Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains
Abstract
:1. Introduction
2. Results
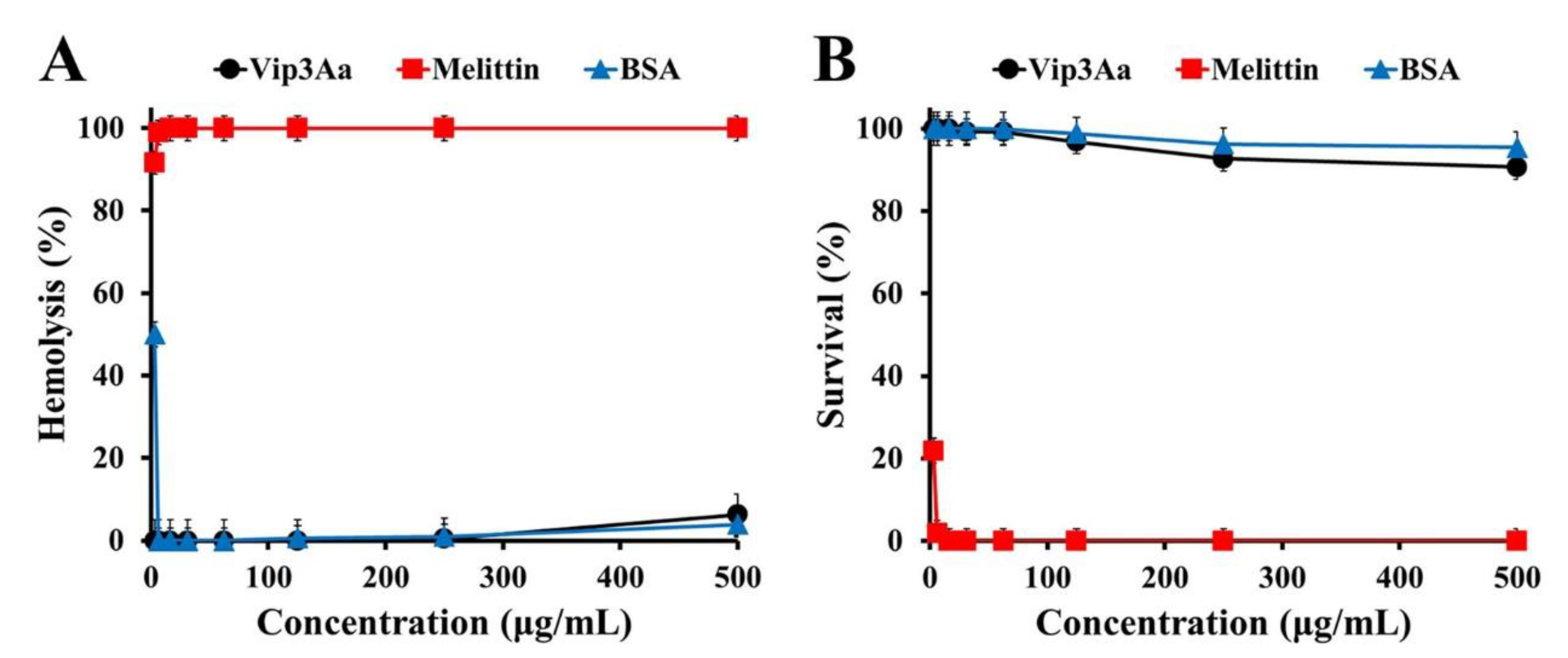
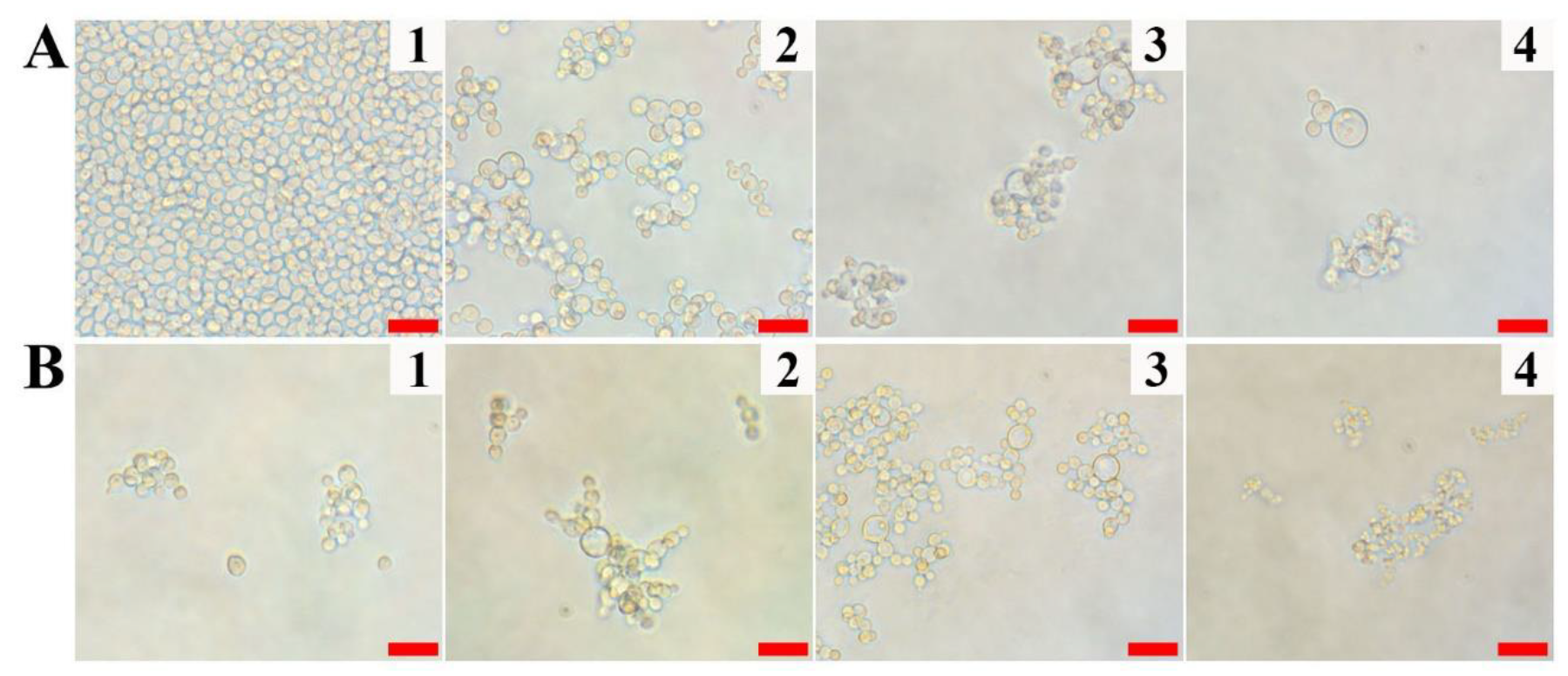
2.1. Antifungal Activity of Vip3Aa Protein against Pathogenic Fungi
2.2. Molecular Mechanism of the Vip3Aa Protein in Fungal Cells
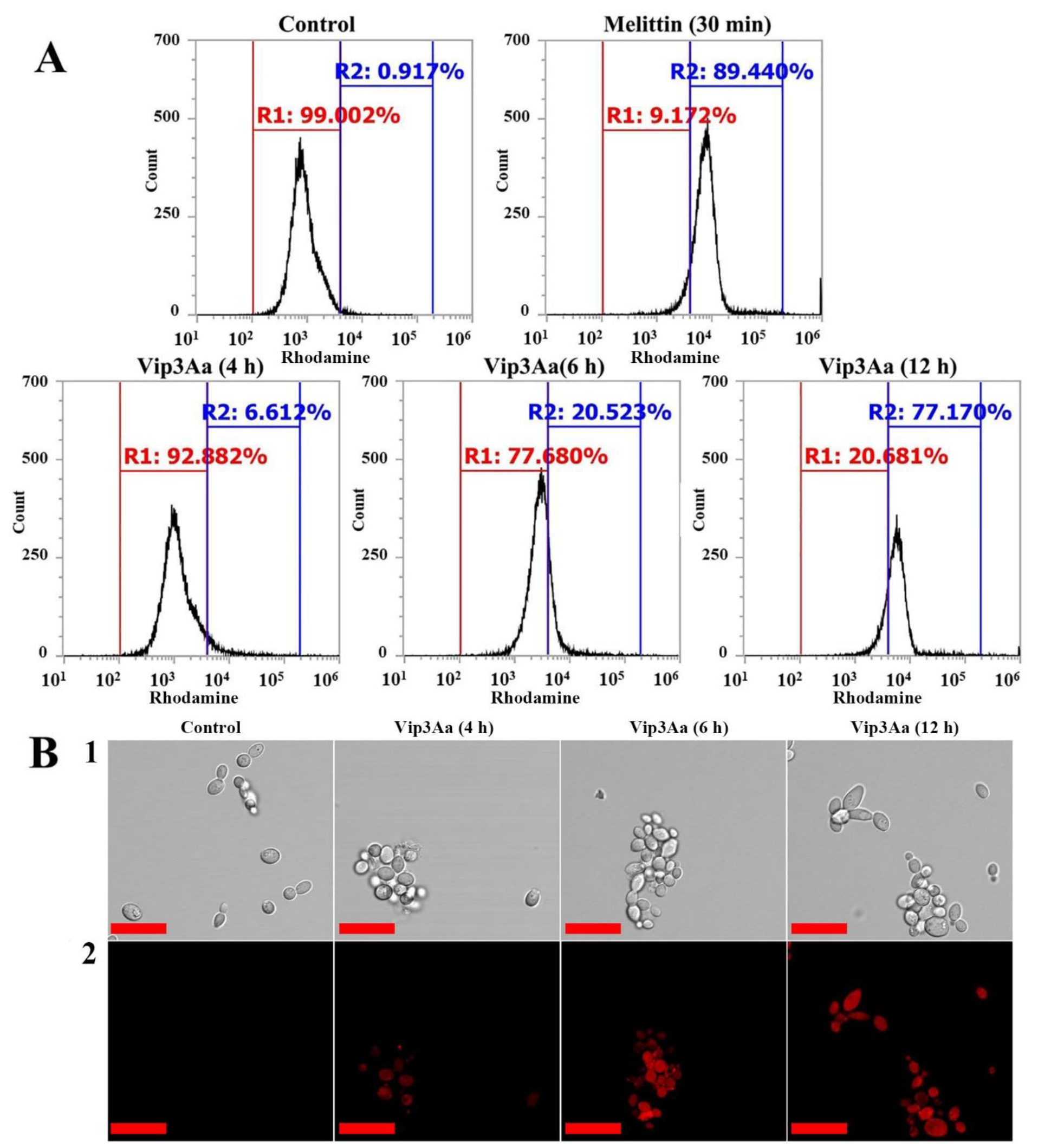
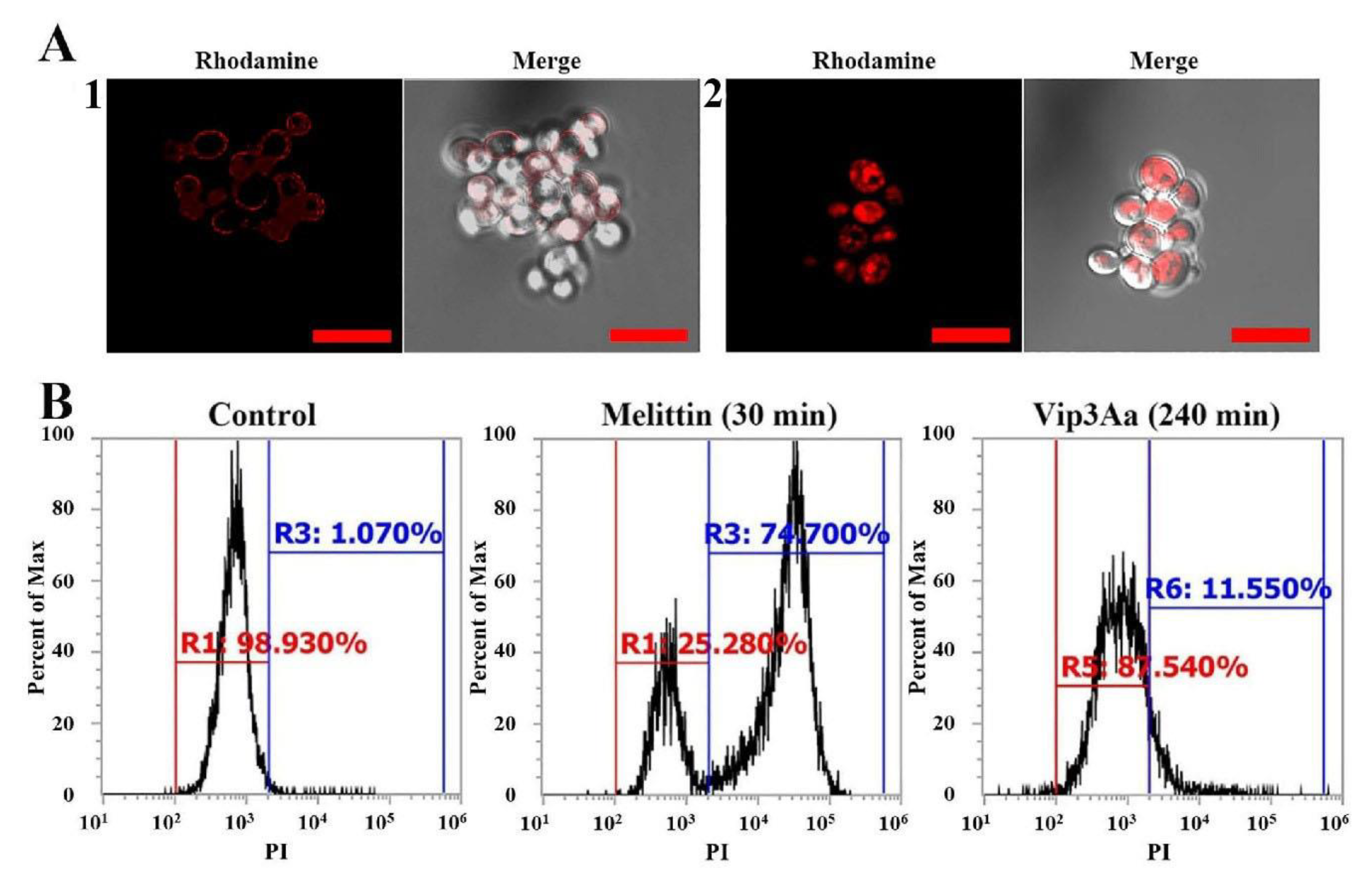
2.3. Intracellular Localization and Molecular Mechanism of Vip3Aa in Fungal Cells
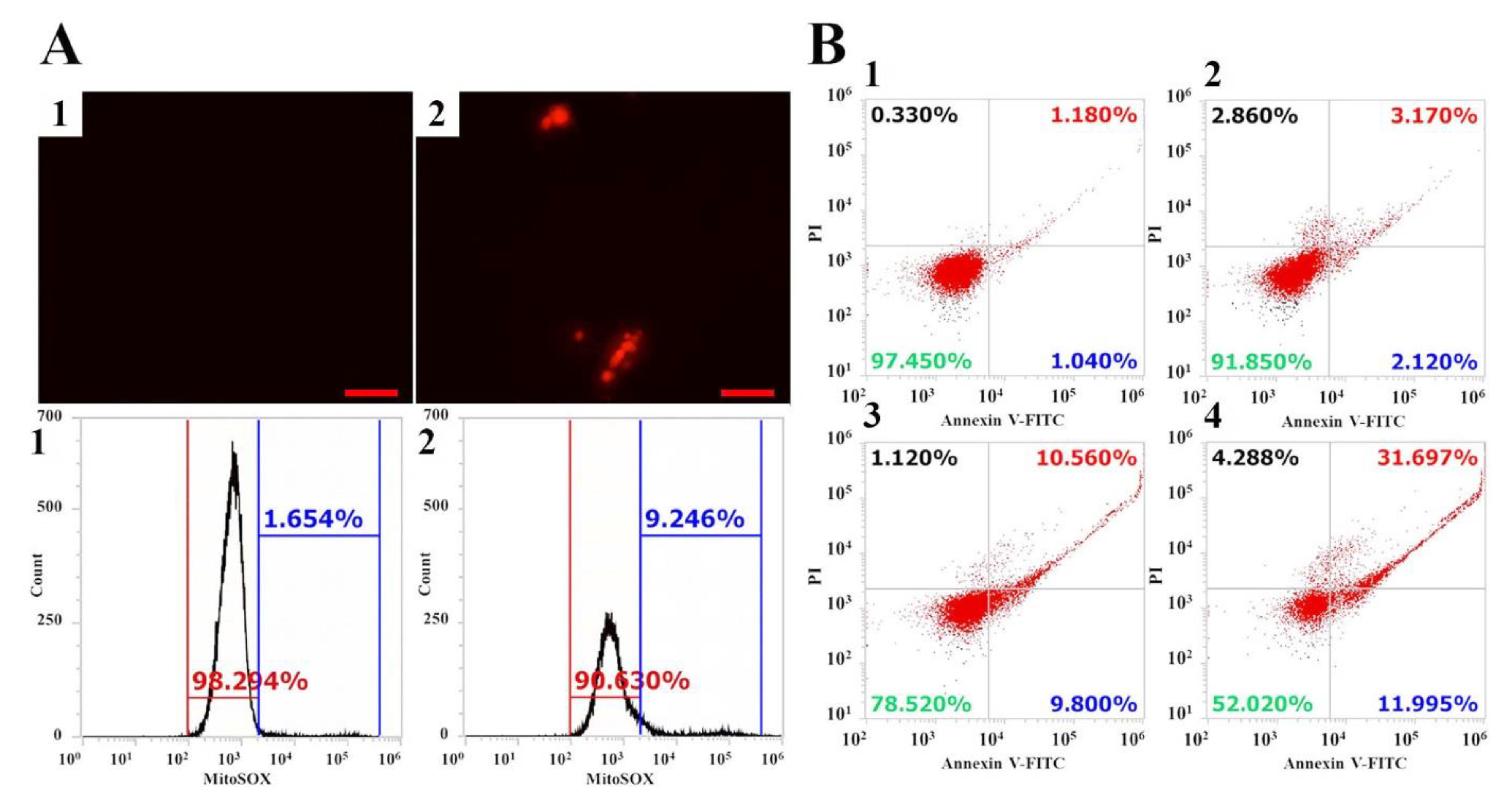
2.4. Intracellular ROS Production and Apoptosis Induction by Vip3Aa
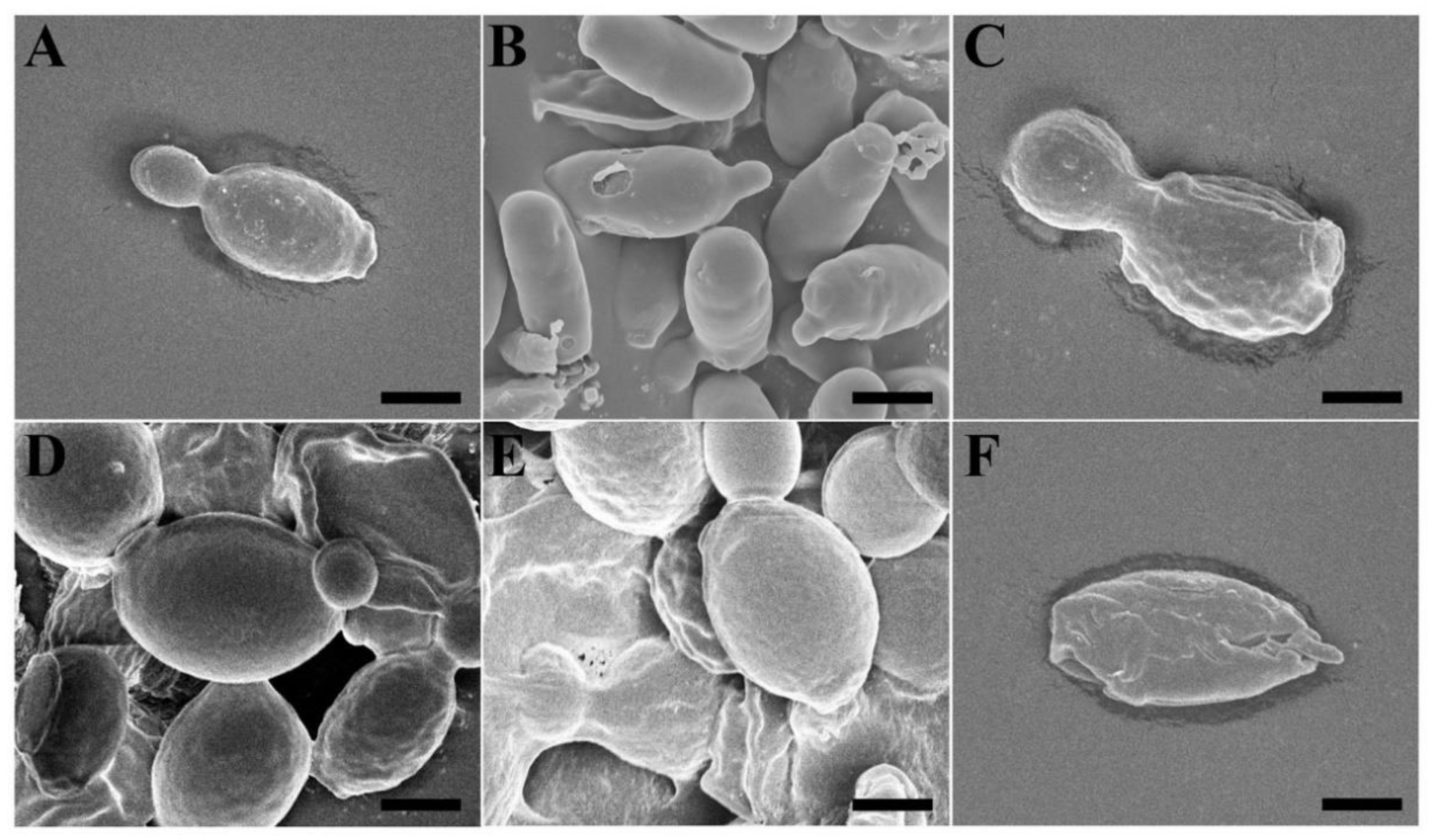
2.5. Morphological Alterations Caused by Vip3Aa in Fungal Cells
3. Materials and Methods
3.1. Materials
3.2. Fungal Cells
3.3. Cloning and Protein Expression of the Vip3Aa Gene in E. coli
3.4. Purification and Structural Analysis of the Vip3Aa Protein
3.5. Antifungal Assay
3.6. Cytotoxicity Assay
3.7. Membrane Integrity Assay Using PI
3.8. Analysis Using CLSM
3.9. MitoSOX Levels
3.10. Apoptosis Measurement
3.11. Analysis Using SEM
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dean, D.H. Biochemical genetics of the bacterial insect-control agent Bacillus thuringiensis: Basic principles and prospects for genetic engineering. Biotechnol. Genet. Eng. Rev. 1984, 2, 341–363. [Google Scholar] [CrossRef] [PubMed]
