Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application
Abstract
1. Introduction
2. Phage Enzymes as the Basis of New Antibacterial Therapies
Roles of Polysaccharide Depolymerases and Lysins during Phage Infection Cycles
3. Phage-Encoded Polysaccharide Depolymerases (PSDs)
3.1. PSD Substrate Diversity
3.2. Plaque Halo Zones
3.3. Roadblocks to PSD-Gene Identification
3.4. Additional PSD Properties
3.5. In Vivo Charateristics
4. Phage-Encoded Peptidoglycan Hydrolases (PGHs)
4.1. Basic Characteristics
4.2. Spectra of Activities and Resistance Evolution
4.3. Structure and Mode of Action of Phage-Derived Peptidoglycan Hydrolases
5. Clinical Trials and Case Studies
5.1. P128: Anti-Staphylococcal Engineered VAPGH
5.1.1. P128 In Vitro Activity Analysis
5.1.2. P128 In Vitro and Ex Vivo Stability and Lack of Cytotoxicity
5.1.3. P128 Resistance
5.1.4. P128 Animal Testing
5.1.5. P128 Clinical Trial
5.2. N-Rephasin® SAL200: Anti-Staphylococcal Recombinant Endolysin
5.2.1. N-Rephasin® SAL200 In Vitro Analysis
5.2.2. N-Rephasin® SAL200 Animal Testing
5.2.3. N-Rephasin® SAL200 Clinical Trial
5.3. CF-301: Anti-Staphylococcal Recombinant Endolysin
5.3.1. CF-301 In Vitro and Ex Vivo Analysis
5.3.2. CF-301 Animal Testing
5.3.3. CF-301 Clinical Trials
5.4. Staphefekt SA.100: Anti-Staphylococcal Engineered Endolysin
5.4.1. Staphefekt SA.100 In Vitro Analysis
5.4.2. Staphefekt SA.100 Clinical Trial
5.4.3. Staphefekt SA.100 Case Study Series
5.5. Development of Enzybiotics Targeting Gram-Negative Bacteria towards Clinical Trials
6. Advantages and Challenges
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Quinn, R. Rethinking antibiotic research and development: World War II and the penicillin collaborative. Am. J. Public Health 2013, 103, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Landecker, H. Antimicrobials before antibiotics: War, peace, and disinfectants. Palgrave Commun. 2019, 5, 45. [Google Scholar] [CrossRef]
- Bud, R. Penicillin: Triumph and Tragedy; Oxford University Press on Demand: New York, NY, USA, 2007. [Google Scholar]
- Kirby, W.M. Extraction of a highly potent penicillin inactivator from penicillin resistant staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.; Rozwadowska-Dowzenko, M. Infection by penicillin-resistant staphylococci. Lancet 1948, 2, 641–644. [Google Scholar] [CrossRef]
- Shaffer, R.K. The challenge of antibiotic-resistant Staphylococcus: Lessons from hospital nurseries in the mid-20th century. Yale J. Biol. Med. 2013, 86, 261–270. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645. [Google Scholar] [CrossRef]
- Steenbergen, J.N.; Alder, J.; Thorne, G.M.; Tally, F.P. Daptomycin: A lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 2005, 55, 283–288. [Google Scholar] [CrossRef]
- Chakravarty, I.; Kundu, K.; Kundu, S. Daptomycin: Discovery, development and perspectives. Battle Against Microb. Pathog. Basic Sci. Technol. Adv. Educ. Progr. 2015, 2, 895–903. [Google Scholar]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef]
- Clokie, M.R.; Kropinski, A.M.; Lavigne, R. Bacteriophages; Springer: New York, NY, USA, 2009. [Google Scholar]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef]
- Catalao, M.J.; Gil, F.; Moniz-Pereira, J.; Sao-Jose, C.; Pimentel, M. Diversity in bacterial lysis systems: Bacteriophages show the way. FEMS Microbiol. Rev. 2013, 37, 554–571. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; García, P.; Rodríguez, A. Potential of the virion-associated peptidoglycan hydrolase HydH5 and its derivative fusion proteins in milk biopreservation. PLoS ONE 2013, 8, e54828. [Google Scholar]
- Rodríguez-Rubio, L.; Gutiérrez, D.; Donovan, D.M.; Martínez, B.; Rodríguez, A.; García, P. Phage lytic proteins: Biotechnological applications beyond clinical antimicrobials. Crit. Rev. Biotechnol. 2016, 36, 542–552. [Google Scholar] [CrossRef]
- Shen, Y.; Mitchell, M.S.; Donovan, D.M.; Nelson, D.C. 15 Phage-based enzybiotics. Bacteriophages Health Dis. 2012, 24, 217. [Google Scholar]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef]
- Pestrak, M.J.; Baker, P.; Dellos-Nolan, S.; Hill, P.J.; da Silva, D.P.; Silver, H.; Lacdao, I.; Raju, D.; Parsek, M.R.; Wozniak, D.J. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob. Agents Chemother. 2019, 63, e00234-19. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, H.; Wang, D.; Zhang, C.; Zhao, K.; Ma, L. Intracellular glycosyl hydrolase PslG shapes bacterial cell fate, signaling, and the biofilm development of Pseudomonas Aeruginosa. bioRxiv 2021. [Google Scholar] [CrossRef]
- Biswas, R.; Voggu, L.; Simon, U.K.; Hentschel, P.; Thumm, G.; Götz, F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 2006, 259, 260–268. [Google Scholar] [CrossRef]
- Qin, Z.; Ou, Y.; Yang, L.; Zhu, Y.; Tolker-Nielsen, T.; Molin, S.; Qu, D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 2007, 153, 2083–2092. [Google Scholar] [CrossRef]
- Porayath, C.; Suresh, M.K.; Biswas, R.; Nair, B.G.; Mishra, N.; Pal, S. Autolysin mediated adherence of Staphylococcus aureus with fibronectin, gelatin and heparin. Int. J. Biol. Macromol. 2018, 110, 179–184. [Google Scholar] [CrossRef]
- Parisien, A.; Allain, B.; Zhang, J.; Mandeville, R.; Lan, C. Novel alternatives to antibiotics: Bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 2008, 104, 1–13. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, ecology, and application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Borysowski, J.; Gorksi, A. Enzybiotics and Their Potential Applications in Medicine; Wiley: New York, NY, USA, 2010; pp. 1–26. [Google Scholar]
- Yang, S.-C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar]
- Oliver, W.T.; Wells, J.E. Lysozyme as an alternative to growth promoting antibiotics in swine production. J. Anim. Sci. Biotechnol. 2015, 6, 35. [Google Scholar] [CrossRef]
- Veiga-Crespo, P.; Ageitos, J.M.; Poza, M.; Villa, T.G. Enzybiotics: A look to the future, recalling the past. J. Pharm. Sci. 2007, 96, 1917–1924. [Google Scholar] [CrossRef]
- Slopek, S.; Weber-Dabrowska, B.; Dabrowski, M.; Kucharewicz-Krukowska, A. Results of bacteriophage treatment of suppurative bacterial infections in the years 1981–1986. Arch. Immunol. Ther. Exp. 1987, 35, 569–583. [Google Scholar]
- Abedon, S.T. Phage-antibiotic combination treatments: Antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.-B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Boucher, F.; Fevre, C.; Perpoint, T.; Chateau, J.; Petitjean, C.; Josse, J.; Chidiac, C.; L’hostis, G.; Leboucher, G. Innovations for the treatment of a complex bone and joint infection due to XDR Pseudomonas aeruginosa including local application of a selected cocktail of bacteriophages. J. Antimicrob. Chemother. 2018, 73, 2901–2903. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophages and Biofilms; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011. [Google Scholar]
- Kutateladze, M.; Adamia, R. Phage therapy experience at the Eliava Institute. Med. Mal. Infect. 2008, 38, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Chanishvili, N. Literature Review of The Practical Application of Bacteriophage Research; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2012. [Google Scholar]
- Weber-Dabrowska, B.; Mulczyk, M.; Gorski, A. Bacteriophage therapy of bacterial infections: An update of our institute’s experience. Arch. Immunol. Exp. 2000, 48, 547–551. [Google Scholar]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóż, P.; Kłak, M.; Wojtasik, E. Clinical aspects of phage therapy. Adv. Virus Res. 2012, 83, 73–121. [Google Scholar]
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. 2021, 21, 427–436. [Google Scholar] [CrossRef]
