Bacteriophage-Derived Depolymerases against Bacterial Biofilm
Abstract
1. Introduction
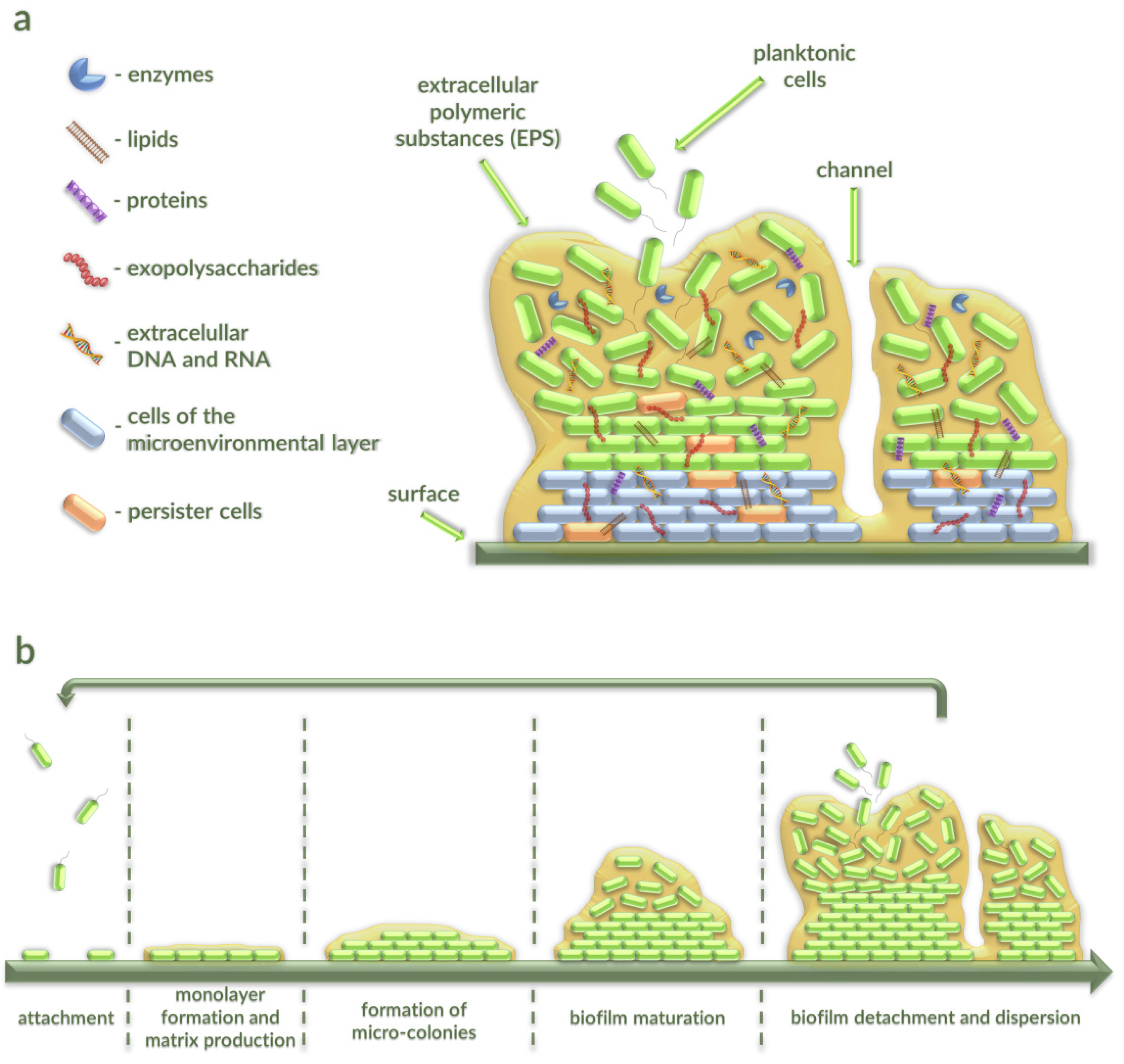
2. Characterization of Bacterial Biofilms
2.1. Stages of Bacterial Biofilm Formation
2.2. The Occurence of Bacterial Biofilms
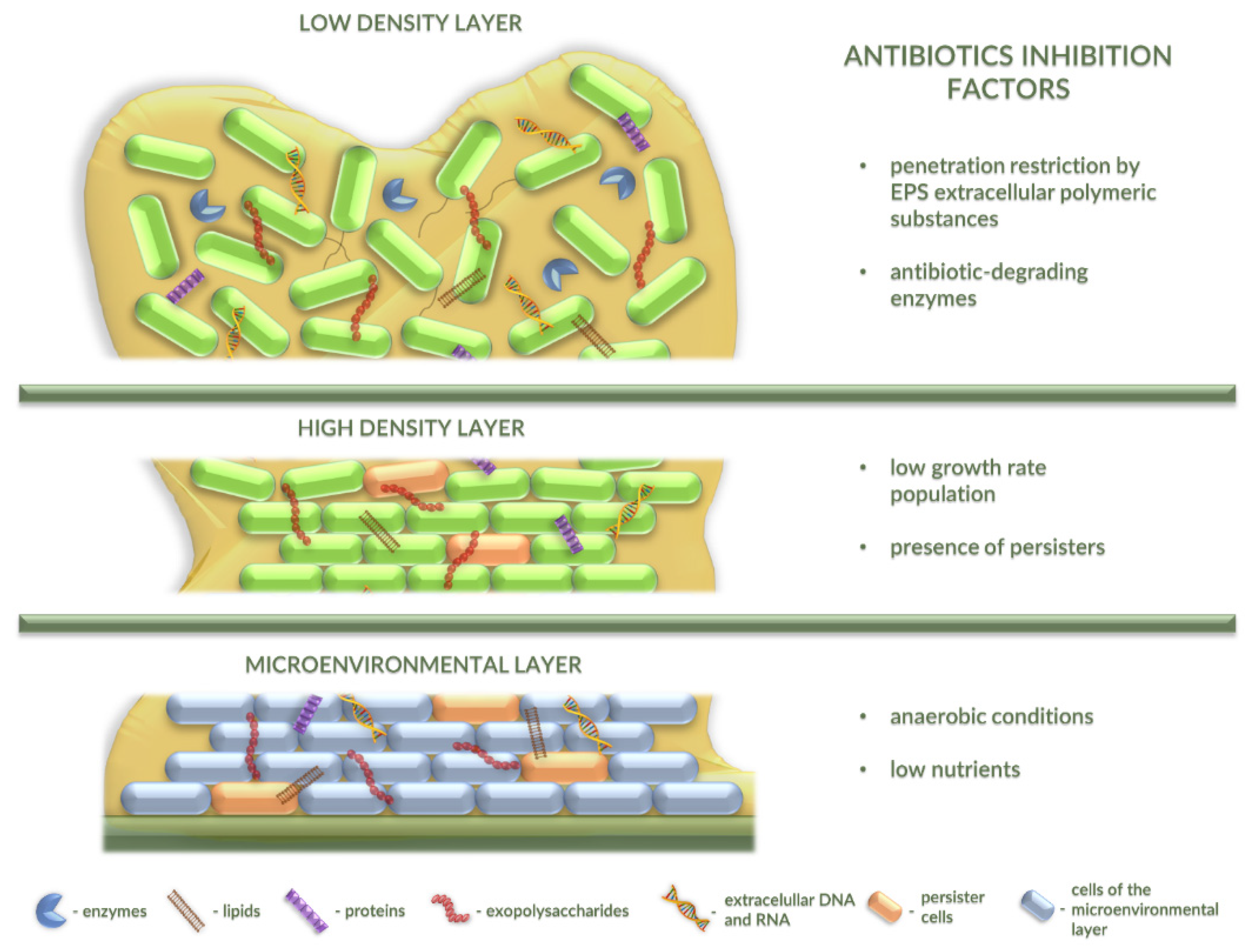
2.3. Antibiotic Resistance in Bacterial Biofilm
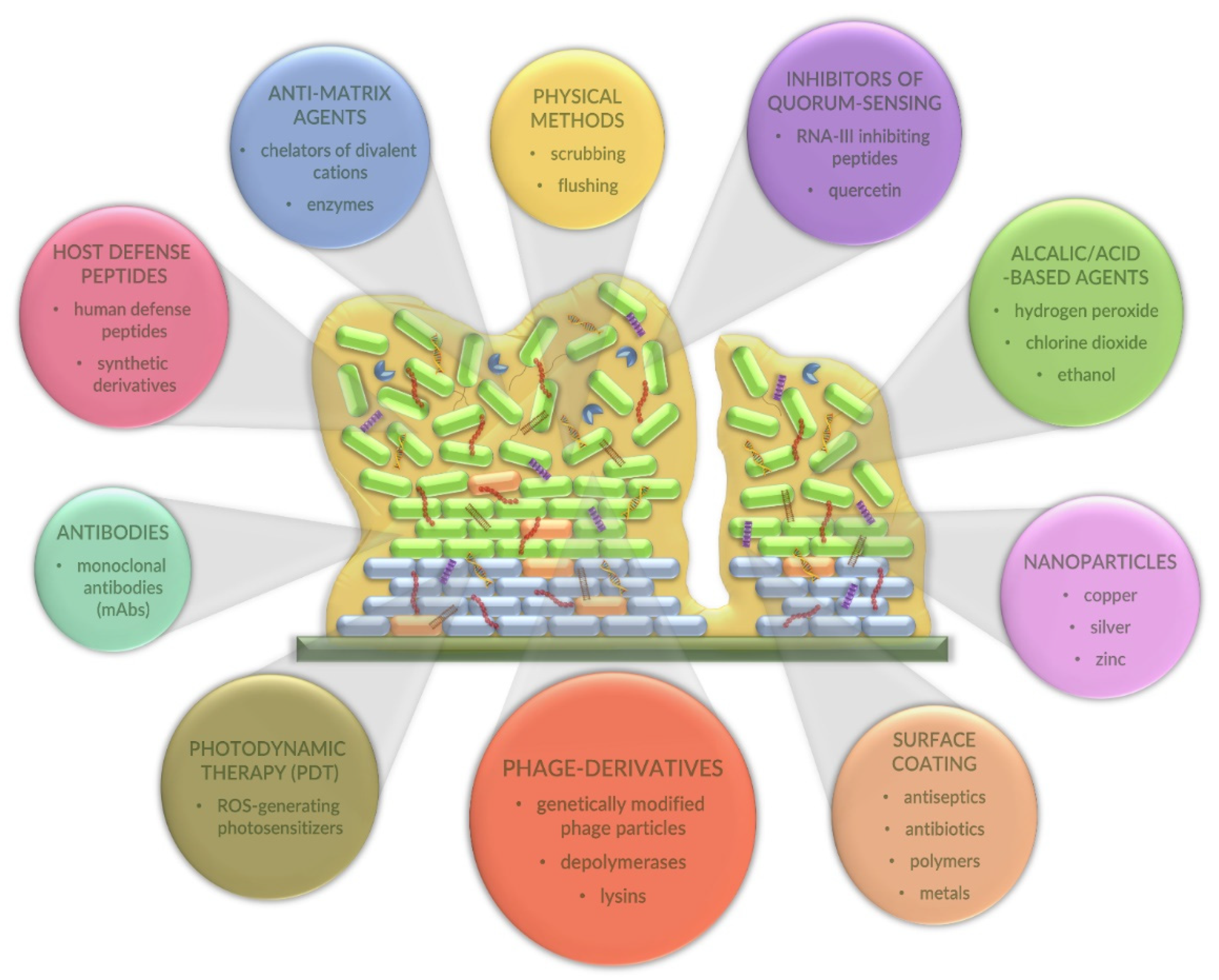
2.4. Alternative Strategies for Combating Bacterial Biofilms
3. Bacteriophage Depolymerases as an Alternative to Antibiotics
The Antibiofilm Activity of Phage Depolymerases: Examples of Applications of Phage Depolymerases against Bacterial Biofilms
4. Combination Therapy of Phage-Derived Depolymerases and Different Therapeutic Agents
4.1. Various Antibiotics
4.2. Bacteriophages
4.3. Chemical Compounds
4.4. Natural Compounds
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. P T 2015, 40, 277–283. [Google Scholar] [PubMed]
- Rode, D.K.H.; Singh, P.K.; Drescher, K. Multicellular and Unicellular Responses of Microbial Biofilms to Stress. Biol. Chem. 2020, 401, 1365–1374. [Google Scholar] [CrossRef]
- Yamasaki, R.; Kawano, A.; Yoshioka, Y.; Ariyoshi, W. Rhamnolipids and Surfactin Inhibit the Growth or Formation of Oral Bacterial Biofilm. BMC Microbiol. 2020, 20, 358. [Google Scholar] [CrossRef]
- Abebe, G. The Role of Bacterial Biofilm in Antibiotic Resistance and Food Contamination. Int. J. Microbiol. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Rasool, F.N.; Saavedra, M.A.; Pamba, S.; Perold, V.; Mmochi, A.J.; Maalim, M.; Simonsen, L.; Buur, L.; Pedersen, R.H.; Syberg, K.; et al. Isolation and Characterization of Human Pathogenic Multidrug Resistant Bacteria Associated with Plastic Litter Collected in Zanzibar. J. Hazard. Mater. 2021, 405, 124591. [Google Scholar] [CrossRef]
- Henrici, A.T. Studies of Freshwater Bacteria: I. A Direct Microscopic Technique. J. Bacteriol. 1933, 25, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Mack, W.N.; Mack, J.P.; Ackerson, A.O. Microbial Film Development in a Trickling Filter. Microb. Ecol. 1975, 2, 215–226. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial Biofilm and Associated Infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Arunasri, K.; Mohan, S.V. Chapter 2.3—Biofilms: Microbial Life on the Electrode Surface. In Microbial Electrochemical Technology; Mohan, S.V., Varjani, S., Pandey, A., Eds.; Biomass, Biofuels and Biochemicals; Elsevier: Amsterdam, The Netherlands, 2019; pp. 295–313. ISBN 978-0-444-64052-9. [Google Scholar]
- Flemming, H.-C.; Wingender, J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Somma, A.D.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Inhibition of Bacterial Biofilm Formation. Bact. Biofilms 2020. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm Formation Mechanisms and Targets for Developing Antibiofilm Agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Carmello, J.C.; de Annunzio, S.R.; Fontana, C.R. Composition, Structure, and Formation of Biofilms Constituted by Periodontopathogenic Microorganisms. Bact. Biofilms 2020. [Google Scholar] [CrossRef]
- Chew, S.C.; Yang, L. Biofilms. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 407–415. ISBN 978-0-12-384953-3. [Google Scholar]
- Rumbaugh, K.P.; Sauer, K. Biofilm Dispersion. Nat. Rev. Microbiol. 2020, 18, 571–586. [Google Scholar] [CrossRef]
- Maurya, A.; Raj, A. 5—Recent advances in the application of biofilm in bioremediation of industrial wastewater and organic pollutants. In Microorganisms for Sustainable Environment and Health; Chowdhary, P., Raj, A., Verma, D., Akhter, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 81–118. ISBN 978-0-12-819001-2. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Otter, J.A.; Vickery, K.; Walker, J.T.; de Lancey Pulcini, E.; Stoodley, P.; Goldenberg, S.D.; Salkeld, J.a.G.; Chewins, J.; Yezli, S.; Edgeworth, J.D. Surface-Attached Cells, Biofilms and Biocide Susceptibility: Implications for Hospital Cleaning and Disinfection. J. Hosp. Infect. 2015, 89, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Taraszkiewicz, A.; Fila, G.; Grinholc, M.; Nakonieczna, J. Innovative Strategies to Overcome Biofilm Resistance. Available online: https://www.hindawi.com/journals/bmri/2013/150653/ (accessed on 16 December 2020).
