Abstract
According to the WHO, P. aeruginosa is one of the antibiotic-resistant bacteria that represent the biggest threat to public health. The aim of the study was to establish the prevalence of antibiotic-resistant P. aeruginosa in the water systems of various healthcare facilities over the course of nine years. A total of 4500 tap water system samples were taken from seventeen healthcare facilities. The culture method was used to detect P. aeruginosa, and the isolates were then tested for antibiotic resistance using the standardised disc diffusion method. Eleven antibiotics from five different classes were tested. P. aeruginosa was found to have contaminated 2.07% (no. 93) of the water samples. The majority of positive samples came from the dental units (30.11%) and the ward kitchens (23.66%). Considering the total isolates, 56.99% (no. 3) were resistant to at least one of the antibiotics tested. A total of 71.43% of P. aeruginosa isolated from water emerging from dental unit handpieces was antibiotic-resistant, with 45% of it resistant to ≥3 classes of antibiotics. Out of the total isolates, 19.35% showed resistance to carbapenems. It would be advisable to systematically screen tap water for opportunistic micro-organisms such as P. aeruginosa, as many countries already do, including this in the Water Safety Plan.
1. Introduction
Healthcare-acquired infections (HAIs), particularly in the critical care setting, have become increasingly common over recent decades, with Gram-negative bacterial infections presenting the highest incidence among them [1]. P. aeruginosa is one of the most frequent and serious causes of HAIs (e.g., respiratory and urinary tract, skin and soft tissues, ear and eye infections, and bacteriemia), which particularly affect immunocompromised patients [2,3,4].
At-risk patients include neonates, patients with deep neutropenia, severely burned patients, patients with invasive devices (e.g., vascular and urinary catheters, endotracheal tubes, ventilators), and patients who have underlying pulmonary disease such as bronchiectasis and cystic fibrosis [5].
The emerging presence of P. aeruginosa multi-drug-resistant (MDR) isolates, resistant to almost all antimicrobials used for hospital patients, has attracted the attention of many researchers in recent decades [6]. P. aeruginosa presents multiple resistance mechanisms, either intrinsic or acquired, frequently with high resistance rates affecting several classes of antibiotics, including carbapenems [7,8].
Various hospital-acquired infection outbreaks due to P. aeruginosa have been reported [9,10,11,12,13]. Different potential environmental reservoirs of this micro-organism have been described in healthcare settings (aerosols, taps, basin and shower drains, respiratory equipment, humidifiers, endoscopes and endoscope washers, hydrotherapy pools, dental units, etc.) [11,14,15,16,17].
In greater detail, water sources and water-related devices are often contaminated with pathogens [18,19,20], which may be responsible for healthcare-associated infections, including P. aeruginosa [21,22,23,24,25,26,27].
It has been estimated that 20% of nosocomial pneumonias are caused by waterborne P. aeruginosa in the US, resulting in a conservative annual mortality of approximately 1400 individuals [28]. Ambrogi et al. [22]. reported a cluster of five cases of infection by P. aeruginosa expressing VIM carbapenemases (VIM-PA) in a nephrology intensive care unit transmitted via hands and associated with contaminated tap water.
Drinking water quality is subject to numerous regulations based on its lifetime health effects in the general population. However, with regard to people with increased susceptibility to infection, insufficiently broad water quality indicators are used (e.g., they do not include opportunistic pathogens), and there is a lack of guidelines covering all healthcare settings [18,21]. Only a small number of European countries (United Kingdom, France) and the US Centers for Disease Control and Prevention have drawn up guidelines for water quality in healthcare facilities [29,30,31]. In Germany and France, environmental surveillance of water systems is an integral part of their infection control programmes. For P. aeruginosa specifically, the target value is <1 CFU/100 mL [30]. In Italy, there is no specific legislation regarding the control of water in healthcare facilities. Only a small number of studies have been carried out to date relating to the environmental surveillance of antibiotic-resistant P. aeruginosa in healthcare facilities, even limited to a specific type of environmental reservoir. The aim of this study was to investigate the prevalence and antimicrobial susceptibility profiles of P. aeruginosa isolates obtained from water samples collected from different healthcare facilities in the Liguria region, Northern Italy, over a nine-year period. Since it was not the aim of the study, we did not carry out a clinical surveillance of P. aeruginosa infections during the monitoring period.
2. Results
Out of the 4500 water samples taken in various healthcare facilities, 2.07% (no. 93) were contaminated with P. aeruginosa. Specifically, only 0.40% of inlet water samples were found to be contaminated with P. aeruginosa.