- Letkiewicz, S.; Międzybrodzki, R.; Kłak, M.; Jończyk, E.; Weber-Dąbrowska, B.; Górski, A. The perspectives of the application of phage therapy in chronic bacterial prostatitis. FEMS Immunol. Med. Microbiol. 2010, 60, 99–112. [Google Scholar] [CrossRef]
- Gupta, P.; Singh, H.S.; Shukla, V.K.; Nath, G.; Bhartiya, S.K. Bacteriophage therapy of chronic nonhealing wound: Clinical study. Int. J. Low. Extrem. Wounds 2019, 18, 171–175. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2020; p. ofaa389. [Google Scholar]
- Tkhilaishvili, T.; Winkler, T.; Müller, M.; Perka, C.; Trampuz, A. Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 64, e00924-19. [Google Scholar] [CrossRef]
- Rogóż, P.; Amanatullah, D.F.; Międzybrodzki, R.; Manasherob, R.; Tikunova, N.V.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Górski, A. Phage therapy in orthopaedic implant-associated infections. In Phage Therapy: A Practical Approach; Springer: Cham, Switzerland, 2019; pp. 189–211. [Google Scholar]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Alves, D.R. Phage therapy in the 21st century: Is there modern, clinical evidence of phage-mediated efficacy? Pharmaceuticals 2021, 14, 1157. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Kutter, E. Bacteriophages: It’s a medicine, Jim, but not as we know it. Lancet Infect. Dis. 2020, 21, 309–311. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; Verbeken, G.; Rose, T.; Jennes, S.; Zizi, M.; Huys, I.; Lavigne, R.; Merabishvili, M.; Vaneechoutte, M.; Buckling, A. Introducing yesterday’s phage therapy in today’s medicine. Future Virol. 2012, 7, 379–390. [Google Scholar] [CrossRef]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The advantages and challenges of using endolysins in a clinical setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. In Advances in Virus Research; Elsevier: Cambridge, CA, USA, 2012; Volume 83, pp. 299–365. [Google Scholar]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef]
- Azeredo, J.; García, P.; Drulis-Kawa, Z. Targeting biofilms using phages and their enzymes. Curr. Opin. Biotechnol. 2021, 68, 251–261. [Google Scholar] [CrossRef]
- Maszewska, A. Phage associated polysaccharide depolymerases-characteristics and application. Postepy Hig. Med. Dosw. 2015, 69, 690–702. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Love, M.J.; Bhandari, D.; Dobson, R.C.; Billington, C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef]
- Divya Ganeshan, S.; Hosseinidoust, Z. Phage therapy with a focus on the human microbiota. Antibiotics 2019, 8, 131. [Google Scholar] [CrossRef]
- Heselpoth, R.D.; Swift, S.M.; Linden, S.B.; Mitchell, M.S.; Nelson, D.C. Enzybiotics: Endolysins and bacteriocins. In Bacteriophages: Biology, Technology, Therapy; Springer: Cham, Switzerland, 2021; pp. 989–1030. [Google Scholar]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-based antibacterials as therapeutics. In Therapeutic Enzymes: Function and Clinical Implications; Springer: Singapore, 2019; pp. 233–253. [Google Scholar]
- Mirski, T.; Lidia, M.; Nakonieczna, A.; Gryko, R. Bacteriophages, phage endolysins and antimicrobial peptides—The possibilities for their common use to combat infections and in the design of new drugs. Ann. Agric. Environ. Med. 2019, 26, 203–209. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Donovan, D.M.; Rodríguez, A.; García, P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013, 39, 427–434. [Google Scholar] [CrossRef]
- Hermoso, J.A.; García, J.L.; García, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472. [Google Scholar] [CrossRef]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Lavigne, R. Breaking barriers: Expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol. 2015, 10, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Tsonos, J.; Vandenheuvel, D.; Briers, Y.; De Greve, H.; Hernalsteens, J.-P.; Lavigne, R. Hurdles in bacteriophage therapy: Deconstructing the parameters. Vet. Microbiol. 2014, 171, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, K.; Gerstmans, H.; Saafan, A.; Dishisha, T.; Briers, Y. The preclinical and clinical progress of bacteriophages and their lytic enzymes: The parts are easier than the whole. Viruses 2019, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H.; Park, B.H. An enzyme produced by a phage-host cell system: II. The properties of the polysaccharide depolymerase. Virology 1956, 2, 719–736. [Google Scholar] [CrossRef]
- Hughes, K.; Sutherland, I.; Clark, J.; Jones, M. Bacteriophage and associated polysaccharide depolymerases–novel tools for study of bacterial biofilms. J. Appl. Microbiol. 1998, 85, 583–590. [Google Scholar] [CrossRef]
- Hughes, K.A.; Sutherland, I.W.; Jones, M.V. Biofilm susceptibility to bacteriophage attack: The role of phage-borne polysaccharide depolymerase. Microbiology 1998, 144 Pt 11, 3039–3047. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Adsorption: Phage acquisition of bacteria. In Bacteriophages: Biology, Technology, Therapy; Springer: Cham, Switzerland, 2021; pp. 93–117. [Google Scholar]
- Rakhuba, D.; Kolomiets, E.; Dey, E.S.; Novik, G. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145. [Google Scholar] [CrossRef]
- Azeredo, J.; Sutherland, I.W. The use of phages for the removal of infectious biofilms. Curr. Pharm. Biotechnol. 2008, 9, 261–266. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Young, R.; Bläsi, U. Holins: Form and function in bacteriophage lysis. FEMS Microbiol. Rev. 1995, 17, 191–205. [Google Scholar] [CrossRef]
- Wang, I.-N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Kongari, R.; Rajaure, M.; Cahill, J.; Rasche, E.; Mijalis, E.; Berry, J.; Young, R. Phage spanins: Diversity, topological dynamics and gene convergence. BMC Bioinform. 2018, 19, 326. [Google Scholar] [CrossRef]
- Delbruck, M. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 1940, 23, 643–660. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Mol. Biol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef]
- Cota-Robles, E.H. Electron microscopy of “lysis from within” of Escherichia coli by coliphage T2. J. Ultrastruct. Res. 1964, 11, 112–122. [Google Scholar] [CrossRef]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 2014, 28, 265–274. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.-S.; Lavigne, R. Learning from bacteriophages-advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef]
- Kim, S.; Oh, D.-B.; Kang, H.A.; Kwon, O. Features and applications of bacterial sialidases. Appl. Microbiol. Biotechnol. 2011, 91, 1–15. [Google Scholar] [CrossRef]
- Yadav, V.; Yadav, P.K.; Yadav, S.; Yadav, K. α-L-Rhamnosidase: A review. Process Biochem. 2010, 45, 1226–1235. [Google Scholar] [CrossRef]
- Murakami, H.; Kuramoto, T.; Mizutani, K.; Nakano, H.; Kitahata, S. Purification and some properties of a new levanase from Bacillus sp. No. 71. Biosci. Biotechnol. Biochem. 1992, 56, 608–613. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miasnikov, A.N. Characterization of a novel endo-levanase and its gene from Bacillus sp. L7. FEMS Microbiol. Lett. 1997, 154, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Beg, Q.; Kapoor, M.; Mahajan, L.; Hoondal, G. Microbial xylanases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2001, 56, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Juturu, V.; Wu, J.C. Microbial exo-xylanases: A mini review. Appl. Biochem. Biotechnol. 2014, 174, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Juturu, V.; Wu, J.C. Microbial xylanases: Engineering, production and industrial applications. Biotechnol. Adv. 2012, 30, 1219–1227. [Google Scholar] [CrossRef]
- Kulkarni, N.; Shendye, A.; Rao, M. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 1999, 23, 411–456. [Google Scholar] [CrossRef]
- Jiménez, E.R. Dextranase in sugar industry: A review. Sugar Tech. 2009, 11, 124–134. [Google Scholar] [CrossRef]