- Liliana, P.L.; Mario, S.; Alejandra, A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 2012, 37, 3–22. [Google Scholar]
- Estruch, J.J.; Warren, G.W.; Mullins, M.A.; Nye, G.J.; Craig, J.A.; Koziel, M.G. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 1996, 93, 5389–5394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, R.; Dalal, M.; Panguluri, S.K.; Jagadish, B.; Mandaokar, A.D.; Singh, A.; Kumar, P.A. Isolation, characterization and expression of a novel vegetative insecticidal protein gene of Bacillus thuringiensis. FEMS Microbiol. Lett. 2005, 243, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.G.; Mullins, M.A.; Warren, G.W.; Koziel, M.G.; Estruch, J.J. The Bacillus thuringiensis vegetative insecticidal protein Vip3A lyses midgut epithelium cells of susceptible insects. Appl. Environ. Microbiol. 1997, 63, 532–536. [Google Scholar] [CrossRef] [Green Version]
- Warren, G.W. Vegetative insecticidal proteins: Novel proteins for control of corn pests. In Advances in Insect Control: Role of Transgenic Plants; Carozzi, N., Koziel, M., Eds.; Taylor & Francis: London, UK, 1997; pp. 109–121. [Google Scholar]
- Estruch, J.J.; Koziel, M.G.; Yu, C.G.; Desai, N.M.; Nye, G.J.; Warren, G.W. Plant Pest Control. Patent WO 9844137, 8 October 1998. [Google Scholar]