Of the isolates, sixty-eight came from the cold water circuit, while the remaining twenty-five were from the hot water circuit. Among the various sampling sites, the highest percentage of positivity (16%) was found in dental units. None of the water samples from taps fitted with absolute filters, such as those in the operating theatres and intensive care units/resuscitation units, showed microbial contamination.
The majority of P. aeruginosa isolates came from the dental units (30.11%) and the water system in the ward kitchens (23.66%).
Considering the total isolates, 56.99% (no. 53) were resistant to at least one of the antibiotics tested; of these, the highest percentages of antibiotic-resistant isolates (37.74% and 28.30%) came from dental units and ward kitchens, respectively (Table 1).

Table 1.
P. aeruginosa isolates in the various water sampling points.
The difference between the number of antibiotic-resistant isolates in the dental units and those found at the other sampling points was borderline significant (Pearson chi2 = 3.4076; p = 0.052).
Water isolates from the neonatology ward, rehabilitation pool, and patient toilets were susceptible to all antibiotics tested.
Figure 1 shows the percentage of isolates for each sampling site. In the dental units (no. 28) and ward kitchens (no. 22), 71.43% and 68.18% of isolates, respectively, showed antibiotic resistance.
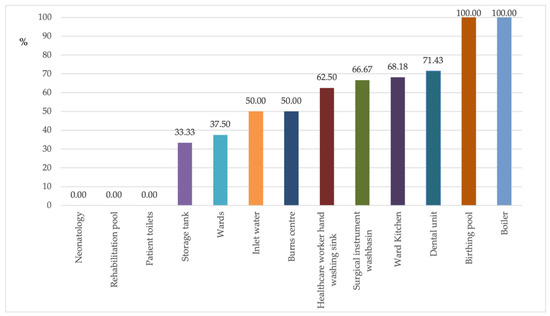
Figure 1.
Percentage of antibiotic-resistant P. aeruginosa in the individual sampling sites.
Of the resistant isolates, only 20.75% were resistant to one antibiotic and 22.64% were resistant to two or three antibiotics. A decreasing percentage of isolates showed resistance to more than three antibiotics. In total, 1.89% of the resistant isolates were resistant to eight of the eleven antibiotics tested (Figure 2) and came from the water circuit of a hospital ward kitchen.
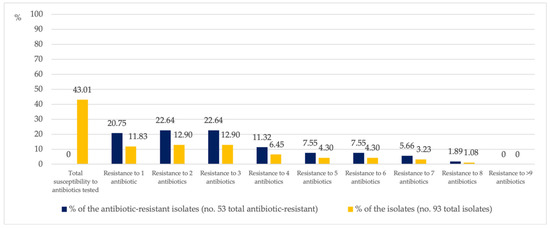
Figure 2.
Percentage of P. aeruginosa isolates simultaneously resistant to between one and eight antibiotics.
The percentage of antibiotic resistant strains has changed over the years, but in a discontinuous way (Figure S1, Supplementary Materials). However, based on the observed data, the probability of occurrence of resistant strains increases by 1.2 over the years (OR = 1.20 [95% CI, 1.01–1.43], p = 0.0351).
Table 2 shows the antibiotic-resistant characteristics relative to the P. aeruginosa isolates.

Table 2.
P. aeruginosa isolates resistant (R) to the various antibiotics tested.
Considering the total number of antibiotic-resistant isolates, Aminoglycosides are the class of antibiotics with the lowest percentage of resistance, with no resistance at all being observed for Amikacin. On the other hand, the highest percentages of resistance were recorded for the Penicillins: Piperacillin (75.47%) and Piperacillin/Tazobactam (69.81%).
Furthermore, almost half of the resistant isolates showed resistance to Cefepime (Cephalosporins) and Levofloxacin (Fluoroquinolones).
2.1. Multiresistant P. aeruginosa
Out of the total isolates, 17 (18.28%) were multiresistant, being resistant to ≥3 groups of antibiotics at the same time (Table 3, grey area).

Table 3.
Percentage of P. aeruginosa isolates resistant to 1, 2, 3, 4, 5 groups of antibiotics tested.
Table 4 shows the antibiotic-resistance profile of the P. aeruginosa isolate and the multiple antimicrobial resistance (MAR) index. P. aeruginosa exhibited twenty-five antibiotic-resistant patterns with the MAR index ranging from 0.09 to 0.73; thirty-one isolates (58.49%) fell into the MAR index >0.2 category.

Table 4.
P. aeruginosa isolate antibiotic-resistance profiles (no. 53) and MAR index.
At the most critical sites in terms of healthcare impact and/or possibility of persistence due to easier biofilm formation, several multiresistant isolates were found. In the water coming from the hand shower in a hospital burns unit, P. aeruginosa was resistant to four classes of antibiotics (seven out of eleven antibiotics tested).