- Desai, J.V.; Mitchell, A.P.; Andes, D.R. Fungal Biofilms, Drug Resistance, and Recurrent Infection. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.C.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M.S. A New Classification Scheme for Periodontal and Peri-Implant Diseases and Conditions—Introduction and Key Changes from the 1999 Classification. J. Clin. Periodontol. 2018, 45, S1–S8. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Bjarnsholt, T. Pseudomonas Aeruginosa Biofilms in Cystic Fibrosis. Future Microbiol. 2010, 5, 1663–1674. [Google Scholar] [CrossRef]
- Reffuveille, F.; de la Fuente-Núñez, C.; Mansour, S.; Hancock, R.E.W. A Broad-Spectrum Antibiofilm Peptide Enhances Antibiotic Action against Bacterial Biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef]
- Malheiro, J.F.; Simões, M. Antimicrobial resistance of biofilms in medical devices. In Biofilms and Implantable Medical Devices: Infection and Control; Woodhead Publishing: Cambridge, UK, 2017; pp. 97–113. ISBN 978-0-08-100382-4. [Google Scholar]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-Associated Infections, Medical Devices and Biofilms: Risk, Tolerance and Control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef]
- Li, X.-H.; Lee, J.-H. Antibiofilm Agents: A New Perspective for Antimicrobial Strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Rafii, F. Antimicrobial Resistance in Clinically Important Biofilms. World J. Pharmacol. 2015, 4, 31. [Google Scholar] [CrossRef]
- Banerjee, A.; Batabyal, K.; Singh, A.D.; Joardar, S.N.; Dey, S.; Isore, D.P.; Sar, T.K.; Dutta, T.K.; Bandyopadhyay, S.; Samanta, I. Multi-Drug Resistant, Biofilm-Producing High-Risk Clonal Lineage of Klebsiella in Companion and Household Animals. Lett. Appl. Microbiol. 2020, 71, 580–587. [Google Scholar] [CrossRef]
- Ciofu, O.; Rojo-Molinero, E.; Macià, M.D.; Oliver, A. Antibiotic Treatment of Biofilm Infections. APMIS 2017, 125, 304–319. [Google Scholar] [CrossRef]
- Beloin, C.; Renard, S.; Ghigo, J.-M.; Lebeaux, D. Novel Approaches to Combat Bacterial Biofilms. Curr. Opin. Pharm. 2014, 18, 61–68. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for Combating Bacterial Biofilms: A Focus on Anti-Biofilm Agents and Their Mechanisms of Action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Achermann, Y.; Goldstein, E.J.C.; Coenye, T.; Shirtliff, M.E. Propionibacterium Acnes: From Commensal to Opportunistic Biofilm-Associated Implant Pathogen. Clin. Microbiol. Rev. 2014, 27, 419–440. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Moser, C.; Wang, H.; Høiby, N.; Song, Z. Strategies for Combating Bacterial Biofilm Infections. Int. J. Oral Sci. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Hengzhuang, W.; Wu, H.; Ciofu, O.; Song, Z.; Høiby, N. In Vivo Pharmacokinetics/Pharmacodynamics of Colistin and Imipenem in Pseudomonas Aeruginosa Biofilm Infection. Antimicrob. Agents Chemother. 2012, 56, 2683–2690. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control. 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Camilli, A.; Bassler, B.L. Bacterial Small-Molecule Signaling Pathways. Science 2006, 311, 1113–1116. [Google Scholar] [CrossRef]
- Chua, S.L.; Yam, J.K.H.; Hao, P.; Adav, S.S.; Salido, M.M.; Liu, Y.; Givskov, M.; Sze, S.K.; Tolker-Nielsen, T.; Yang, L. Selective Labelling and Eradication of Antibiotic-Tolerant Bacterial Populations in Pseudomonas Aeruginosa Biofilms. Nat. Commun. 2016, 7, 10750. [Google Scholar] [CrossRef]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple Roles for Enterococcus Faecalis Glycosyltransferases in Biofilm-Associated Antibiotic Resistance, Cell Envelope Integrity, and Conjugative Transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef]