- Gupta, M.; Kumar, H.; Kaur, S. Vegetative insecticidal protein (Vip): A potential contender from Bacillus thuringiensis for efficient management of various detrimental agricultural pests. Front. Microbiol. 2021, 12, 659736. [Google Scholar] [CrossRef]
- Han, S.; Craig, A.J.; Putnam, C.; Carozzi, N.B.; Tainer, A.J. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Genet. 1999, 6, 932–936. [Google Scholar] [CrossRef]
- Leuber, M.; Orlik, F.; Schiffler, B.; Sickmann, A.A.; Benz, R. Vegetative insecticidal protein (Vip1Ac) of Bacillus thuringiensis HD201: Evidence for oligomer and channel formation. Biochemistry 2006, 45, 283–288. [Google Scholar] [CrossRef]
- Lee, M.K.; Walters, F.S.; Hart, H.; Palekar, N.; Chen, J.-S. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab δ-Endotoxin. Appl. Environ. Microbiol. 2003, 69, 4648–4657. [Google Scholar] [CrossRef] [Green Version]
- Dow, J. Extremely high pH in biological systems: A model for carbonate transport. Am. J. Physiol. Integr. Comp. Physiol. 1984, 246, R633–R636. [Google Scholar] [CrossRef]
- Dow, J. pH gradients in lepidopteran midgut. J. Exp. Biol. 1992, 172, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Doss, V.A.; Kumar, K.A.; Jayakumar, R.; Sekar, V. Cloning and expression of the vegetative insecticidal protein (vip3V) gene of Bacillus thuringiensis in Escherichia coli. Protein Expr. Purif. 2002, 26, 82–88. [Google Scholar] [CrossRef]