- Jiménez, E.R. The dextranase along sugar-making industry. Biotecnol. Apl. 2005, 22, 20–27. [Google Scholar]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef]
- Sutherland, I.W. Polysaccharide lyases. FEMS Microbiol. Rev. 1995, 16, 323–347. [Google Scholar] [CrossRef]
- Michaud, P.; Da Costa, A.; Courtois, B.; Courtois, J. Polysaccharide lyases: Recent developments as biotechnological tools. Crit. Rev. Biotechnol. 2003, 23, 233–266. [Google Scholar] [CrossRef]
- Singh, S.K.; Bharati, A.P.; Singh, N.; Pandey, P.; Joshi, P.; Singh, K.; Mitra, K.; Gayen, J.R.; Sarkar, J.; Akhtar, M.S. The prophage-encoded hyaluronate lyase has broad substrate specificity and is regulated by the N-terminal domain. J. Biol. Chem. 2014, 289, 35225–35236. [Google Scholar] [CrossRef]
- Cornelissen, A.; Ceyssens, P.J.; T’Syen, J.; Van Praet, H.; Noben, J.P.; Shaburova, O.V.; Krylov, V.N.; Volckaert, G.; Lavigne, R. The T7-related Pseudomonas putida phage phi15 displays virion-associated biofilm degradation properties. PLoS ONE 2011, 6, e18597. [Google Scholar] [CrossRef]
- Cornelissen, A.; Ceyssens, P.-J.; Krylov, V.N.; Noben, J.-P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, C.-G.; Lee, E.Y. Alginate lyase: Structure, property, and application. Biotechnol. Bioprocess Eng. 2011, 16, 843. [Google Scholar] [CrossRef]
- Scholl, D.; Rogers, S.; Adhya, S.; Merril, C.R. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 2001, 75, 2509–2515. [Google Scholar] [CrossRef]
- Olszak, T.; Shneider, M.M.; Latka, A.; Maciejewska, B.; Browning, C.; Sycheva, L.V.; Cornelissen, A.; Danis-Wlodarczyk, K.; Senchenkova, S.N.; Shashkov, A.S.; et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci. Rep. 2017, 7, 16302. [Google Scholar] [CrossRef]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B. Bacteriophages and phage-derived proteins--application approaches. Curr. Med. Chem. 2015, 22, 1757–1773. [Google Scholar] [CrossRef]
- Harper, D.R.; Parracho, H.M.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Sutherland, I.W.; Hughes, K.A.; Skillman, L.C.; Tait, K. The interaction of phage and biofilms. FEMS Microbiol. Lett. 2004, 232, 1–6. [Google Scholar] [CrossRef]
- Santos, S.B.; Carvalho, C.M.; Sillankorva, S.; Nicolau, A.; Ferreira, E.C.; Azeredo, J. The use of antibiotics to improve phage detection and enumeration by the double-layer agar technique. BMC Microbiol. 2009, 9, 148. [Google Scholar] [CrossRef]
- Fischer, D.; Eisenberg, D. Finding families for genomic ORFans. Bioinformatics 1999, 15, 759–762. [Google Scholar] [CrossRef]
- Siew, N.; Fischer, D. Structural biology sheds light on the puzzle of genomic ORFans. J. Mol. Biol. 2004, 342, 369–373. [Google Scholar] [CrossRef]
- Wagemans, J.; Blasdel, B.G.; Van den Bossche, A.; Uytterhoeven, B.; De Smet, J.; Paeshuyse, J.; Cenens, W.; Aertsen, A.; Uetz, P.; Delattre, A.S. Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa. Cell. Microbiol. 2014, 16, 1822–1835. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-F.; Wang, X.-F.; Tang, H. Predicting bacteriophage enzymes and hydrolases by using combined features. Front. Bioeng. Biotechnol. 2020, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Yang, W.; Tang, H.; Feng, P.-M.; Huang, J.; Chen, W.; Lin, H. PHYPred: A tool for identifying bacteriophage enzymes and hydrolases. Virol. Sin. 2016, 31, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Dills, M.; Young, M.J. The human gut phage community and its implications for health and disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.K.; Maurice, C.F. Ménage à trois in the human gut: Interactions between host, bacteria and phages. Nat. Rev. Microbiol. 2017, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, S.; Avellaneda-Franco, L.; Barr, J.J. Phages to shape the gut microbiota? Curr. Opin. Biotechnol. 2021, 68, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Muniesa, M. Phages in the human body. Front. Microbiol. 2017, 8, 566. [Google Scholar] [CrossRef]
- McCallum, K.L.; Laakso, D.H.; Whitfield, C. Use of a bacteriophage-encoded glycanase enzyme in the generation of lipopolysaccharide O side chain deficient mutants of Escherichia coli O9: K30 and Klebsiella O1: K20: Role of O and K antigens in resistance to complement-mediated serum killing. Can. J. Microbiol. 1989, 35, 994–999. [Google Scholar] [CrossRef]
- Smith, H.W.; Huggins, M. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. Microbiology 1982, 128, 307–318. [Google Scholar] [CrossRef]
- Lin, H.; Paff, M.L.; Molineux, I.J.; Bull, J.J. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front. Microbiol. 2017, 8, 2257. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef]
- Manzanares, P.; Vallés, S.; Ramòn, D.; Orejas, M. α-L-Rhamnosidases: Old and new insights. In Industrial Enzymes; Springer: Dordrecht, The Netherlands, 2007; pp. 117–140. [Google Scholar]
- Sieiro, C.; García-Fraga, B.; López-Seijas, J.; da Silva, A.F.; Villa, T.G. Microbial pectic enzymes in the food and wine industry. In Food Industrial Processes-Methods and Equipment; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar]
- Born, Y.; Fieseler, L.; Klumpp, J.; Eugster, M.R.; Zurfluh, K.; Duffy, B.; Loessner, M.J. The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ. Microbiol. 2014, 16, 2168–2180. [Google Scholar] [CrossRef]
- Biziulevičius, G.A.; Biziulevičienė, G.; Kazlauskaitė, J. A list of enzyme preparations covered by the term enzybiotics should not be restricted to bacteriophage-encoded peptidoglycan hydrolases (lysins). J. Pharm. Pharmacol. 2008, 60, 531–532. [Google Scholar] [CrossRef]
- Kim, W.-S.; Geider, K. Characterization of a viral EPS-depolymerase, a potential tool for control of fire blight. Phytopathology 2000, 90, 1263–1268. [Google Scholar] [CrossRef]
- Scorpio, A.; Tobery, S.A.; Ribot, W.J.; Friedlander, A.M. Treatment of experimental anthrax with recombinant capsule depolymerase. Antimicrob. Agents Chemother. 2008, 52, 1014–1020. [Google Scholar] [CrossRef]
- Gutierrez, D.; Briers, Y.; Rodriguez-Rubio, L.; Martinez, B.; Rodriguez, A.; Lavigne, R.; Garcia, P. Role of the Pre-neck appendage protein (Dpo7) from phage vB_SepiS-phiIPLA7 as an anti-biofilm agent in Staphylococcal Species. Front. Microbiol. 2015, 6, 1315. [Google Scholar] [CrossRef]
- Oliveira, H.; Costa, A.R.; Ferreira, A.; Konstantinides, N.; Santos, S.B.; Boon, M.; Noben, J.P.; Lavigne, R.; Azeredo, J. Functional analysis and antivirulence properties of a new depolymerase from a myovirus that infects Acinetobacter baumannii capsule K45. J. Virol. 2019, 93, e01163-18. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Łątka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 2016, 8, 324. [Google Scholar] [CrossRef]
- Mushtaq, N.; Redpath, M.B.; Luzio, J.P.; Taylor, P.W. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 2004, 48, 1503–1508. [Google Scholar] [CrossRef]
- Mushtaq, N.; Redpath, M.B.; Luzio, J.P.; Taylor, P.W. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J. Antimicrob. Chemother. 2005, 56, 160–165. [Google Scholar] [CrossRef]
- Bansal, S.; Soni, S.K.; Harjai, K.; Chhibber, S. Aeromonas punctata derived depolymerase that disrupts the integrity of Klebsiella pneumoniae capsule: Optimization of depolymerase production. J. Basic Microbiol. 2014, 54, 711–720. [Google Scholar] [CrossRef]
- Kim, I.-G.; Lee, M.-S.; Jin, T.-E.; Hwang, B.-K.; Lee, J.-H.; Suh, S.-C.; Rhim, S.-L. Inhibitory effect of bacteriophage EPS-depolymerase on growth of Asian pear blight pathogen Erwinia pyrifoliae. J. Microbiol. Biotechnol. 2004, 14, 872–876. [Google Scholar]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef]
- Lin, H.; Paff, M.; Molineux, I.; Bull, J. Antibiotic therapy using phage depolymerases: Robustness across a range of conditions. Viruses 2018, 10, 622. [Google Scholar] [CrossRef]