Another critical control point was the healthcare workers’ hand-washing sink, where 20% of resistant isolates (no. = 5) were multidrug-resistant (resistant to four classes of antibiotics). Out of the antibiotic-resistant isolates in the water within the internal circuit of dental units, 45% were resistant to ≥3 classes of antibiotics (Figure 3).
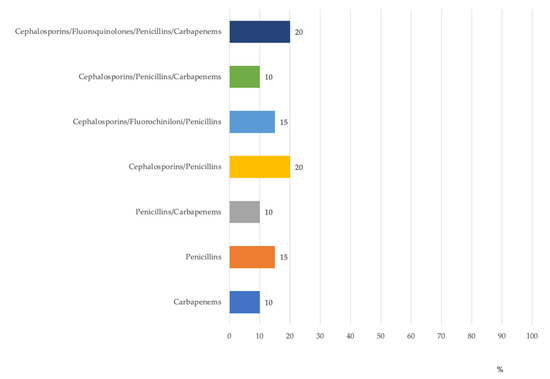
Figure 3.
Percentage distribution of antibiotic-resistant P. aeruginosa isolates in dental units by antibiotic groups.
2.2. Carbapenem Resistance
19.35% of the total P. aeruginosa isolates were found to be resistant to carbapenems. Among the antibiotic-resistant isolates (no. 53), carbapenem resistance was 33.96%. Regarding the latter, Figure 4 shows the percentage of carbapenem-resistant isolates (R) and of antibiotic-resistant but carbapenem-susceptible isolates (AR) on the basis of the sampling point.
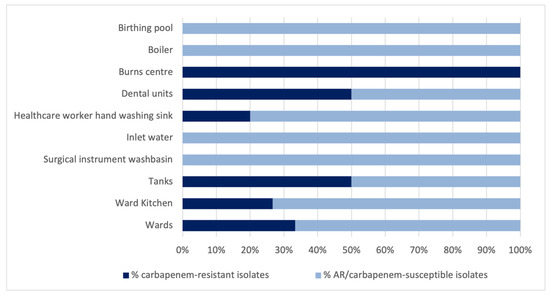
Figure 4.
Percentage distribution of carbapenem-resistant P. aeruginosa isolates in the various sampling points.
In the dental units, ten P. aeruginosa isolates showed resistance to carbapenems. The difference between the number of carbapenem-resistant isolates in the dental units and those found at the other sampling points was borderline significant (Pearson chi2 = 3.6837; p = 0.053).
3. Discussion
Antimicrobial resistance is now one of the most important public health problems in both human and veterinary medicine and is set to become one of the major health challenges of the coming decades.
To tackle the problem of antibiotic resistance, the WHO has promoted the “One Health” strategy, integrating all relevant sectors, from human and veterinary use to food, agricultural and environmental safety. In Italy, where the phenomenon of antibiotic resistance is among the highest in Europe, at levels almost always above average, the Ministry of Health has published the “National Plan to Combat Antimicrobial Resistance (PNCAR) 2017–2020” [32] which addresses the problem from a general, human, and veterinary medicine perspective.
The spread of multidrug-resistant P. aeruginosa is particularly concerning. Over recent years, the worldwide spread of so-called “high-risk clones” of multidrug-resistant or extensively drug-resistant (MDR/XDR) P. aeruginosa has become a public health threat that needs to be urgently and decisively studied and managed [3].
According to the WHO [7], Pseudomonas aeruginosa is one of the antibiotic-resistant bacteria that represents the biggest threat to public health. It is intrinsically resistant to most antimicrobial agents due to its selective ability to prevent various antibiotic molecules from penetrating its outer membrane or extruding them if they enter the cell. It also exhibits acquired resistance mechanisms.
The ECDC’s European Antimicrobial Resistance Surveillance Network (EARS-Net) reported that, for 2019, 31.8% of P. aeruginosa isolates in the EU/EEA were resistant to at least one of the antimicrobial groups under regular surveillance (piperacillin + tazobactam, fluoroquinolones, ceftazidime, aminoglycosides, and carbapenems). The highest EU/EEA population-weighted mean resistance percentage in 2019 was reported for fluoroquinolones (18.9%), followed by piperacillin + tazobactam (16.9%), carbapenems (16.5%), ceftazidime (14.3%), and aminoglycosides (11.5%) [8].
ECDC data from 2018 [33], which is more detailed for individual countries in the EU/EEA, showed P. aeruginosa combined resistance (resistance to three or more antimicrobial groups among piperacillin ± tazobactam, ceftazidime, fluoroquinolones, aminoglycosides and carbapenems) in the EU/EEA of 12.8%, with considerable variation between countries, from 0% in Iceland to 49.4% in Romania. In Italy, 14.9% of 2006 P. aeruginosa clinical isolates were multiresistant.