- Chen, J.; Yu, J.; Tang, L.; Tang, M.; Shi, Y.; Pang, Y. Comparison of the expression of Bacillus thuringiensis full-length and N-terminally truncated vip3A gene in Escherichia coli. J. Appl. Microbiol. 2003, 95, 310–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Xu, N.; Huang, X.; Wang, W.; Cheng, J.; Wu, K.; Shen, Z. Bacillus thuringiensis Vip3 mutant proteins: Insecticidal activity and trypsin sensitivity. Biocontrol Sci. Technol. 2007, 17, 699–708. [Google Scholar] [CrossRef]
- Selvapandiyan, A.; Arora, N.; Rajagopal, R.; Jalali, S.K.; Venkatesan, T.; Singh, S.P.; Bhatnagar, R.K. Toxicity analysis of N- and C-Terminus-deleted vegetative insecticidal protein from Bacillus thuringiensis. Appl. Environ. Microbiol. 2001, 67, 5855–5858. [Google Scholar] [CrossRef] [Green Version]
- Bel, Y.; Banyuls, N.; Chakroun, M.; Escriche, B.; Ferré, J. Insights into the structure of the Vip3Aa insecticidal protein by protease digestion analysis. Toxins 2017, 9, 131. [Google Scholar] [CrossRef] [Green Version]