- Niemann, H.; Kwiatkowski, B.; Westphal, U.; Stirm, S. Klebsiella serotype 25 capsular polysaccharide: Primary structure and depolymerization by a bacteriophage-borne glycanase. J. Bacteriol. 1977, 130, 366–374. [Google Scholar] [CrossRef]
- Altmann, F.; Christian, R.; Czerny, T.; Nimmich, W.; März, L. Bacteriophage-associated glycan hydrolases specific for Escherichia coli capsular serotype K12. Eur. J. Biochem. 1990, 189, 307–312. [Google Scholar] [CrossRef]
- Cheng, Q.; Fischetti, V.A. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 2007, 74, 1284–1291. [Google Scholar] [CrossRef]
- Pastagia, M.; Euler, C.; Chahales, P.; Fuentes-Duculan, J.; Krueger, J.G.; Fischetti, V.A. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and-sensitive Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2011, 55, 738–744. [Google Scholar] [CrossRef]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. PlyPH, a bacteriolytic enzyme with a broad pH range of activity and lytic action against Bacillus anthracis. J. Bacteriol. 2006, 188, 2711–2714. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P.; et al. A novel tail-associated O91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e00145-20. [Google Scholar] [CrossRef] [PubMed]
- Jado, I.; López, R.; García, E.; Fenoll, A.; Casal, J.; García, P. Phage lytic enzymes as therapy for antibiotic-resistant Streptococcus pneumoniae infection in a murine sepsis model. J. Antimicrob. Chemother. 2003, 52, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xu, W.; Lei, L.; Huang, J.; Feng, X.; Sun, C.; Du, C.; Zuo, J.; Li, Y.; Du, T. LysGH15, a novel bacteriophage lysin, protects a murine bacteremia model efficiently against lethal methicillin-resistant Staphylococcus aureus infection. J. Clin. Microbiol. 2011, 49, 111–117. [Google Scholar] [CrossRef]
- Gupta, R.; Prasad, Y. P-27/HP endolysin as antibacterial agent for antibiotic resistant Staphylococcus aureus of human infections. Curr. Microbiol. 2011, 63, 39. [Google Scholar] [CrossRef]
- K Chan, B.; T Abedon, S. Bacteriophages and their enzymes in biofilm control. Curr. Pharmaceut. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef]
- Waseh, S.; Hanifi-Moghaddam, P.; Coleman, R.; Masotti, M.; Ryan, S.; Foss, M.; MacKenzie, R.; Henry, M.; Szymanski, C.M.; Tanha, J. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: Prospects of a novel therapy against bacterial infections. PLoS ONE 2010, 5, e13904. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, E.; Yang, L.; Song, J.; Wu, B. Therapeutic application of bacteriophage PHB02 and its putative depolymerase against Pasteurella multocida capsular type A in mice. Front. Microbiol. 2018, 9, 1678. [Google Scholar] [CrossRef]
- Wang, C.; Li, P.; Niu, W.; Yuan, X.; Liu, H.; Huang, Y.; An, X.; Fan, H.; Zhangxiang, L.; Mi, L.; et al. Protective and therapeutic application of the depolymerase derived from a novel KN1 genotype of Klebsiella pneumoniae bacteriophage in mice. Res. Microbiol. 2019, 170, 156–164. [Google Scholar] [CrossRef]
- Pan, Y.-J.; Lin, T.-L.; Lin, Y.-T.; Su, P.-A.; Chen, C.-T.; Hsieh, P.-F.; Hsu, C.-R.; Chen, C.-C.; Hsieh, Y.-C.; Wang, J.-T. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047. [Google Scholar] [CrossRef]
- Shahed-Al-Mahmud, M.; Roy, R.; Sugiokto, F.G.; Islam, M.N.; Lin, M.-D.; Lin, L.-C.; Lin, N.-T. Phage φAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics 2021, 10, 279. [Google Scholar] [CrossRef]
- Liu, Y.; Leung, S.S.Y.; Guo, Y.; Zhao, L.; Jiang, N.; Mi, L.; Li, P.; Wang, C.; Qin, Y.; Mi, Z.; et al. The capsule depolymerase Dpo48 rescues Galleria mellonella and mice from Acinetobacter baumannii systemic infections. Front. Microbiol. 2019, 10, 545. [Google Scholar] [CrossRef]
- Oliveira, H.; Mendes, A.; Fraga, A.G.; Ferreira, A.; Pimenta, A.I.; Mil-Homens, D.; Fialho, A.M.; Pedrosa, J.; Azeredo, J. K2 capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. Appl. Environ. Microbiol. 2019, 85, e00934-00919. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E. Phage-encoded endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins-extending their application to tissues and the bloodstream. Curr. Opin. Biotechnol. 2021, 68, 51–59. [Google Scholar] [CrossRef]
- Linden, S.B.; Alreja, A.B.; Nelson, D.C. Application of bacteriophage-derived endolysins to combat streptococcal disease: Current State and perspectives. Curr. Opin. Biotechnol. 2021, 68, 213–220. [Google Scholar] [CrossRef]
- Nachimuthu, R.; Madurantakam Royam, M.; Manohar, P.; Leptihn, S. Application of bacteriophages and endolysins in aquaculture as a biocontrol measure. Biol. Control 2021, 160, 104678. [Google Scholar] [CrossRef]
- Tišáková, L.; Godány, A. Bacteriophage endolysins and their use in biotechnological processes. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 164–170. [Google Scholar]
- Lai, M.-J.; Lin, N.-T.; Hu, A.; Soo, P.-C.; Chen, L.-K.; Chen, L.-H.; Chang, K.-C. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both gram-positive and gram-negative bacteria. Appl. Microbiol. Biotechnol. 2011, 90, 529–539. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Oh, M.-D.; Choi, Y.-J.; Lee, W.J.; Kong, J.-C.; Seol, J.G.; Kang, S.H. Antibacterial properties of a pre-formulated recombinant phage endolysin, SAL-1. Int. J. Antimicrob. Agents 2013, 41, 156–161. [Google Scholar] [CrossRef]
- Huang, G.; Shen, X.; Gong, Y.; Dong, Z.; Zhao, X.; Shen, W.; Wang, J.; Hu, F.; Peng, Y. Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect. Dis. 2014, 14, 681. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Chang, W.-L.; Gutiérrez, D.; Lavigne, R.; Martínez, B.; Rodríguez, A.; Govers, S.K.; Aertsen, A.; Hirl, C.; Biebl, M. ‘Artilysation’of endolysin λSa2lys strongly improves its enzymatic and antibacterial activity against streptococci. Sci. Rep. 2016, 6, 35382. [Google Scholar] [CrossRef]
- Skorynina, A.V.; Piligrimova, E.G.; Kazantseva, O.A.; Kulyabin, V.A.; Baicher, S.D.; Ryabova, N.A.; Shadrin, A.M. Bacillus-infecting bacteriophage Izhevsk harbors thermostable endolysin with broad range specificity. PLoS ONE 2020, 15, e0242657. [Google Scholar] [CrossRef]
- Letrado, P.; Corsini, B.; Díez-Martínez, R.; Bustamante, N.; Yuste, J.E.; García, P. Bactericidal synergism between antibiotics and phage endolysin Cpl-711 to kill multidrug-resistant Pneumococcus. Future Microbiol. 2018, 13, 1215–1223. [Google Scholar] [CrossRef]
- Shen, Y.; Barros, M.; Vennemann, T.; Gallagher, D.T.; Yin, Y.; Linden, S.B.; Heselpoth, R.D.; Spencer, D.J.; Donovan, D.M.; Moult, J. A bacteriophage endolysin that eliminates intracellular streptococci. Elife 2016, 5, e13152. [Google Scholar] [CrossRef]
- Loessner, M.J. Bacteriophage endolysins-current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int. J. Med. Microbiol. 2010, 300, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Loessner, M.J. Bacteriophage endolysins: Applications for food safety. Curr. Opin. Biotechnol. 2016, 37, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Environ. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y. Bacteriophage-derived endolysins applied as potent biocontrol agents to enhance food safety. Microorganisms 2020, 8, 724. [Google Scholar] [CrossRef]
- Lai, W.C.B.; Chen, X.; Ho, M.K.Y.; Xia, J.; Leung, S.S.Y. Bacteriophage-derived endolysins to target gram-negative bacteria. Int. J. Pharm. 2020, 589, 119833. [Google Scholar] [CrossRef]
- Caflisch, K.M.; Suh, G.A.; Patel, R. Biological challenges of phage therapy and proposed solutions: A literature review. Expert Rev. Anti-Infect. Ther. 2019, 17, 1011–1041. [Google Scholar] [CrossRef]