- Brackman, G.; Breyne, K.; De Rycke, R.; Vermote, A.; Van Nieuwerburgh, F.; Meyer, E.; Van Calenbergh, S.; Coenye, T. The Quorum Sensing Inhibitor Hamamelitannin Increases Antibiotic Susceptibility of Staphylococcus Aureus Biofilms by Affecting Peptidoglycan Biosynthesis and EDNA Release. Sci. Rep. 2016, 6, 20321. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Wang, L.; Zhang, H.; Zhu, M.; Zhang, P.; Zhu, X. Extracellular Polymeric Substances Affect the Responses of Multi-Species Biofilms in the Presence of Sulfamethizole. Environ. Pollut. 2018, 235, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Billings, N.; Birjiniuk, A.; Samad, T.S.; Doyle, P.S.; Ribbeck, K. Material Properties of Biofilms-a Review of Methods for Understanding Permeability and Mechanics. Rep. Prog. Phys. 2015, 78, 036601. [Google Scholar] [CrossRef]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.L.; Parsek, M.R. The Pel Polysaccharide Can Serve a Structural and Protective Role in the Biofilm Matrix of Pseudomonas Aeruginosa. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Miyaue, S.; Suzuki, E.; Komiyama, Y.; Kondo, Y.; Morikawa, M.; Maeda, S. Bacterial Memory of Persisters: Bacterial Persister Cells Can Retain Their Phenotype for Days or Weeks After Withdrawal from Colony-Biofilm Culture. Front. Microbiol. 2018, 9, 1396. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial Persister Cell Formation and Dormancy. Appl. Environ. Microbiol 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.-F. Molecular Mechanisms of Biofilm-Based Antibiotic Resistance and Tolerance in Pathogenic Bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Papadovasilaki, M.; Oberthür, D.; Gessmann, R.; Sarrou, I.; Betzel, C.; Scoulica, E.; Petratos, K. Biophysical and Enzymatic Properties of Aminoglycoside Adenylyltransferase AadA6 from Pseudomonas Aeruginosa. Biochem. Biophys. Rep. 2015, 4, 152–157. [Google Scholar] [CrossRef]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of Antibiotic Penetration, Oxygen Limitation, and Low Metabolic Activity to Tolerance of Pseudomonas Aeruginosa Biofilms to Ciprofloxacin and Tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Shang, W.; Yang, Y.; Zhou, R.; Rao, X. Fighting Mixed-Species Microbial Biofilms With Cold Atmospheric Plasma. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-Species Biofilms: Living with Friendly Neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Govaert, M.; Smet, C.; Walsh, J.L.; Van Impe, J.F.M. Dual-Species Model Biofilm Consisting of Listeria Monocytogenes and Salmonella Typhimurium: Development and Inactivation with Cold Atmospheric Plasma (CAP). Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P. A Novel Tail-Associated O91-Specific Polysaccharide Depolymerase from a Podophage Reveals Lytic Efficacy of Shiga Toxin-Producing Escherichia Coli. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef]
- Bayston, R.; Fisher, L.E.; Weber, K. An Antimicrobial Modified Silicone Peritoneal Catheter with Activity against both Gram-Positive and Gram-Negative Bacteria. Biomaterials 2009, 30, 3167–3173. [Google Scholar] [CrossRef]
- Cyphert, E.L.; von Recum, H.A. Emerging Technologies for Long-Term Antimicrobial Device Coatings: Advantages and Limitations. Exp. Biol Med. 2017, 242, 788–798. [Google Scholar] [CrossRef]
- Kart, D.; Kustimur, A.S.; Sağıroğlu, M.; Kalkancı, A. Evaluation of Antimicrobial Durability and Anti-Biofilm Effects in Urinary Catheters against Enterococcus Faecalis Clinical Isolates and Reference Strains. Balk. Med. J. 2017, 34, 546–552. [Google Scholar] [CrossRef]
- Marques, D.M.; de Oliveira, V.C.; Souza, M.T.; Zanotto, E.D.; Issa, J.P.M.; Watanabe, E. Biomaterials for Orthopedics: Anti-Biofilm Activity of a New Bioactive Glass Coating on Titanium Implants. Biofouling 2020, 36, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Thakur, S.; Gupta, A.; Kaur, G.; Pandey, O.P. Antibacterial and Anticancerous Drug Loading Kinetics for (10-x)CuO-XZnO-20CaO-60SiO2-10P2O5 (2 ≤ x ≤ 8) Mesoporous Bioactive Glasses. J. Mater. Sci. Mater. Med. 2017, 28, 11. [Google Scholar] [CrossRef]
- Barbera, L.; Plano, L.M.D.; Franco, D.; Gattuso, G.; Guglielmino, S.P.P.; Lando, G.; Notti, A.; Parisi, M.F.; Pisagatti, I. Antiadhesive and Antibacterial Properties of Pillar [5] Arene-Based Multilayers. Chem. Commun. 2018, 54, 10203–10206. [Google Scholar] [CrossRef] [PubMed]