- Banyuls, N.; Hernández-Martínez, P.; Quan, Y.; Ferré, J. Artefactual band patterns by SDS-PAGE of the Vip3Af protein in the presence of proteases mask the extremely high stability of this protein. Int. J. Biol. Macromol. 2018, 120, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Zack, M.D.; Sopko, M.S.; Frey, M.L.; Wang, X.; Tan, S.Y.; Arruda, J.M.; Letherer, T.T.; Narva, K.E. Functional characterization of Vip3Ab1 and Vip3Bc1: Two novel insecticidal proteins with differential activity against lepidopteran pests. Sci. Rep. 2017, 7, 11112. [Google Scholar] [CrossRef] [Green Version]
- Quan, Y.; Ferré, J. Quan structural domains of the Bacillus thuringiensis Vip3Af protein unraveled by Tryptic digestion of Alanine Mutants. Toxins 2019, 11, 368. [Google Scholar] [CrossRef] [Green Version]
- Shao, E.; Zhang, A.; Yan, Y.; Wang, Y.; Jia, X.; Sha, L.; Guan, X.; Wang, P.; Huang, Z. Oligomer formation and insecticidal activity of Bacillus thuringiensis Vip3Aa toxin. Toxins 2020, 12, 274. [Google Scholar] [CrossRef] [Green Version]
- Chakroun, M.; Banyuls, N.; Bel, Y.; Escriche, B.; Ferré, J. Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 329–350. [Google Scholar] [CrossRef] [Green Version]
- Palma, L.; Scott, D.J.; Harris, G.; Din, S.-U.; Williams, T.L.; Roberts, O.J.; Young, M.T.; Caballero, P.; Berry, C. The Vip3Ag4 insecticidal protoxin from Bacillus thuringiensis adopts a tetrameric configuration that is maintained on proteolysis. Toxins 2017, 9, 165. [Google Scholar] [CrossRef]