- Broendum, S.S.; Buckle, A.M.; McGowan, S. Catalytic diversity and cell wall binding repeats in the phage-encoded endolysins. Mol. Microbiol. 2018, 110, 879–896. [Google Scholar] [CrossRef]
- Dunne, M.; Mertens, H.D.; Garefalaki, V.; Jeffries, C.M.; Thompson, A.; Lemke, E.A.; Svergun, D.I.; Mayer, M.J.; Narbad, A.; Meijers, R. The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor. PLoS Pathog. 2014, 10, e1004228. [Google Scholar] [CrossRef]
- Donovan, D.M.; Foster-Frey, J.; Dong, S.; Rousseau, G.M.; Moineau, S.; Pritchard, D.G. The cell lysis activity of the Streptococcus agalactiae bacteriophage B30 endolysin relies on the cysteine, histidine-dependent amidohydrolase/peptidase domain. Appl. Environ. Microbiol. 2006, 72, 5108–5112. [Google Scholar] [CrossRef]
- Park, S.; Jun, S.Y.; Kim, C.-H.; Jung, G.M.; Son, J.S.; Jeong, S.T.; Yoon, S.J.; Lee, S.Y.; Kang, S.H. Characterisation of the antibacterial properties of the recombinant phage endolysins AP50-31 and LysB4 as potent bactericidal agents against Bacillus anthracis. Sci. Rep. 2018, 8, 18. [Google Scholar] [CrossRef]
- Yang, D.; Chen, Y.; Sun, E.; Hua, L.; Peng, Z.; Wu, B. Characterization of a lytic bacteriophage vB_EfaS_PHB08 harboring endolysin Lys08 against Enterococcus faecalis biofilms. Microorganisms 2020, 8, 1332. [Google Scholar] [CrossRef]
- Lim, J.-A.; Shin, H.; Kang, D.-H.; Ryu, S. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 2012, 163, 233–241. [Google Scholar] [CrossRef]
- Gong, P.; Cheng, M.; Li, X.; Jiang, H.; Yu, C.; Kahaer, N.; Li, J.; Zhang, L.; Xia, F.; Hu, L. Characterization of Enterococcus faecium bacteriophage IME-EFm5 and its endolysin LysEFm5. Virology 2016, 492, 11–20. [Google Scholar] [CrossRef]
- Walmagh, M.; Boczkowska, B.; Grymonprez, B.; Briers, Y.; Drulis-Kawa, Z.; Lavigne, R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl. Microbiol. Biotechnol. 2013, 97, 4369–4375. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.D.; Oliveira, H.; Faustino, A.; Sillankorva, S. Characterization of MSlys, the endolysin of Streptococcus pneumoniae phage MS1. Biotechnol. Rep. 2020, 28, e00547. [Google Scholar] [CrossRef] [PubMed]
- Son, B.; Kong, M.; Lee, Y.; Ryu, S. Development of a novel chimeric endolysin, Lys109 with enhanced lytic activity against Staphylococcus aureus. Front. Microbiol. 2021, 11, 3490. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Gerstmans, H.; Thorpe, S.; Mesnage, S.; Lavigne, R.; Briers, Y. DUF3380 domain from a Salmonella phage endolysin shows potent N-acetylmuramidase activity. Appl. Environ. Microbiol. 2016, 82, 4975–4981. [Google Scholar] [CrossRef]
- Gutierrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- De Wit, J.; Totte, J.E.; van Mierlo, M.M.; van Veldhuizen, J.; van Doorn, M.B.; Schuren, F.H.; Willemsen, S.P.; Pardo, L.M.; Pasmans, S. Endolysin treatment against Staphylococcus aureus in adults with atopic dermatitis: A randomized controlled trial. J. Allergy Clin. Immunol. 2019, 144, 860–863. [Google Scholar] [CrossRef]
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int. J. Antimicrob. Agents 2020, 55, 105844. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.-P. Engineered endolysin-based “Artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 2014, 5, e01379-14. [Google Scholar] [CrossRef]
- Thummeepak, R.; Kitti, T.; Kunthalert, D.; Sitthisak, S. Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage ØABP-01 endolysin (LysABP-01) in combination with colistin. Front. Microbiol. 2016, 7, 1402. [Google Scholar] [CrossRef]
- García, P.; Rodríguez, L.; Rodríguez, A.; Martínez, B. Food biopreservation: Promising strategies using bacteriocins, bacteriophages and endolysins. Trends Food Sci. Technol. 2010, 21, 373–382. [Google Scholar] [CrossRef]
- Gerstmans, H.; Rodríguez-Rubio, L.; Lavigne, R.; Briers, Y. From endolysins to Artilysin® s: Novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 2016, 44, 123–128. [Google Scholar] [CrossRef]
- Morita, M.; Tanji, Y.; Orito, Y.; Mizoguchi, K.; Soejima, A.; Unno, H. Functional analysis of antibacterial activity of Bacillus amyloliquefaciens phage endolysin against Gram-negative bacteria. FEBS Lett. 2001, 500, 56–59. [Google Scholar] [CrossRef]
- Briers, Y.; Schmelcher, M.; Loessner, M.J.; Hendrix, J.; Engelborghs, Y.; Volckaert, G.; Lavigne, R. The high-affinity peptidoglycan binding domain of Pseudomonas phage endolysin KZ144. Biochem. Biophys. Res. Commun. 2009, 383, 187–191. [Google Scholar] [CrossRef]
- Peng, S.-Y.; You, R.-I.; Lai, M.-J.; Lin, N.-T.; Chen, L.-K.; Chang, K.-C. Highly potent antimicrobial modified peptides derived from the Acinetobacter baumannii phage endolysin LysAB2. Sci. Rep. 2017, 7, 11477. [Google Scholar] [CrossRef]
- Blasco, L.; Ambroa, A.; Trastoy, R.; Bleriot, I.; Moscoso, M.; Fernández-Garcia, L.; Perez-Nadales, E.; Fernández-Cuenca, F.; Torre-Cisneros, J.; Oteo-Iglesias, J.; et al. In vitro and in vivo efficacy of combinations of colistin and different endolysins against clinical strains of multi-drug resistant pathogens. Sci. Rep. 2020, 10, 7163. [Google Scholar] [CrossRef]
- Singh, P.K.; Donovan, D.M.; Kumar, A. Intravitreal injection of the chimeric phage endolysin Ply187 protects mice from Staphylococcus aureus endophthalmitis. Antimicrob. Agents Chemother. 2014, 58, 4621–4629. [Google Scholar] [CrossRef]
- Briers, Y.; Peeters, L.M.; Volckaert, G.; Lavigne, R. The lysis cassette of bacteriophage фKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage 2011, 1, 25–30. [Google Scholar] [CrossRef]
- Kim, S.; Jin, J.-S.; Choi, Y.-J.; Kim, J. LysSAP26, a new recombinant phage endolysin with a broad spectrum antibacterial activity. Viruses 2020, 12, 1340. [Google Scholar] [CrossRef]
- Obeso, J.M.; Martínez, B.; Rodríguez, A.; García, P. Lytic activity of the recombinant staphylococcal bacteriophage ΦH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 2008, 128, 212–218. [Google Scholar] [CrossRef]
- Briers, Y.; Volckaert, G.; Cornelissen, A.; Lagaert, S.; Michiels, C.W.; Hertveldt, K.; Lavigne, R. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages φKZ and EL. Mol. Microbiol. 2007, 65, 1334–1344. [Google Scholar] [CrossRef]
- Kong, M.; Sim, J.; Kang, T.; Nguyen, H.H.; Park, H.K.; Chung, B.H.; Ryu, S. A novel and highly specific phage endolysin cell wall binding domain for detection of Bacillus cereus. Eur. Biophys. J. 2015, 44, 437–446. [Google Scholar] [CrossRef]
- Guo, M.; Feng, C.; Ren, J.; Zhuang, X.; Zhang, Y.; Zhu, Y.; Dong, K.; He, P.; Guo, X.; Qin, J. A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 293. [Google Scholar] [CrossRef]
- Legotsky, S.A.; Vlasova, K.Y.; Priyma, A.D.; Shneider, M.M.; Pugachev, V.G.; Totmenina, O.D.; Kabanov, A.V.; Miroshnikov, K.A.; Klyachko, N.L. Peptidoglycan degrading activity of the broad-range Salmonella bacteriophage S-394 recombinant endolysin. Biochimie 2014, 107, 293–299. [Google Scholar] [CrossRef]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef]
- Shen, Y.; Köller, T.; Kreikemeyer, B.; Nelson, D.C. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin. J. Antimicrob. Chemother. 2013, 68, 1818–1824. [Google Scholar] [CrossRef]
- Kashani, H.H.; Schmelcher, M.; Sabzalipoor, H.; Hosseini, E.S.; Moniri, R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: Current status of research and novel delivery strategies. Clin. Microbiol. Rev. 2018, 31, e00071-17. [Google Scholar]
- Khatibi, P.A.; Roach, D.R.; Donovan, D.M.; Hughes, S.R.; Bischoff, K.M. Saccharomyces cerevisiae expressing bacteriophage endolysins reduce Lactobacillus contamination during fermentation. Biotechnol. Biofuels 2014, 7, 104. [Google Scholar] [CrossRef]
- Oliveira, H.; Vilas Boas, D.; Mesnage, S.; Kluskens, L.D.; Lavigne, R.; Sillankorva, S.; Secundo, F.; Azeredo, J. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Front. Microbiol. 2016, 7, 208. [Google Scholar] [CrossRef]