- Waryah, C.B.; Wells, K.; Ulluwishewa, D.; Chen-Tan, N.; Gogoi-Tiwari, J.; Ravensdale, J.; Costantino, P.; Gökçen, A.; Vilcinskas, A.; Wiesner, J.; et al. In Vitro Antimicrobial Efficacy of Tobramycin against Staphylococcus Aureus Biofilms in Combination with or Without DNase I and/or Dispersin B: A Preliminary Investigation. Microb. Drug Resist. 2017, 23, 384–390. [Google Scholar] [CrossRef]
- Cavaliere, R.; Ball, J.L.; Turnbull, L.; Whitchurch, C.B. The Biofilm Matrix Destabilizers, EDTA and DNaseI, Enhance the Susceptibility of Nontypeable Hemophilus Influenzae Biofilms to Treatment with Ampicillin and Ciprofloxacin. Microbiologyopen 2014, 3, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Nithyanand, P.; Beema Shafreen, R.M.; Muthamil, S.; Karutha Pandian, S. Usnic Acid Inhibits Biofilm Formation and Virulent Morphological Traits of Candida Albicans. Microbiol. Res. 2015, 179, 20–28. [Google Scholar] [CrossRef]
- De la Fuente-Núñez, C.; Cardoso, M.H.; de Souza Cândido, E.; Franco, O.L.; Hancock, R.E.W. Synthetic Antibiofilm Peptides. Biochim. Biophys. Acta 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Rampioni, G.; Leoni, L.; Williams, P. The Art of Antibacterial Warfare: Deception through Interference with Quorum Sensing-Mediated Communication. Bioorg. Chem. 2014, 55, 60–68. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharm. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Gaglione, R.; Cesaro, A.; Dell’Olmo, E.; Di Girolamo, R.; Tartaglione, L.; Pizzo, E.; Arciello, A. Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. Int. J. Mol. Sci. 2020, 21, 2049. [Google Scholar] [CrossRef]
- Shahrour, H.; Ferrer-Espada, R.; Dandache, I.; Bárcena-Varela, S.; Sánchez-Gómez, S.; Chokr, A.; Martinez-de-Tejada, G. AMPs as Anti-Biofilm Agents for Human Therapy and Prophylaxis. Adv. Exp. Med. Biol. 2019, 1117, 257–279. [Google Scholar] [CrossRef]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.W.; Rehm, B.H.A.; Hancock, R.E.W. Human Host Defense Peptide LL-37 Prevents Bacterial Biofilm Formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.-C.; Huang, C.-T. Effects of Quorum-Sensing Deficiency on Pseudomonas Aeruginosa Biofilm Formation and Antibiotic Resistance. J. Antimicrob. Chemother. 2002, 49, 309–314. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, C. Exploiting Quorum Sensing Interfering Strategies in Gram-Negative Bacteria for the Enhancement of Environmental Applications. Front. Microbiol. 2016, 6. [Google Scholar] [CrossRef]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin Influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Kunimura, K.; Suzuki, J.-I.; Kodera, Y. Antimicrobial Properties of Hydrophobic Compounds in Garlic: Allicin, Vinyldithiin, Ajoene and Diallyl Polysulfides. Exp. Med. 2020, 19, 1550–1553. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Zhong, N. Quorum Sensing Inhibition and Anti-Biofilm Activity of Traditional Chinese Medicines. Food Saf. Some Glob. Trends 2018. [Google Scholar] [CrossRef]
- Hook, A.L.; Chang, C.-Y.; Yang, J.; Luckett, J.; Cockayne, A.; Atkinson, S.; Mei, Y.; Bayston, R.; Irvine, D.J.; Langer, R.; et al. Combinatorial Discovery of Polymers Resistant to Bacterial Attachment. Nat. Biotechnol. 2012, 30, 868–875. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Sobala, M.; Bruhn-Olszewska, B.; Cashel, M.; Potrykus, K. Methylobacterium extorquens RSH Enzyme Synthesizes (p)ppGpp and pppApp in Vitro and In Vivo, and Leads to Discovery of pppApp Synthesis in Escherichia coli. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Morones-Ramirez, J.R.; Winkler, J.A.; Spina, C.S.; Collins, J.J. Silver Enhances Antibiotic Activity against Gram-Negative Bacteria. Sci. Transl. Med. 2013, 5, 190ra81. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Núñez, C.; Mansour, S.C.; Wang, Z.; Jiang, L.; Breidenstein, E.B.M.; Elliott, M.; Reffuveille, F.; Speert, D.P.; Reckseidler-Zenteno, S.L.; Shen, Y.; et al. Anti-Biofilm and Immunomodulatory Activities of Peptides that Inhibit Biofilms Formed by Pathogens Isolated from Cystic Fibrosis Patients. Antibiotics 2014, 3, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kim, Y.