- Park, S.C.; Kim, J.Y.; Shin, S.O.; Jeong, C.Y.; Kim, M.H.; Shin, S.Y.; Cheong, G.W.; Park, Y.; Hahm, K.S. Investigation of toroidal pore and oligomerization by melittin using Transmission electron mcroscopy. Biochem. Biophys. Res. Commun. 2004, 321, 631–637. [Google Scholar] [CrossRef]
- Liao, C.; Selvan, M.E.; Zhao, J.; Slimovitch, J.L.; Schneebeli, S.; Shelley, M.; Shelley, J.C.; Li, J. Melittin aggregation in aqueous solutions: Insight from molecular dynamics simulations. J. Phys. Chem. B 2015, 119, 10390–10398. [Google Scholar] [CrossRef]
- Yang, Z.; Zheng, J.; Chan, C.-F.; Wong, I.L.; Heater, B.S.; Chow, L.M.; Lee, M.M.; Chan, M.K. Targeted delivery of antimicrobial peptide by Cry protein crystal to treat intramacrophage infection. Biomaterials 2019, 217, 119286. [Google Scholar] [CrossRef] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-C.; Kim, H.; Kim, J.-Y.; Kim, H.; Cheong, G.-W.; Lee, J.R.; Jang, M.-K. Improved Cell Selectivity of Pseudin-2 via substitution in the Leucine-Zipper motif: In Vitro and In Vivo Antifungal Activity. Antibiotics 2020, 9, 921. [Google Scholar] [CrossRef]
- Park, S.-C.; Kim, I.R.; Kim, J.-Y.; Lee, Y.; Yoo, S.-H.; Jung, J.H.; Cheong, G.-W.; Lee, S.Y.; Jang, M.-K.; Lee, J.R. Functional characterization of a rice Thioredoxin protein OsTrxm and its cysteine mutant variant with antifungal activity. Antioxidants 2019, 8, 598. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-Y.; Park, S.-C.; Noh, G.; Kim, H.; Yoo, S.-H.; Kim, I.R.; Lee, J.R.; Jang, M.-K. Antifungal effect of a chimeric peptide Hn-Mc against pathogenic fungal strains. Antibiotics 2020, 9, 454. [Google Scholar] [CrossRef]
- Chen, R.-C.; Lan, C.-Y. Human antimicrobial peptide hepcidin 25-induced Apoptosis in Candida albicans. Microorganism 2020, 8, 585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Lee, D.G. The influence of the N-terminal region of antimicrobial peptide pleurocidin on fungal apoptosis. J. Microbiol. Biotechnol. 2013, 23, 1386–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dantas, A.D.S.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherz-Shouval, R.; Elazar, Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007, 17, 422–427. [Google Scholar] [CrossRef] [PubMed]

| Fungal Strains | Vip3Aa | Melittin | ||
|---|---|---|---|---|
| µM (µg/mL) | ||||
| IC50 | MIC | IC50 | MIC | |
| Yeast | ||||
| C. albicans | 0.7 (62.5) | 1.4 (125) | 11 (31.3) | 22 (62.5) |
| C. krusei | 0.7 (62.5) | 1.4 (125) | 11 (31.3) | 22 (62.5) |
| C. parapsilosis | 1.4 (125) | 2.8 (250) | 22 (62.5) | 44 (125) |
| C. tropicalis | 1.4 (125) | 2.8 (250) | 22 (62.5) | 44 (125) |