Because P. aeruginosa is one of the main agents of nosocomial infections and is increasingly resistant to antibiotics, environmental reservoirs in hospital settings are of great concern.
To date, some studies have been conducted on the circulation and/or the prevalence of multidrug-resistant P. aeruginosa in healthcare facility environmental reservoirs (e.g., water) [24,25,26], but they are still few in number.
Our study has revealed the presence of P. aeruginosa in different healthcare water samples (2.07%), including resistant strains. The percentage of positivity for P. aeruginosa found in this study is within the range of prevalence observed in similar investigations [18,25,26].
Out of the P. aeruginosa isolates analysed, 56.99% (no. = 53) were resistant to at least one of the antibiotics tested, of which 37.74% and 28.30% were found in dental units and ward kitchens, respectively.
P. aeruginosa exhibited twenty-five antibiotic-resistant patterns with the MAR index ranging from 0.09 to 0.73. Thirty-one isolates (58.49%) fell into the MAR index >0.2 category. The MAR index is a good tool for health risk assessment which identifies whether isolates are from a region of high or low antibiotic use. A MAR index >0.2 indicates a “high-risk” source of contamination [34,35].
Out of the total isolates, 18.28% were multiresistant, being resistant to ≥3 groups of antibiotics. In the most critical sites in terms of healthcare impact and/or possibility of persistence due to biofilm formation, several multiresistant isolates were found. Twenty percent of P. aeruginosa isolates from healthcare workers’ handwashing tap water were resistant to four classes of antibiotics.
In water from the hand shower of a hospital burns unit, P. aeruginosa was resistant to seven out of eleven antibiotics tested, belonging to four distinct classes. Infections caused by MDR bacteria act as a risk factor for mortality in burns patients. Out of all MDR bacteria, P. aeruginosa proved the most significant because this bacterium showed the most growth on the moist surface of burn wounds and is highly pathogenic in immunocompromised patients [36].
Considering the other critical control points monitored in our study, it emerged that 71.43% of P. aeruginosa isolated from water flowing from dental unit handpieces was antibiotic-resistant, and 45% of this was resistant to ≥3 classes of antibiotics.
An important characteristic of P. aeruginosa is its ability to form biofilms as an adaptation to adverse environmental conditions [37,38,39]. In dental units, the water conduit can be composed of approximately 6 m of narrow-bore flexible polyurethane or PVC plastic tubing (1/16 in. or 2 mm diameter), which encourages biofilm formation of a wide variety of microorganisms [39,40,41], including antibiotic-resistant P. aeruginosa. This microorganism can be responsible for infections in immunocompromised patients treated at dental units harbouring these organisms [42].
The circulation of carbapenem-resistant strains of P. aeruginosa is particularly concerning as infections sustained by these strains are difficult to treat, both because there are often no adequately effective and safe therapeutic options available and because the associated mortality rate is higher than for infections with carbapenem-sensitive P. aeruginosa [43].
In Italy, the percentage of invasive isolates with resistance to carbapenems (imipenem or/and meropenem) was between 10% and <25% in 2019 [8].
In our study, 19.35% of the total P. aeruginosa water isolates were found to be resistant to carbapenems. Among the antibiotic-resistant isolates (no. 53), carbapenem resistance was 33.96%.
The data collected in this study highlight the importance of environmental surveillance of antibiotic-resistant and MDR microorganisms, together with the adoption of measures to prevent environmental contamination.
According to WHO guidelines [7], measures to prevent the transmission of multiresistant P. aeruginosa in healthcare facilities should include at least the following: hand hygiene (with the appropriate use of alcohol-based solutions), contact precautions, patient isolation (single room or cohort), environmental cleanliness, and surveillance.
In order to specifically mitigate the risks of water contamination by microorganisms responsible for care-related infections, the implementation of a Water Safety Plan (WSP) is essential. The main elements of this plan should comprise active infection surveillance, the adoption of disinfection procedures or other water treatments, the maintenance of water networks, and the scheduling of periodic checks of the water withdrawn at the most significant points of the tap water system [44,45].
In light of the findings that emerged from this study and international scientific evidence, it would be advisable to systematically screen tap water for opportunistic micro-organisms such as P. aeruginosa, as many countries already do, including this in the Water Safety Plan.
Understanding potential environmental reservoirs of infectious bacterial species and the role that water and water-related devices play as reservoirs for antimicrobial-resistant bacteria is crucial to prevent HAIs.