-G.; Gwon, G.; Wood, T.K.; Lee, J. Halogenated Indoles Eradicate Bacterial Persister Cells and Biofilms. Amb Express 2016, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Prateeksha, N.; Upreti, D.K.; Singh, B.R.; Defoirdt, T.; Gupta, V.K.; De Souza, A.O.; Singh, H.B.; Barreira, J.C.M.; Ferreira, I.C.F.R.; et al. Bactericidal, Quorum Quenching and Anti-Biofilm Nanofactories: A New Niche for Nanotechnologists. Crit. Rev. Biotechnol. 2017, 37, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Markowska, K.; Grudniak, A.M.; Wolska, K.I. Silver Nanoparticles as an Alternative Strategy against Bacterial Biofilms. Acta Biochim. Pol. 2013, 60, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Stensberg, M.C.; Wei, Q.; McLamore, E.S.; Porterfield, D.M.; Wei, A.; Sepúlveda, M.S. Toxicological Studies on Silver Nanoparticles: Challenges and Opportunities in Assessment, Monitoring and Imaging. Nanomedicine 2011, 6, 879–898. [Google Scholar] [CrossRef]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of Bacteriophages versus Phage Enzymes to Combat and Cure Bacterial Infections: An Ambitious and Also a Realistic Application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef] [PubMed]
- Ferriol-González, C.; Domingo-Calap, P. Phages for Biofilm Removal. Antibiotics 2020, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Doub, J.B. Bacteriophage Therapy for Clinical Biofilm Infections: Parameters That Influence Treatment Protocols and Current Treatment Approaches. Antibiotics 2020, 9, 799. [Google Scholar] [CrossRef]
- Tursi, S.A.; Puligedda, R.D.; Szabo, P.; Nicastro, L.K.; Miller, A.L.; Qiu, C.; Gallucci, S.; Relkin, N.R.; Buttaro, B.A.; Dessain, S.K.; et al. Salmonella Typhimurium Biofilm Disruption by a Human Antibody That Binds a Pan-Amyloid Epitope on Curli. Nat. Commun. 2020, 11, 1007. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.Q.; Estellés, A.; Li, L.; Abdelhady, W.; Gonzales, R.; Bayer, A.S.; Tenorio, E.; Leighton, A.; Ryser, S.; Kauvar, L.M. A Human Biofilm-Disrupting Monoclonal Antibody Potentiates Antibiotic Efficacy in Rodent Models of Both Staphylococcus Aureus and Acinetobacter Baumannii Infections. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Lyu, Z.; Shang, Y.; Wang, X.; Wu, Y.; Zheng, J.; Liu, H.; Gong, T.; Ye, L.; Qu, D. Monoclonal Antibodies Specific to the Extracellular Domain of Histidine Kinase YycG of Staphylococcus Epidermidis Inhibit Biofilm Formation. Front. Microbiol. 2020, 11, 1839. [Google Scholar] [CrossRef] [PubMed]
- Zagami, R.; Franco, D.; Pipkin, J.D.; Antle, V.; De Plano, L.; Patanè, S.; Guglielmino, S.; Monsù Scolaro, L.; Mazzaglia, A. Sulfobutylether-β-Cyclodextrin/5,10,15,20-Tetrakis(1-Methylpyridinium-4-Yl)Porphine Nanoassemblies with Sustained Antimicrobial Phototherapeutic Action. Int. J. Pharm. 2020, 585, 119487. [Google Scholar] [CrossRef]
- Yuan, Z.; Tao, B.; He, Y.; Mu, C.; Liu, G.; Zhang, J.; Liao, Q.; Liu, P.; Cai, K. Remote Eradication of Biofilm on Titanium Implant via Near-Infrared Light Triggered Photothermal/Photodynamic Therapy Strategy. Biomaterials 2019, 223, 119479. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, J.; Liu, X.; Cui, Z.; Yang, X.; Zhu, S.; Li, Z.; Yuan, X.; Zheng, Y.; Yeung, K.W.K.; et al. Rapid Biofilm Eradication on Bone Implants Using Red Phosphorus and Near-Infrared Light. Adv. Mater. 2018, 30, e1801808. [Google Scholar] [CrossRef]
- Matusiak, D. Urinary Tract Infections Caused by Proteus Mirabilis—Role of the Biofilm and the Encrustation of the Urological Catheter. Postepy Mikrobiol. 2014, 53, 173–181. [Google Scholar]
- Pokrowiecki, R.; Tyski, S.; Zaleska, M. Infections Associated with Implantable Biomaterials. Postepy Mikrobiol. 2014, 53, 123–134. [Google Scholar]