| Molds | ||||
| Colletotrichum gloeosporioides | 5.6 (500) | 11.1 (1000) | 88 (250) | 176 (500) |
| F. graminearum | 5.6 (500) | 11.1 (1000) | 88 (250) | 176 (500) |
| F. solani | 2.8 (250) | 5.6 (500) | 88 (250) | 88 (250) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-C.; Kim, J.-Y.; Lee, J.-K.; Lim, H.S.; Son, H.; Yoo, S.-H.; Mun, S.-E.; Jang, M.-K.; Lee, J.R. Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains. Antibiotics 2021, 10, 1558. https://doi.org/10.3390/antibiotics10121558
Park S-C, Kim J-Y, Lee J-K, Lim HS, Son H, Yoo S-H, Mun S-E, Jang M-K, Lee JR. Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains. Antibiotics. 2021; 10(12):1558. https://doi.org/10.3390/antibiotics10121558
Chicago/Turabian StylePark, Seong-Cheol, Jin-Young Kim, Jong-Kook Lee, Hye Song Lim, Hyosuk Son, Su-Hyang Yoo, Seong-Eun Mun, Mi-Kyeong Jang, and Jung Ro Lee. 2021. "Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains" Antibiotics 10, no. 12: 1558. https://doi.org/10.3390/antibiotics10121558
APA StylePark, S.-C., Kim, J.-Y., Lee, J.-K., Lim, H. S., Son, H., Yoo, S.-H., Mun, S.-E., Jang, M.-K., & Lee, J. R. (2021). Antifungal Mechanism of Vip3Aa, a Vegetative Insecticidal Protein, against Pathogenic Fungal Strains. Antibiotics, 10(12), 1558. https://doi.org/10.3390/antibiotics10121558