A limitation of our study was the possible underestimation of microbial contamination of water leaving taps fitted with POU filters (e.g., in the operating theatre and intensive care/resuscitation unit) upstream from the taps, as sampling in these cases was carried out without removing the filter.
Various authors have highlighted how prolonged use of point-of-use filters may create a water flow slowdown and retrograde contamination [46]. Therefore, sampling the water after the removal of the absolute filter and allowing the water to flow would have led to possible environmental dispersion of potentially pathogenic microorganisms concentrated in the end of the taps, constituting a possible health risk for severely debilitated patients.
Another limitation of the study was the lack of surveillance of cases of nosocomial P. aeruginosa infection in the healthcare facilities examined during the nine years of monitoring. The use of molecular biology on clinical and environmental isolates could have made it possible to highlight clonal relationships between patient and tap water isolates and thus the potential water-based origin of the infections. On the other hand, the opposite route of transmission cannot be excluded. Some scientific evidence has shown that transmissions of P. aeruginosa can occur both from tap to patient and from patient to tap [47].
4. Materials and Methods
4.1. Setting
Between January 2011 and January 2020, a total of 4500 water samples were collected from seventeen healthcare facilities in Northern Italy (region of Liguria): eight hospitals, three nursing homes, and six outpatient clinics.
Various points of the healthcare facility water systems were sampled: inlet water, storage tanks, birthing pools, boilers, rehabilitation pools, and various critical points such as medical wards, neonatology, operating theatres (surgical instrument washing sink, surgical scrub sink), intensive care/resuscitation, hand washing for health workers, burn centre, dental units, ward kitchens, patient toilets, etc.
Water sampling was carried out as part of the routine surveillance plan of water quality in health care facilities. The sampling was conducted every six months (in spring/summer and in autumn/winter) both on the cold and hot water circuits for a total of about 500 samples per year. The hot water circuit was equipped with chlorination systems within the healthcare facilities, while the cold water circuit did not undergo any additional disinfection with respect to disinfection already applied within the water supply system.
4.2. Water Sampling and Microbiological Analysis
The water was drawn from taps, showers or by immersion (in the case of storage tanks). In the case of taps, the sampling was carried out after removing the diffuser head (when present), flushing the tap, and letting the water run for 1–3 min.
In taps equipped with absolute filters, water was taken without dismantling them and therefore downstream of them.
In the case of dental units, the water was taken from handpieces. From each sampling point, water was collected in sterile disposable plastic (polyethylene) bottles containing sodium thiosulphate to inhibit the action of residual chlorine in the sampled water. The samples were transported in heat-insulated containers under refrigerated conditions and analysed within two hours of their arrival at the laboratory.
The samples were analysed for the detection of Pseudomonas aeruginosa using a standard method based on the membrane filtration technique (UNI EN ISO 16266) [48]. Briefly, the water sample (100 mL) was filtered through a cellulose ester membrane (0.45 μm porosity, 47 mm diameter); the membrane was then placed on Pseudomonas CN agar medium (Liofilchem, Roseto degli Abruzzi (TE), Italy), which is a selective medium for P. aeruginosa, and subsequently cultured at 36 ± 2 °C for 44 ± 4 h before colony counting.
Blue/green pyocyanin-producing colonies were counted as confirmed P. aeruginosa. Fluorescent non-pyocyanin-producing or reddish-brown colonies were recorded as presumptive P. aeruginosa and subjected to confirmation tests according to ISO 16266.
4.3. Antibiotic Susceptibility Testing
The P. aeruginosa isolates were gradually frozen at −80 °C as they were collected, and then simultaneously revitalised and tested for antibiotic resistance in 2020.
The P. aeruginosa isolates were tested using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standardised disc diffusion method.
These tests were performed on Mueller−Hinton agar using the Kirby−Bauer disc diffusion technique. All plates were incubated at 35 °C ± 1 °C for 16 ± 18 h. Zones of inhibition around the disk were measured and interpreted as proposed by the latest EUCAST breakpoint criteria.
Eleven antibiotics from five different classes were tested: Amikacin 30 µg, Gentamicin 10 µg, Tobramycin 10 µg, Cefepime 30 µg, Ceftazidime 10 µg, Ciprofloxacin 5 µg, Levofloxacin 5 µg, Piperacillin 30 µg, Piperacillin/Tazobactam (36 µg), Imipenem 10 µg, Meropenem 10 µg.
Pseudomonas aeruginosa ATTC 27853 was used for quality control.
The Multiple Antimicrobial Resistance (MAR) index has been calculated as the ratio between the number of antibiotics that an isolate is resistant to and the total number of antibiotics the organism is exposed to.