- Górski, A.; Borysowski, J.; Międzybrodzki, R. Phage Therapy: Towards a Successful Clinical Trial. Antibiotics 2020, 9, 827. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-Encoded Depolymerases: Their Diversity and Biotechnological Applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-Encoded Virion-Associated Enzymes to Overcome the Carbohydrate Barriers during the Infection Process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; São-José, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10, 396. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and Phage-Derived Proteins--Application Approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef]
- Pan, Y.-J.; Lin, T.-L.; Chen, Y.-Y.; Lai, P.-H.; Tsai, Y.-T.; Hsu, C.-R.; Hsieh, P.-F.; Lin, Y.-T.; Wang, J.-T. Identification of Three Podoviruses Infecting Klebsiella Encoding Capsule Depolymerases That Digest Specific Capsular Types. Microb. Biotechnol. 2019, 12, 472–486. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Latka, A.; Berisio, R.; Squeglia, F.; Maciejewska, B.; Briers, Y.; Drulis-Kawa, Z. Phage-Borne Depolymerases Decrease Klebsiella Pneumoniae Resistance to Innate Defense Mechanisms. Front. Microbiol. 2018, 9, 2517. [Google Scholar] [CrossRef]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage Tailspike Proteins as Molecular Probes for Sensitive and Selective Bacterial Detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Bacteriophages and Their Enzymes in Biofilm Control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Zuppi, M.; Tozzoli, R.; Chiani, P.; Quiros, P.; Martinez-Velazquez, A.; Michelacci, V.; Muniesa, M.; Morabito, S. Investigation on the Evolution of Shiga Toxin-Converting Phages Based on Whole Genome Sequencing. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms 2020, 8, 1293. [Google Scholar] [CrossRef]
- Brown-Jaque, M.; Calero-Cáceres, W.; Espinal, P.; Rodríguez-Navarro, J.; Miró, E.; González-López, J.J.; Cornejo, T.; Hurtado, J.C.; Navarro, F.; Muniesa, M. Antibiotic Resistance Genes in Phage Particles Isolated from Human Faeces and Induced from Clinical Bacterial Isolates. Int. J. Antimicrob. Agents 2018, 51, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Abedon, S.T. Pharmacologically Aware Phage Therapy: Pharmacodynamic and Pharmacokinetic Obstacles to Phage Antibacterial Action in Animal and Human Bodies. Microbiol. Mol. Biol. Rev. 2019, 83. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Reuter, M.; Kruger, D.H. Approaches to Optimize Therapeutic Bacteriophage and Bacteriophage-Derived Products to Combat Bacterial Infections. Virus Genes 2020, 56, 136–149. [Google Scholar] [CrossRef]
- Deveau, H.; Van Calsteren, M.-R.; Moineau, S. Effect of Exopolysaccharides on Phage-Host Interactions in Lactococcus Lactis. Appl. Environ. Microbiol. 2002, 68, 4364–4369. [Google Scholar] [CrossRef]
- Jung, J.-H.; Choi, N.-Y.; Lee, S.-Y. Biofilm Formation and Exopolysaccharide (EPS) Production by Cronobacter Sakazakii Depending on Environmental Conditions. Food Microbiol. 2013, 34, 70–80. [Google Scholar] [CrossRef]
- Lu, T.K.; Collins, J.J. Dispersing Biofilms with Engineered Enzymatic Bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced Biofilm Formation and Increased Resistance to Antimicrobial Agents and Bacterial Invasion Are Caused by Synergistic Interactions in Multispecies Biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef]
- Kifelew, L.G.; Mitchell, J.G.; Speck, P. Mini-Review: Efficacy of Lytic Bacteriophages on Multispecies Biofilms. Biofouling 2019, 35, 472–481. [Google Scholar] [CrossRef]
- Kay, M.K.; Erwin, T.C.; McLean, R.J.C.; Aron, G.M. Bacteriophage Ecology in Escherichia Coli and Pseudomonas Aeruginosa Mixed-Biofilm Communities. Appl. Environ. Microbiol. 2011, 77, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.R.; Parracho, H.M.R.T.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and Biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Teplitski, M.; Ritchie, K. How Feasible Is the Biological Control of Coral Diseases? Trends Ecol. Evol. 2009, 24, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Sutherland, I.W.; Jones, M.V. Biofilm Susceptibility to Bacteriophage Attack: The Role of Phage-Borne Polysaccharide Depolymerase. Microbiology 1998, 144, 3039–3047. [Google Scholar] [CrossRef] [PubMed]