- Totté, J.E.; van Doorn, M.B.; Pasmans, S.G. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical endolysin Staphefekt SA. 100: A report of 3 cases. Case Rep. Dermatol. 2017, 9, 19–25. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Camp, M.J.; Pohl, C.S.; Donovan, D.M. Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl. Microbiol. Biotechnol. 2015, 99, 8475–8486. [Google Scholar] [CrossRef]
- García, P.; Martínez, B.; Rodríguez, L.; Rodríguez, A. Synergy between the phage endolysin LysH5 and nisin to kill Staphylococcus aureus in pasteurized milk. Int. J. Food Microbiol. 2010, 141, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Gerstmans, H.; Criel, B.; Briers, Y. Synthetic biology of modular endolysins. Biotechnol. Adv. 2018, 36, 624–640. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Thiagarajan, V.; Walmagh, M.; Sillankorva, S.; Lavigne, R.; Neves-Petersen, M.T.; Kluskens, L.D.; Azeredo, J. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against gram-negative pathogens in presence of weak acids. PLoS ONE 2014, 9, e108376. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Lavigne, R. Use of bacteriophage endolysin EL188 and outer membrane permeabilizers against Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 110, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- King, H.; Ajay Castro, S.; Pohane, A.A.; Scholte, C.M.; Fischetti, V.A.; Korotkova, N.; Nelson, D.C.; Dorfmueller, H.C. Molecular basis for recognition of the group A carbohydrate backbone by the PlyC streptococcal bacteriophage endolysin. Biochem. J. 2021, 478, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Resch, G.; Moreillon, P.; Fischetti, V.A. PEGylating a bacteriophage endolysin inhibits its bactericidal activity. AMB Express 2011, 1, 29. [Google Scholar] [CrossRef]
- Schuch, R.; Pelzek, A.J.; Nelson, D.C.; Fischetti, V.A. The PlyB endolysin of bacteriophage vB_BanS_Bcp1 exhibits broad-spectrum bactericidal activity against Bacillus cereus sensu lato isolates. Appl. Environ. Microbiol. 2019, 85, e00003-19. [Google Scholar] [CrossRef]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef]
- Linden, S.B.; Zhang, H.; Heselpoth, R.D.; Shen, Y.; Schmelcher, M.; Eichenseher, F.; Nelson, D.C. Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 741–752. [Google Scholar] [CrossRef]
- Shang, X.; Nelson, D.C. Contributions of net charge on the PlyC endolysin CHAP domain. Antibiotics 2019, 8, 70. [Google Scholar] [CrossRef]
- Linden, S.B.; Scholte, C.M.; Vander Elst, N.; Moyes, K.M.; Nelson, D.C. Development of the PlyC endolysin as a bovine mastitis therapeutic for lactating dairy cows. In Proceedings of the 100th Annual conference of Research Workers in Animal Diseases (CRWAD 2019), Chicago, IL, USA, 2–5 November 2019. [Google Scholar]
- Heselpoth, R.D.; Yin, Y.; Moult, J.; Nelson, D.C. Increasing the stability of the bacteriophage endolysin PlyC using rationale-based FoldX computational modeling. Protein Eng. Des. Sel. 2015, 28, 85–92. [Google Scholar] [CrossRef]
- Linden, S.B.; Vander Elst, N.; Nelson, D.C. The PlySs9 endolysin contains unique catalytic domains and is a potential therapeutic against Streptococcus suis. In Proceedings of the 100th Annual conference of Research Workers in Animal Diseases (CRWAD 2019), Chicago, IL, USA, 2–5 November 2019. [Google Scholar]
- Heselpoth, R.D.; Owens, J.M.; Nelson, D.C. Quantitative analysis of the thermal stability of the gamma phage endolysin PlyG: A biophysical and kinetic approach to assaying therapeutic potential. Virology 2015, 477, 125–132. [Google Scholar] [CrossRef]
- Scholte, C.; Nelson, D.; Garcia, M.; Linden, S.; Elsasser, T.; Kahl, S.; Qu, Y.; Moyes, K. Recombinant bacteriophage endolysin PlyC is nontoxic and does not alter blood neutrophil oxidative response in lactating dairy cows. J. Dairy Sci. 2018, 101, 6419–6423. [Google Scholar] [CrossRef]
- Harhala, M.; Nelson, D.C.; Miernikiewicz, P.; Heselpoth, R.D.; Brzezicka, B.; Majewska, J.; Linden, S.B.; Shang, X.; Szymczak, A.; Lecion, D. Safety studies of pneumococcal endolysins Cpl-1 and Pal. Viruses 2018, 10, 638. [Google Scholar] [CrossRef]
- Jasim, H.N.; Hafidh, R.R.; Abdulamir, A.S. Formation of therapeutic phage cocktail and endolysin to highly multi-drug resistant Acinetobacter baumannii: In vitro and in vivo study. Iran. J. Basic Med. Sci. 2018, 21, 1100. [Google Scholar]
- Basit, A.; Qadir, S.; Qureshi, S.; Rehman, S.U. Cloning and expression analysis of fused holin-endolysin from RL bacteriophage; Exhibits broad activity against multi drug resistant pathogens. Enzym. Microb. Technol. 2021, 149, 109846. [Google Scholar] [CrossRef]
- Donovan, D.; Becker, S.; Dong, S.; Baker, J.; Foster-Frey, J.; Pritchard, D. Peptidoglycan hydrolase enzyme fusions for treating multi-drug resistant pathogens. Biotech. Int. 2009, 6–10. [Google Scholar]
- Ali, M.R.; Abdulamir, A.S.; Kadhim, S.R. Extraction, purification and therapeutic use of bacteriophage endolysin against multi-drug resistant Staphylococcus aureus: In-vivo and in-vitro study. J. Contemp. Med. Sci. 2018, 4. [Google Scholar]
- Haddad Kashani, H.; Fahimi, H.; Dasteh Goli, Y.; Moniri, R. A novel chimeric endolysin with antibacterial activity against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 290. [Google Scholar] [CrossRef]
- Khan, F.M.; Gondil, V.S.; Li, C.; Jiang, M.; Li, J.; Yu, J.; Wei, H.; Yang, H. A novel Acinetobacter baumannii bacteriophage endolysin LysAB54 with high antibacterial activity against multiple Gram-negative microbes. Front. Cell. Infect. Microbiol. 2021, 11, 70. [Google Scholar] [CrossRef]
- Mayer, M.J.; Garefalaki, V.; Spoerl, R.; Narbad, A.; Meijers, R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J. Bacteriol. 2011, 193, 5477–5486. [Google Scholar] [CrossRef]
- Schleimer, N.; Kaspar, U.; Knaack, D.; von Eiff, C.; Molinaro, S.; Grallert, H.; Idelevich, E.A.; Becker, K. In vitro activity of the bacteriophage endolysin HY-133 against Staphylococcus aureus small-colony variants and their corresponding wild types. Int. J. Mol. Sci. 2019, 20, 716. [Google Scholar] [CrossRef]
- Knaack, D.; Idelevich, E.A.; Schleimer, N.; Molinaro, S.; Kriegeskorte, A.; Peters, G.; Becker, K. Bactericidal activity of bacteriophage endolysin HY-133 against Staphylococcus aureus in comparison to other antibiotics as determined by minimum bactericidal concentrations and time-kill analysis. Diagn. Microbiol. Infect. Dis. 2019, 93, 362–368. [Google Scholar] [CrossRef]
- Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. J. Microbiol. Biotechnol. 2014, 24, 803–811. [Google Scholar] [CrossRef]
- Zhang, H.; Buttaro, B.A.; Fouts, D.E.; Sanjari, S.; Evans, B.S.; Stevens, R.H. Bacteriophage φEf11 ORF28 endolysin, a multifunctional lytic enzyme with properties distinct from all other identified Enterococcus faecalis phage endolysins. Appl. Environ. Microbiol. 2019, 85, e00555-00519. [Google Scholar] [CrossRef]
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812. [Google Scholar] [CrossRef]
- Rahimzadeh, G.; Gill, P.; Rezai, M.S. Endolysins of bacteriophages as an anti-methicillin resistant staphylococcus aureus infection in children: A narrative review. J. Pediatr. Rev. 2018, 6, 36–43. [Google Scholar] [CrossRef]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile lytic enzyme fusion proteins that target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef]
- Muharram, M.M.; Abulhamd, A.T.; Aldawsari, M.F.; Alqarni, M.H.; Labrou, N.E. Development of Staphylococcus enzybiotics: The Ph28 gene of Staphylococcus epidermidis phage PH15 is a two-domain endolysin. Antibiotics 2020, 9, 148. [Google Scholar] [CrossRef]
- Domenech, M.; García, E.; Moscoso, M. In vitro destruction of Streptococcus pneumoniae biofilms with bacterial and phage peptidoglycan hydrolases. Antimicrob. Agents Chemother. 2011, 55, 4144–4148. [Google Scholar] [CrossRef]