4.4. Statistical Analysis
The data were processed using the statistical programme STATA SE14TM (StataCorp, College Station, Texas, USA). Descriptive statistics were performed. The collected information was summarised using frequency and percentage for qualitative data. Differences between antibiotics resistance isolates were evaluated by means of the non-parametric chi-square test and Fisher exact test. A p value less than 0.05 was considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10121500/s1, Figure S1: P. aeruginosa antibiotic resistant isolates and total of P. aeruginosa isolates in the years of observation.
Author Contributions
Conceptualization, M.L.C. and A.M.S.; methodology, M.S. and A.M.S.; software, E.S. and G.O.; validation, M.L.C.; formal analysis, M.S. and B.C.; investigation, E.S. and G.O.; resources, M.L.C.; data curation, M.S. and B.C.; writing—original draft preparation, M.L.C. and A.M.S.; writing—review and editing, M.L.C., M.S., and A.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on motivated request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Feretzakis, G.; Loupelis, E.; Sakagianni, A.; Skarmoutsou, N.; Michelidou, S.; Velentza, A.; Martsoukou, M.; Valakis, K.; Petropoulou, S.; Koutalas, E. A 2-Year Single-Centre Audit on Antibiotic Resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae Strains from an Intensive Care Unit and Other Wards in a General Public Hospital in Greece. Antibiotics 2019, 8, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogiel, T.; Prażyńska, M.; Kwiecińska-Piróg, J.; Mikucka, A.; Gospodarek-Komkowska, E. Carbapenem-Resistant Pseudomonas aeruginosa Strains-Distribution of the Essential Enzymatic Virulence Factors Genes. Antibiotics 2021, 10, 8. [Google Scholar] [CrossRef]
- Spagnolo, A.M.; Sartini, M.; Cristina, M.L. Pseudomonas aeruginosa in the healthcare facility setting. Rev. Med. Microbiol. 2021, 32, 169–175. [Google Scholar] [CrossRef]
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Bédard, E.; Prévost, M.; Déziel, E. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiologyopen 2016, 5, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Pappa, O.; Apostolos, V.; Galanis, A.; Vantarakis, G.; Mavridou, A. Antibiotic resistance profiles of Pseudomonas aeruginosa isolated from various Greek aquatic environments. FEMS Microbiol. Ecol. 2016, 92, fiw042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization (WHO). Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Healthcare Facilities. Available online: https://apps.who.int/iris/bitstream/handle/10665/259462/9789241550178-eng.pdf;jsessionid=1D1BF01EC76D852B0261FA36B6A4A582?sequence=1 (accessed on 30 August 2021).
- ECDC. Antimicrobial Resistance in the EU/EEA (EARS-Net). Annual Epidemiological Report for 2019. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2019 (accessed on 1 August 2021).
- Pitten, F.A.; Panzig, B.; Schröder, G.; Tietze, K.; Kramer, A. Transmission of a multiresistant Pseudomonas aeruginosa strain at a German University Hospital. J. Hosp. Infect. 2001, 47, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Climo, M.W.; Pastor, A.; Wong, E.S. An outbreak of Pseudomonas aeruginosa related to contaminated urodynamic equipment. Infect. Contr. Hosp. Epidemiol. 1997, 18, 509–510. [Google Scholar] [CrossRef]
- Ferroni, A.; Nguyen, L.; Pron, B.; Quesne, G.; Brusset, M.C.; Berche, P. Outbreak of nosocomial urinary tract infections due to Pseudomonas aeruginosa in a paediatric surgical unit associated with tap-water contamination. J. Hosp. Inf. 1998, 39, 301–307. [Google Scholar] [CrossRef]
- Moolenaar, R.L.; Crutcher, J.M.; San Joaquzin, V.H.; Sewell, L.V.; Hutwagner, L.C.; Carson, L.A.; Robinson, D.A.; Smithee, L.M.; Jarvis, W.R. A prolonged outbreak of Pseudomonas aeruginosa in a neonatal intensive care unit: Did staff fingernails play a role in disease transmission? Infect. Contr. Hosp. Epidemiol. 2000, 21, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Bicking Kinsey, C.; Koirala, S.; Solomon, B.; Rosenberg, J.; Robinson, B.F.; Neri, A.; Laufer Halpin, A.; Arduino, M.J.; Moulton-Meissner, H.; Noble-Wang, J.; et al. Pseudomonas aeruginosa outbreak in a neonatal intensive care unit attributed to hospital tap water. Infect. Contr. Hosp. Epidemiol. 2017, 38, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Döring, G.; Ulrich, M.; Müller, W.; Bitzer, J.; Schmidt-Koenig, L.; Münst, L.; Grupp, H.; Wolz, C.; Stern, M.; Botzenhart, K. Generation of Pseudomonas aeruginosa aerosols during handwashing from contaminated sink drains, transmission to hands of hospital personnel, and its prevention by use of a new heating device. Zentralbl. Hyg. Umweltmed. 1991, 191, 494–505. [Google Scholar] [PubMed]