- Skillman, L.C.; Sutherland, I.W.; Jones, M.V. The Role of Exopolysaccharides in Dual Species Biofilm Development. J. Appl Microbiol 1998, 85 (Suppl. 1), 13S–18S. [Google Scholar] [CrossRef]
- Sillankorva, S.; Neubauer, P.; Azeredo, J. Phage Control of Dual Species Biofilms of Pseudomonas Fluorescens and Staphylococcus Lentus. Biofouling 2010, 26, 567–575. [Google Scholar] [CrossRef]
- Briandet, R.; Lacroix-Gueu, P.; Renault, M.; Lecart, S.; Meylheuc, T.; Bidnenko, E.; Steenkeste, K.; Bellon-Fontaine, M.-N.; Fontaine-Aupart, M.-P. Fluorescence Correlation Spectroscopy to Study Diffusion and Reaction of Bacteriophages inside Biofilms. Appl. Environ. Microbiol. 2008, 74, 2135–2143. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chhibber, S. Restricting Ciprofloxacin-Induced Resistant Variant Formation in Biofilm of Klebsiella Pneumoniae B5055 by Complementary Bacteriophage Treatment. J. Antimicrob. Chemother. 2009, 64, 1212–1218. [Google Scholar] [CrossRef]
- Wingender, J.; Neu, T.R.; Flemming, H.-C. What are Bacterial Extracellular Polymeric Substances? In Microbial Extracellular Polymeric Substances: Characterization, Structure and Function; Wingender, J., Neu, T.R., Flemming, H.-C., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–19. ISBN 978-3-642-60147-7. [Google Scholar]
- Gutiérrez, D.; Briers, Y.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Role of the Pre-Neck Appendage Protein (Dpo7) from Phage VB_SepiS-PhiIPLA7 as an Anti-Biofilm Agent in Staphylococcal Species. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Hernandez-Morales, A.C.; Lessor, L.L.; Wood, T.L.; Migl, D.; Mijalis, E.M.; Cahill, J.; Russell, W.K.; Young, R.F.; Gill, J.J. Genomic and Biochemical Characterization of Acinetobacter Podophage Petty Reveals a Novel Lysis Mechanism and Tail-Associated Depolymerase Activity. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Liu, Y.; Wang, C.; He, T.; Gao, S.; Xing, S.; Huang, Y.; Fan, H.; Zhang, X.; Yu, W.; et al. Identification of a Lytic Pseudomonas Aeruginosa Phage Depolymerase and Its Anti-Biofilm Effect and Bactericidal Contribution to Serum. Virus Genes 2019, 55, 394–405. [Google Scholar] [CrossRef]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli Biofilm: Development and Therapeutic Strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef]
- Guo, Z.; Huang, J.; Yan, G.; Lei, L.; Wang, S.; Yu, L.; Zhou, L.; Gao, A.; Feng, X.; Han, W.; et al. Identification and Characterization of Dpo42, a Novel Depolymerase Derived from the Escherichia Coli Phage VB_EcoM_ECOO78. Front. Microbiol. 2017, 8, 1460. [Google Scholar] [CrossRef] [PubMed]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm Formation and Antibiotic Resistance of Klebsiella Pneumoniae Isolated from Clinical Samples in a Tertiary Care Hospital, Klaten, Indonesia. BMC Proc. 2019, 13. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella Pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chhibber, S. Structural Changes Induced by a Lytic Bacteriophage Make Ciprofloxacin Effective against Older Biofilm of Klebsiella Pneumoniae. Biofouling 2010, 26, 729–737. [Google Scholar] [CrossRef]
- Bansal, S.; Harjai, K.; Chhibber, S. Aeromonas Punctata Derived Depolymerase Improves Susceptibility of Klebsiella Pneumoniae Biofilm to Gentamicin. BMC Microbiol. 2015, 15. [Google Scholar] [CrossRef]
- Latka, A.; Drulis-Kawa, Z. Advantages and Limitations of Microtiter Biofilm Assays in the Model of Antibiofilm Activity of Klebsiella Phage KP34 and Its Depolymerase. Sci. Rep. 2020, 10, 20338. [Google Scholar] [CrossRef]
- Tait, K.; Skillman, L.C.; Sutherland, I.W. The Efficacy of Bacteriophage as a Method of Biofilm Eradication. Biofouling 2002, 18, 305–311. [Google Scholar] [CrossRef]
- Chai, Z.; Wang, J.; Tao, S.; Mou, H. Application of Bacteriophage-Borne Enzyme Combined with Chlorine Dioxide on Controlling Bacterial Biofilm. LWT Food Sci. Technol. 2014, 59, 1159–1165. [Google Scholar] [CrossRef]
- Chhibber, S.; Nag, D.; Bansal, S. Inhibiting Biofilm Formation by Klebsiella Pneumoniae B5055 Using an Iron Antagonizing Molecule and a Bacteriophage. BMC Microbiol. 2013, 13, 174. [Google Scholar] [CrossRef]