- Landlinger, C.; Tisakova, L.; Oberbauer, V.; Schwebs, T.; Muhammad, A.; Latka, A.; Van Simaey, L.; Vaneechoutte, M.; Guschin, A.; Resch, G. Engineered phage endolysin eliminates Gardnerella biofilm without damaging beneficial bacteria in bacterial vaginosis ex vivo. Pathogens 2021, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Pennone, V.; Sanz-Gaitero, M.; O’connor, P.; Coffey, A.; Jordan, K.; van Raaij, M.J.; McAuliffe, O. Inhibition of L. monocytogenes biofilm formation by the amidase domain of the phage vB_LmoS_293 endolysin. Viruses 2019, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, Y.; Wang, J.; Zhao, Y.; Zhong, Q.; Li, G.; Fu, Z.; Lu, S. Phage endolysin LysP108 showed promising antibacterial potential against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2021, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, H.-H.; Duc, H.M.; Masuda, Y.; Honjoh, K.-i.; Miyamoto, T. Endolysin LysSTG2: Characterization and application to control Salmonella Typhimurium biofilm alone and in combination with slightly acidic hypochlorous water. Food Microbiol. 2021, 98, 103791. [Google Scholar] [CrossRef]
- López, R.; García, E.; García, P.; García, J.L. The pneumococcal cell wall degrading enzymes: A modular design to create new lysins? Microb. Drug Resist. 1997, 3, 199–211. [Google Scholar] [CrossRef]
- Briers, Y.; Walmagh, M.; Grymonprez, B.; Biebl, M.; Pirnay, J.-P.; Defraine, V.; Michiels, J.; Cenens, W.; Aertsen, A.; Miller, S. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 3774–3784. [Google Scholar] [CrossRef]
- Defraine, V.; Schuermans, J.; Grymonprez, B.; Govers, S.K.; Aertsen, A.; Fauvart, M.; Michiels, J.; Lavigne, R.; Briers, Y. Efficacy of artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016, 60, 3480–3488. [Google Scholar] [CrossRef]
- Theuretzbacher, U.; Piddock, L.J. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 2019, 26, 61–72. [Google Scholar] [CrossRef]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.-I.; Muraoka, A.; Sugai, M.; Hiramatsu, K. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage ϕMR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef]
- Kim, N.-H.; Park, W.B.; Cho, J.E.; Choi, Y.J.; Choi, S.J.; Jun, S.Y.; Kang, C.K.; Song, K.-H.; Choe, P.G.; Bang, J.-H. Effects of phage endolysin SAL200 combined with antibiotics on Staphylococcus aureus infection. Antimicrob. Agents Chemother. 2018, 62, e00731-18. [Google Scholar] [CrossRef]
- Becker, S.C.; Foster-Frey, J.; Donovan, D.M. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008, 287, 185–191. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Becker, S.C.; Camp, M.J.; Donovan, D.M. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl. Environ. Microbiol. 2012, 78, 2297–2305. [Google Scholar] [CrossRef]
- Loeffler, J.; Fischetti, V. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and-resistant Streptococcus pneumoniae strains. Antimicrob. Agents Chemother. 2003, 47, 375–377. [Google Scholar] [CrossRef]
- Djurkovic, S.; Loeffler, J.M.; Fischetti, V.A. Synergistic killing of Streptococcus pneumoniae with the bacteriophage lytic enzyme Cpl-1 and penicillin or gentamicin depends on the level of penicillin resistance. Antimicrob. Agents Chemother. 2005, 49, 1225–1228. [Google Scholar] [CrossRef]
- Daniel, A.; Euler, C.; Collin, M.; Chahales, P.; Gorelick, K.J.; Fischetti, V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 1603–1612. [Google Scholar] [CrossRef]
- Vouillamoz, J.; Entenza, J.M.; Giddey, M.; Fischetti, V.A.; Moreillon, P.; Resch, G. Bactericidal synergism between daptomycin and the phage lysin Cpl-1 in a mouse model of pneumococcal bacteraemia. Int. J. Antimicrob. Agents 2013, 42, 416–421. [Google Scholar] [CrossRef]
- Park, D.-W.; Park, J.-H. Characterization of Endolysin LysECP26 Derived from rV5-like Phage vB_EcoM-ECP26 for Inactivation of Escherichia coli O157: H7. J. Microbiol. Biotechnol. 2020, 30, 1552–1558. [Google Scholar] [CrossRef]
- Entenza, J.; Loeffler, J.; Grandgirard, D.; Fischetti, V.; Moreillon, P. Therapeutic effects of bacteriophage Cpl-1 lysin against Streptococcus pneumoniae endocarditis in rats. Antimicrob. Agents Chemother. 2005, 49, 4789–4792. [Google Scholar] [CrossRef]
- Witzenrath, M.; Schmeck, B.; Doehn, J.M.; Tschernig, T.; Zahlten, J.; Loeffler, J.M.; Zemlin, M.; Müller, H.; Gutbier, B.; Schütte, H. Systemic use of the endolysin Cpl-1 rescues mice with fatal pneumococcal pneumonia. Crit. Care Med. 2009, 37, 642–649. [Google Scholar] [CrossRef]
- Cassino, C.; Murphy, M.; Boyle, J.; Rotolo, J.; Wittekind, M. Results of the first in human study of lysin CF-301 evaluating the safety, tolerability and pharmacokinetic profile in healthy volunteers. In Proceedings of the 26th European Congress of Clinical Microbiology and Infectious Diseases, Amsterdam, The Netherlands, 8–11 April 2016; pp. 151–152. [Google Scholar]
- Jandourek, A.; Boyle, J.; Cassino, C.; Wittekind, M.; Kirby, H. Long term immunology results of a phase 1 placebo controlled dose escalating study to examine the safety of CF-301 in human volunteers. In Proceedings of the 27th ECCMID, Vienna, Austria, 22 April 2017; pp. 22–25. [Google Scholar]
- Jandourek, A.; Boyle, J.; Murphy, G.; Cassino, C. Inflammatory markers in a phase 1 placebo controlled dose escalating study of intravenous doses of CF-301 in human subjects. In Proceedings of the ASM Microbe, New Orleans, LA, USA, 2 June 2017; p. 2. [Google Scholar]
- Ghahramani, P.; Khariton, T.; Jones, S.; Murphy, J.; Boyle, G.; Jandourek, A.; Cassino, C. Population pharmacokinetic-pharmacodynamic assessment of cardiac safety endpoints for CF-301, a first-in-class antibacterial lysin. In Proceedings of the ASM Microbe, New Orleans, LA, USA, 3 June 2017. [Google Scholar]
- Rotolo, J.A.; Ramirez, R.A.; Schuch, R.; Machacek, M.; Khariton, T.; Ghahramani, P.; Wittekind, M. PK-PD driver of efficacy for CF-301, a novel anti-staphylococcal lysin: Implications for human target dose. In Proceedings of the ASM Microbe, Boston, MA, USA, 18 June 2016; pp. 16–20. [Google Scholar]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Choi, Y.-J.; Koh, W.S.; Moon, K.S.; Kang, S.H. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob. Agents Chemother. 2014, 58, 2084–2088. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jung, G.M.; Yoon, S.J.; Youm, S.Y.; Han, H.Y.; Lee, J.H.; Kang, S.H. Pharmacokinetics of the phage endolysin-based candidate drug SAL 200 in monkeys and its appropriate intravenous dosing period. Clin. Exp. Pharmacol. Physiol. 2016, 43, 1013–1016. [Google Scholar] [CrossRef]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef] [PubMed]
- Tamai, E.; Yoshida, H.; Sekiya, H.; Nariya, H.; Miyata, S.; Okabe, A.; Kuwahara, T.; Maki, J.; Kamitori, S. X-ray structure of a novel endolysin encoded by episomal phage phiSM 101 of C lostridium perfringens. Mol. Microbiol. 2014, 92, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, T.; Lo Curto, R.; Minniti, E.; Narbad, A.; Mayer, M.J. Application of Lactobacillus johnsonii expressing phage endolysin for control of Clostridium perfringens. Lett. Appl. Microbiol. 2014, 59, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Wernicki, A.; Nowaczek, A.; Urban-Chmiel, R. Bacteriophage therapy to combat bacterial infections in poultry. Virol. J. 2017, 14, 179. [Google Scholar] [CrossRef]
- Zhang, H.; Bao, H.; Billington, C.; Hudson, J.A.; Wang, R. Isolation and lytic activity of the Listeria bacteriophage endolysin LysZ5 against Listeria monocytogenes in soya milk. Food Microbiol. 2012, 31, 133–136. [Google Scholar] [CrossRef]