- Spagnolo, A.M.; Sartini, M.; Cristina, M.L. Microbial contamination of dental unit waterlines and potential risk of infection: A narrative review. Pathogens 2020, 9, 651. [Google Scholar] [CrossRef]
- Tuvo, B.; Totaro, M.; Cristina, M.L.; Spagnolo, A.M.; Di Cave, D.; Profeti, S.; Baggiani, A.; Privitera, G.; Casini, B. Prevention and Control of Legionella and Pseudomonas spp. Colonization in Dental Units. Pathogens 2020, 9, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristina, M.L.; Spagnolo, A.M.; Orlando, P.; Perdelli, F. The role of the environment in the spread of emerging pathogens in at-risk hospital wards. Rev. Med. Microbiol. 2013, 24, 104–112. [Google Scholar] [CrossRef]
- Cristina, M.L.; Spagnolo, A.M.; Casini, B.; Baggiani, A.; Del Giudice, P.; Brusaferro, S.; Poscia, A.; Moscato, U.; Perdelli, F.; Orlando, P. The impact of aerators on water contamination by emerging gram-negative opportunists in at-risk hospital departments. Infect. Control. Hosp. Epidemiol. 2014, 35, 122–129. [Google Scholar] [CrossRef]
- Montagna, M.T.; Cristina, M.L.; De Giglio, O.; Spagnolo, A.M.; Napoli, C.; Cannova, L.; Deriu, M.G.; Delia, S.A.; Giuliano, A.; Guida, M.; et al. Serological and molecular identification of Legionella spp. isolated from water and surrounding air samples in Italian healthcare facilities. Environ. Res. 2016, 146, 47–50. [Google Scholar] [CrossRef]
- Montagna, M.T.; De Giglio, O.; Napoli, C.; Diella, G.; Rutigliano, S.; Agodi, A.; Auxilia, F.; Baldovin, T.; Bisetto, F.; Arnoldo, L.; et al. Control and prevention measures for legionellosis in hospitals: A cross-sectional survey in Italy. Environ. Res. 2018, 166, 55–60. [Google Scholar] [CrossRef]
- Anaissie, E.J.; Penzak, S.R.; Dignani, M.C. The hospital water supply as a source of nosocomial infections: A plea for action. Arch. Intern. Med. 2002, 162, 1483–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrogi, V.; Cavalié, L.; Mantion, B.; Ghiglia, M.J.; Cointault, O.; Dubois, D.; Prère, M.F.; Levitzki, N.; Kamar, N.; Malavaud, S. Transmission of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nephrology-transplant intensive care unit with potential link to the environment. J. Hosp. Infect. 2016, 92, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, A.M.; Orlando, P.; Perdelli, F.; Cristina, M.L. Hospital water and prevention of waterborne infections. Rev. Med. Microbiol. 2016, 27, 25–32. [Google Scholar] [CrossRef]
- Borges, C.R.M.; Lascowski, K.M.S.; Filho, N.R.; Pelayo, J.S. Microbiological quality of water and dialysate in a haemodialysis unit in Ponta Grossa-PR, Brazil. J. Appl. Microbiol. 2007, 103, 1791–1797. [Google Scholar] [CrossRef]
- Lefebvre, A.; Bertrand, X.; Quantin, C.; Vanhems, P.; Lucet, J.C.; Nuemi, G.; Astruc, K.; Chavanet, P.; Aho-Glélé, L.S. Association between Pseudomonas aeruginosa positive water samples and healthcare-associated cases: Nine-year study at one university hospital. J. Hosp. Infect. 2017, 96, 238–243. [Google Scholar] [CrossRef]
- Schiavano, G.F.; Carloni, E.; Andreoni, F.; Magi, S.; Chironna, M.; Brandi, G.; Amagliani, G. Prevalence and antibiotic resistance of Pseudomonas aeruginosa in water samples in central Italy and molecular characterization of oprD in imipenem resistant isolates. PLoS ONE 2017, 12, e0189172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsay, K.A.; Wardell, S.J.T.; Patrick, W.M.; Brockway, B.; Reid, D.W.; Winstanley, C.; Bell, S.C.; Lamont, I.L. Genomic and phenotypic comparison of environmental and patient-derived isolates of Pseudomonas aeruginosa suggest that antimicrobial resistance is rare within the environment. J. Med. Microbiol. 2019, 68, 1591–1595. [Google Scholar] [CrossRef]
- Hayward, C.; Ross, K.E.; Brown, M.H.; Whiley, H. Water as a Source of Antimicrobial Resistance and Healthcare-Associated Infections. Pathogens 2020, 9, 667. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for Environmental Infection Control in Health-Care Facilities. Recommendations from CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). Available online: https://www.cdc.gov/infectioncontrol/guidelines/environmental/index.html (accessed on 30 August 2021).