- Chhibber, S.; Bansal, S.; Kaur, S. Disrupting the Mixed-Species Biofilm of Klebsiella Pneumoniae B5055 and Pseudomonas Aeruginosa PAO Using Bacteriophages Alone or in Combination with Xylitol. Microbiology 2015, 161, 1369–1377. [Google Scholar] [CrossRef]
- Oliveira, A.; Ribeiro, H.G.; Silva, A.C.; Silva, M.D.; Sousa, J.C.; Rodrigues, C.F.; Melo, L.D.R.; Henriques, A.F.; Sillankorva, S. Synergistic Antimicrobial Interaction between Honey and Phage against Escherichia Coli Biofilms. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Azeredo, J.; Sutherland, I. The Use of Phages for the Removal of Infectious Biofilms. Curr. Pharm. Biotechnol. 2008, 9, 261–266. [Google Scholar] [CrossRef]
- Hanlon, G.W.; Denyer, S.P.; Olliff, C.J.; Ibrahim, L.J. Reduction in Exopolysaccharide Viscosity as an Aid to Bacteriophage Penetration through Pseudomonas Aeruginosa Biofilms. Appl. Environ. Microbiol. 2001, 67, 2746–2753. [Google Scholar] [CrossRef]
- Maszewska, A. Fagowe depolimerazy polisacharydów—charakterystyka i zastosowanie. Postepy Hig. Med. Dosw. 2015, 69, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Hancock, V.; Dahl, M.; Klemm, P. Abolition of Biofilm Formation in Urinary Tract Escherichia Coli and Klebsiella Isolates by Metal Interference through Competition for Fur. Appl. Environ. Microbiol. 2010, 76, 3836–3841. [Google Scholar] [CrossRef] [PubMed]
| Enzyme Name | Agent Used | Biofilm Type | Results (with Regard to the Action of the Agent Alone) | Reference |
|---|---|---|---|---|
| Depolymerase produced by lytic bacteriophage KPO1K2 | Ciprofloxacin | Klebsiella pneumoniae strain B5055 | Biofilm eradication more pronounced | [124,133] |
| Depolymerase produced by lytic bacteriophage KPO1K2 | Gentamycin | Klebsiella pneumoniae strain B5055 | Reduction in bacterial counts of young biofilm (up to 4 days) | [134] |
| Depolymerase Dep42 produced by lytic bacteriophage SH-KP152226 | Polymyxin | Klebsiella pneumoniae strain 2226 | Reduction in bacterial counts | [132] |
| Depolymerase KP34p57 produced by lytic bacteriophage KP34 | Ciprofloxacin | Klebsiella pneumoniae strain 77 | Reduction in colony counts | [135] |
| Depolymerase KP34p57 produced by lytic bacteriophage KP34 | Depolymerase-nonbearing phage KP15 | Klebsiella pneumoniae strain 77 | Reduction in colony counts | [135] |
| Depolymerase KP34p57 produced by lytic bacteriophage KP34 | Ciprofloxacin together with depolymerase-nonbearing phage KP15 | Klebsiella pneumoniae strain 77 | Reduction in colony counts | [135] |
| Depolymerase produced by lytic bacteriophage φEnt | Disinfectant | Enterobacter agglomerans strain Ent | Biofilm reduction more effective | [136] |
| Depolymerase obtained from the phage that infects Klebsiella strains | Chlorine dioxide | Klebsiella sp. | Reduction in biofilm-residing cells | [137] |
| Depolymerase produced by lytic bacteriophage KPO1K2 | Cobalt sulfate | Klebsiella pneumoniae strain B5055 | Reduction in the bacterial number | [138] |
| Depolymerase produced by lytic bacteriophage KPO1K2 | Xylitol | Pseudomonas aeruginosa PAO, Klebsiella pneumoniae strain B5055 | Biofilm reduction more effective | [139] |
| Phage EC3a bearing the depolymerase activity | Honey | Escherichia coli CECT 434 | More efficient antibiofilm activity | [140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175. https://doi.org/10.3390/antibiotics10020175
Topka-Bielecka G, Dydecka A, Necel A, Bloch S, Nejman-Faleńczyk B, Węgrzyn G, Węgrzyn A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics. 2021; 10(2):175. https://doi.org/10.3390/antibiotics10020175
Chicago/Turabian StyleTopka-Bielecka, Gracja, Aleksandra Dydecka, Agnieszka Necel, Sylwia Bloch, Bożena Nejman-Faleńczyk, Grzegorz Węgrzyn, and Alicja Węgrzyn. 2021. "Bacteriophage-Derived Depolymerases against Bacterial Biofilm" Antibiotics 10, no. 2: 175. https://doi.org/10.3390/antibiotics10020175
APA StyleTopka-Bielecka, G., Dydecka, A., Necel, A., Bloch, S., Nejman-Faleńczyk, B., Węgrzyn, G., & Węgrzyn, A. (2021). Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics, 10(2), 175. https://doi.org/10.3390/antibiotics10020175








