escherichia coli from domestic pets and assessment of associated risk markers using a generalized linear mixed model. Prev. Vet. Med. 2014, 117, 28–39. [Google Scholar] [CrossRef]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic Resistance and Virulence Traits in Vancomycin-Resistant Enterococci (VRE) and Extended-Spectrum-Lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from Humans and Pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef]
- Dhanji, H.; Doumith, M.; Rooney, P.J.; O’Leary, M.C.; Loughrey, A.C.; Hope, R. Molecular epidemiology of fluoroquinolone-resistant ST131 Escherichia coli producing CTX-M extended-spectrum β-lactamases in nursing homes in Belfast, UK. J. Antimicrob. Chemother. 2011, 66, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Carvalho, J.; Castro, A.; Chenouf, N.; Igrejas, G.; Torres, C.; Poeta, P. ESBL-producing Escherichia coli from hospitalized patients with bacteraemia: A reservoir for antibiotic-resistance genes. In II Jornadas de Jóvenes Investigadores; INNOVA: Salamanca, Spain, 2019; p. CSP1. [Google Scholar]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable High Rates of Extended-Spectrum-Beta-Lactamase-Producing Escherichia coli in Birds of Prey from Germany and Mongolia. PLoS ONE 2013, 7, e53039. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El Garch, F.; Sauget, M.; Hocquet, D.; LeChaudee, D.; Woehrle, F.; Bertrand, X. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin. Microbiol. Infect. 2017, 23, 51.e51–51.e54. [Google Scholar] [CrossRef] [PubMed]
- Solà-Ginés, M.; Cameron-Veas, K.; Badiola, I.; Dolz, R.; Majó, N.; Dahbi, G.; Viso, S.; Mora, A.; Blanco, J.; Piedra-Carrasco, N.; et al. Diversity of Multi-Drug Resistant Avian Pathogenic Escherichia coli (APEC) Causing Outbreaks of Colibacillosis in Broilers during 2012 in Spain. PLoS ONE 2015, 10, e0143191. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Pimenta, P.; Cunha, R.; Martins, C.; Igrejas, G.; Torres, C.; Poeta, P. Diversity of CTX-M beta-lactamases in Escherichia coli isolated from dogs in Portugal. In Proceedings of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Amsterdam, The Netherlands, 13–16 April 2019; p. 189. [Google Scholar]
- Carvalho, I.; Tejedor-Junco, M.T.; González-Martín, M.; Corbera, J.A.; Suárez-Pérez, A.; Silva, V.; Igrejas, G.; Torres, C.; Poeta, P. Molecular Diversity of Extended-Spectrum β-lactamase-producing Escherichia coli from vultures in Canary Islands. Environ. Microbiol. Rep. 2020, 12, 540–547. [Google Scholar] [CrossRef]
- Johan, K.; Hilde, R.; Wesley, S.; Hong, Y.; Tryggve, N.; Åsa, M. Rapid Increase in Carriage Rates of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases in Healthy Preschool Children, Sweden. Emerg. Infect. Dis 2018, 24, 1874. [Google Scholar] [CrossRef]
- Nadia Safia Chenouf, I.C.; Messaï, C.R.; Ruiz-Ripa, L.; Mama, O.M.; Titouche, Y.; Zitouni, A.; Hakem, A.; Torres, C. Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae from Broiler Liver in the Center of Algeria, with Detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli. Microb. Drug Resist. 2020, 27, 268–276. [Google Scholar] [CrossRef]
- Ellis, S.J.; Crossman, L.C.; McGrath, C.J.; Chattaway, M.A.; Hölken, J.M.; Brett, B.; Bundy, L.; Kay, G.L.; Wain, J.; Schüller, S. Identification and characterisation of enteroaggregative Escherichia coli subtypes associated with human disease. Sci. Rep. 2020, 10, 7475. [Google Scholar] [CrossRef]
- Papouskova, A.; Masarikova, M.; Valcek, A.; Senk, D.; Cejkova, D.; Jahodarova, E.; Cizek, A. Genomic analysis of Escherichia coli strains isolated from diseased chicken in the Czech Republic. BMC Vet. Res. 2020, 16, 189. [Google Scholar] [CrossRef] [PubMed]
- Manageiro, V.; Romao, R.; Moura, I.B.; Sampaio, D.A.; Vieira, L.; Ferreira, E.; Canica, M. Molecular Epidemiology and Risk Factors of Carbapenemase-Producing Enterobacteriaceae Isolates in Portuguese Hospitals: Results From European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE). Front. Microbiol. 2018, 9, 2834. [Google Scholar] [CrossRef] [PubMed]
- CLSI (Clinical and Laboratory Standards Institute). Performed Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement: Wayne, PA, USA, 2019. [Google Scholar]
- Mirelis, B.; Rivera, A.; Miró, E.; Mesa, R.J.; Navarro, F.; Coll, P. A simple phenotypic method for differentiation between acquired and chromosomal AmpC beta-lactamases in Escherichia coli. Enferm. Infecc. Microbiol. Clin. 2006, 24, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.S.; Quigley, M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981, 114, 193–197. [Google Scholar] [CrossRef]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Porres-Osante, N.; Azcona-Gutiérrez, J.M.; Rojo-Bezares, B.; Undabeitia, E.; Torres, C.; Sáenz, Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J. Antimicrob. Chemother. 2014, 69, 1792–1795. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 2019, 318, 108478. [Google Scholar] [CrossRef]
- Pérez-Moreno, M.O.; Estepa, V.; Sáenz, Y.; Cortell-Ortolá, M.; Fort-Gallifa, I.; Ruiz, J.; Torres, C. Intrahospitalary dissemination of Klebsiella pneumoniae carrying bla(DHA-1) and qnrB4 genes within a novel complex class 1 integron. Diagn. Microbiol. Infect. Dis. 2012, 73, 210–211. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Abdelwahab, S.; Hasanen, A.; Safwat, D. Multidrug resistant Egyptian isolates of Acinetobacter baumannii. Am. J. Sci. 2011, 7, 1013–1019. [Google Scholar]
- Adams-Haduch, J.M.; Paterson, D.L.; Sidjabat, H.E.; Pasculle, A.W.; Potoski, B.A.; Muto, C.A.; Harrison, L.H.; Doi, Y. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at a tertiary medical center in Pennsylvania. Antimicrob. Agents Chemother. 2008, 52, 3837–3843. [Google Scholar] [CrossRef] [PubMed]
- Vinué, L.; Sáenz, Y.; Somalo, S.; Escudero, E.; Moreno, M.A.; Ruiz-Larrea, F.; Torres, C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. Antimicrob. Chemother. 2008, 62, 934–937. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).