- Ministère de la Santé e des Solidarités. L’eau dans les établissements de santé. Available online: http://www.sante.gouv.fr/IMG/pdf/Guide_technique_de_l_eau_dans_les_etablissements_de_sante_-_edition_2005.pdf (accessed on 3 June 2021).
- Department of Health. Health Technical Memorandum 04-01: Safe water in Healthcare Premises 2016; The Stationery Office: London, UK, 2016.
- Piano Nazionale di Contrasto dell’Antimicrobico-Resistenza (PNCAR) 2017–2020. Available online: https://www.salute.gov.it/imgs/C_17_pubblicazioni_2660_allegato.pdf (accessed on 3 June 2021).
- ECDC. Surveillance of Antimicrobial Resistance in Europe 2018. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2018 (accessed on 13 October 2020).
- Davis, R.; Brown, P.D. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016, 65, 261–271. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Panghal, M.; Singh, K.; Kadyan, S.; Chaudary, U.; Yadav, J.P. The analysis of distribution of multidrug resistant Pseudomonas and Bacillus species from burn patients and burn ward environment. Burns 2015, 41, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respir. Cell. Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Spagnolo, A.M.; Sartini, M.; Cave, D.D.; Casini, B.; Tuvo, B.; Cristina, M.L. Evaluation of Microbiological and Free-Living Protozoa Contamination in Dental Unit Waterlines. Int. J. Environ. Res. Public Health 2019, 16, 2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, D.; Mercier, A.; Gravouil, K.; Lesobre, J.; Delafont, V.; Bousseau, A.; Verdon, J.; Imbert, C. Pyrosequencing analysis of bacterial diversity in dental unit waterlines. Water Res. 2015, 81, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Lizzadro, J.; Mazzotta, M.; Girolamini, L.; Dormi, A.; Pellati, T.; Cristino, S. Comparison between Two Types of Dental Unit Waterlines: How Evaluation of Microbiological Contamination Can Support Risk Containment. Int. J. Environ. Res. Public Health 2019, 16, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Oliveira, A.C.; Maluta, R.P.; Stella, A.E.; Rigobelo, E.C.; Marin, J.M.; Ávila, F.A. Isolation of Pseudomonas aeruginosa strains from dental office environments and units in Barretos, state of São Paulo, Brazil, and analysis of their susceptibility to antimicrobial drugs. Braz. J. Microbiol. 2008, 39, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Persoon, M.C.; Voor, A.F.; Wielders, C.C.H.; Gommers, D.; Vos, M.C.; Severin, J.A. Mortality associated with carbapenem-susceptible and Verona Integron-encoded Metallo-β-lactamase-positive Pseudomonas aeruginosa bacteremia. Antimicrob. Resist. Infect. Control 2020, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Water Safety Plan Manual: Step-by-Step Risk Management for Drinking-Water Suppliers. Available online: https://apps.who.int/iris/bitstream/handle/10665/75141/9789241562638_eng.pdf?sequence=1&isAllowed=y (accessed on 30 August 2021).
- Casini, B.; Buzzigoli, A.; Cristina, M.L.; Spagnolo, A.M.; Del Giudice, P.; Brusaferro, S.; Poscia, A.; Moscato, U.; Valentini, P.; Baggiani, A.; et al. Long-term effects of hospital water network disinfection on Legionella and other waterborne bacteria in an Italian university hospital. Infect. Control Hosp. Epidemiol. 2014, 35, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Vonberg, R.P.; Sohr, D.; Bruderek, J.; Gastmeier, P. Impact of a silver layer on the membrane of tap water filters on the microbiological quality of filtered water. BMC Infect. Dis. 2008, 8, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, S.; Sigge, A.; Wiedeck, H.; Trautmann, M. Analysis of transmission pathways of Pseudomonas aeruginosa between patients and tap water outlets. Crit. Care Med. 2002, 30, 2222–2228. [Google Scholar] [CrossRef]
- UNI EN ISO 16266: 2008 Water Quality. Detection and Enumeration of Pseudomonas Aeruginosa. Method by Membrane Filtration; Technical committee ISO/TC 147, Subcommittee SC 4; International Organization for Standardization: Geneva, Switzerland, 2008. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).