- Van Nassau, T.J.; Lenz, C.A.; Scherzinger, A.S.; Vogel, R.F. Combination of endolysins and high pressure to inactivate Listeria monocytogenes. Food Microbiol. 2017, 68, 81–88. [Google Scholar] [CrossRef]
- Chang, Y.; Kim, M.; Ryu, S. Characterization of a novel endolysin LysSA11 and its utility as a potent biocontrol agent against Staphylococcus aureus on food and utensils. Food Microbiol. 2017, 68, 112–120. [Google Scholar] [CrossRef]
- Villa, T.G.; Feijoo-Siota, L.; Rama, J.L.R.; Sánchez-Pérez, A.; de Miguel-Bouzas, T. Chapter 40—Enzybiotics: Application in food packaging. In Antimicrobial Food Packaging; Barros-Velázquez, J., Ed.; Academic Press: San Diego, CA, USA, 2016; pp. 491–502. [Google Scholar]
- Roach, D.R.; Khatibi, P.A.; Bischoff, K.M.; Hughes, S.R.; Donovan, D.M. Bacteriophage-encoded lytic enzymes control growth of contaminating Lactobacillus found in fuel ethanol fermentations. Biotechnol. Biofuels 2013, 6, 20. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of bacteriophages in the agro-food sector: A long way toward approval. Front. Cell. Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef]
- Hausbeck, M.; Bell, J.; Medina-Mora, C.; Podolsky, R.; Fulbright, D. Effect of bactericides on population sizes and spread of Clavibacter michiganensis subsp. michiganensis on tomatoes in the greenhouse and on disease development and crop yield in the field. Phytopathology 2000, 90, 38–44. [Google Scholar] [CrossRef]
- De Vries, J.; Harms, K.; Broer, I.; Kriete, G.; Mahn, A.; Düring, K.; Wackernagel, W. The bacteriolytic activity in transgenic potatoes expressing a chimeric T4 lysozyme gene and the effect of T4 lysozyme on soil-and phytopathogenic bacteria. Syst. Appl. Microbiol. 1999, 22, 280–286. [Google Scholar] [CrossRef]
- Timbermont, L.; Haesebrouck, F.; Ducatelle, R.; Van Immerseel, F. Necrotic enteritis in broilers: An updated review on the pathogenesis. Avian Pathol. 2011, 40, 341–347. [Google Scholar] [CrossRef]
- Junjappa, R.P.; Desai, S.N.; Roy, P.; Narasimhaswamy, N.; Raj, J.R.M.; Durgaiah, M.; Vipra, A.; Bhat, U.R.; Satyanarayana, S.K.; Shankara, N. Efficacy of anti-staphylococcal protein P128 for the treatment of canine pyoderma: Potential applications. Vet. Res. Commun. 2013, 37, 217–228. [Google Scholar] [CrossRef]
- Angelopoulou, A.; Warda, A.K.; Hill, C.; Ross, R.P. Non-antibiotic microbial solutions for bovine mastitis–live biotherapeutics, bacteriophage, and phage lysins. Crit. Rev. Microbiol. 2019, 45, 564–580. [Google Scholar] [CrossRef]
- Donovan, D.M.; Lardeo, M.; Foster-Frey, J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006, 265, 133–139. [Google Scholar] [CrossRef]
- Fan, J.; Zeng, Z.; Mai, K.; Yang, Y.; Feng, J.; Bai, Y.; Sun, B.; Xie, Q.; Tong, Y.; Ma, J. Preliminary treatment of bovine mastitis caused by Staphylococcus aureus, with trx-SA1, recombinant endolysin of S. aureus bacteriophage IME-SA1. Vet. Microbiol. 2016, 191, 65–71. [Google Scholar] [CrossRef]
- Zduńczyk, S.; Janowski, T. Bacteriophages and associated endolysins in therapy and prevention of mastitis and metritis in cows: Current knowledge. Anim. Reprod. Sci. 2020, 218, 106504. [Google Scholar] [CrossRef]
- Vander Elst, N.; Meyer, E. Potential therapeutic application of bacteriophages and phage-derived endolysins as alternative treatment of bovine mastitis. Vlaams Diergeneeskd. Tijdschr. 2018, 87, 181–187. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Lu, C. Purified recombinant phage lysin LySMP: An extensive spectrum of lytic activity for swine streptococci. Curr. Microbiol. 2009, 58, 609–615. [Google Scholar] [CrossRef]
- Hoopes, J.T.; Stark, C.J.; Kim, H.A.; Sussman, D.J.; Donovan, D.M.; Nelson, D.C. Use of a bacteriophage lysin, PlyC, as an enzyme disinfectant against Streptococcus equi. Appl. Environ. Microbiol. 2009, 75, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Nelson, D.; Fischetti, V.A. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 2002, 418, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Sundarrajan, S.; Raghupatil, J.; Vipra, A.; Narasimhaswamy, N.; Saravanan, S.; Appaiah, C.; Poonacha, N.; Desai, S.; Nair, S.; Bhatt, R.N. Bacteriophage-derived CHAP domain protein, P128, kills Staphylococcus cells by cleaving interpeptide cross-bridge of peptidoglycan. Microbiology 2014, 160, 2157–2169. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ragland, S.A.; Humbert, M.V.; Christodoulides, M.; Criss, A.K. Neisseria gonorrhoeae employs two protein inhibitors to evade killing by human lysozyme. PLoS Pathog. 2018, 14, e1007080. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Akinbi, H.T.; Standish, A.J.; Weiser, J.N. Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus Pneumoniae. PLoS Pathog. 2008, 4, e1000241. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008, 32, 287–306. [Google Scholar] [CrossRef]
- Guariglia-Oropeza, V.; Helmann, J.D. Bacillus subtilis σV confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 2011, 193, 6223–6232. [Google Scholar] [CrossRef]
- Gründling, A.; Missiakas, D.M.; Schneewind, O. Staphylococcus aureus mutants with increased lysostaphin resistance. J. Bacteriol. 2006, 188, 6286–6297. [Google Scholar] [CrossRef]
- DeHart, H.P.; Heath, H.E.; Heath, L.S.; LeBlanc, P.A.; Sloan, G.L. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl. Environ. Microbiol. 1995, 61, 1475–1479. [Google Scholar] [CrossRef]
- Davis, K.M.; Weiser, J.N. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 2011, 79, 562–570. [Google Scholar] [CrossRef]
- Lukacik, P.; Barnard, T.J.; Keller, P.W.; Chaturvedi, K.S.; Seddiki, N.; Fairman, J.W.; Noinaj, N.; Kirby, T.L.; Henderson, J.P.; Steven, A.C. Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc. Natl. Acad. Sci. USA 2012, 109, 9857–9862. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Briers, Y. Lysins breaking down the walls of gram-negative bacteria, no longer a no-go. Curr. Opin. Biotechnol. 2021, 68, 15–22. [Google Scholar] [CrossRef]
- Briers, Y.; Miller, S.; Lavigne, R. Artilysins Are a Novel Class of Enzyme-Based Antibacterials That Quickly Kill (Multidrug-Resistant) Pseudomonas Aeruginosa and Their Persisters: From Concept to Application. Available online: https://www.semanticscholar.org/paper/Artilysins-are-a-novel-class-of-enzyme-based-that-Briers-Miller/bcd3a549fa7c897ef05fdc1012387b73c4e14f19 (accessed on 2 December 2021).
- Antonova, N.P.; Vasina, D.V.; Lendel, A.M.; Usachev, E.V.; Makarov, V.V.; Gintsburg, A.L.; Tkachuk, A.P.; Gushchin, V.A. Broad bactericidal activity of the Myoviridae bacteriophage lysins LysAm24, LysECD7, and LysSi3 against Gram-negative ESKAPE pathogens. Viruses 2019, 11, 284. [Google Scholar] [CrossRef]
- Díez-Martínez, R.; de Paz, H.; Bustamante, N.; García, E.; Menéndez, M.; García, P. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob. Agents Chemother. 2013, 57, 5355–5365. [Google Scholar] [CrossRef]
- Ciepluch, K.; Skrzyniarz, K.; Barrios-Gumiel, A.; Quintana, S.; Sánchez-Nieves, J.; de la Mata, F.J.; Maciejewska, B.; Drulis-Kawa, Z.; Arabski, M. Dendronized silver nanoparticles as bacterial membrane permeabilizers and their interactions with P. aeruginosa lipopolysaccharides, lysozymes, and phage-derived endolysins. Front. Microbiol. 2019, 10, 2771. [Google Scholar] [CrossRef]
- Gerstmans, H.; Grimon, D.; Gutiérrez, D.; Lood, C.; Rodríguez, A.; van Noort, V.; Lammertyn, J.; Lavigne, R.; Briers, Y. A VersaTile-driven platform for rapid hit-to-lead development of engineered lysins. Sci. Adv. 2020, 6, eaaz1136. [Google Scholar] [CrossRef]
- Briers, Y. Phage lysins as simple as Lego. Caspid Tail 2020, 79. [Google Scholar]
- De Maesschalck, V.; Gutiérrez, D.; Paeshuyse, J.; Lavigne, R.; Briers, Y. Advanced engineering of third-generation lysins and formulation strategies for clinical applications. Crit. Rev. Microbiol. 2020, 46, 548–564. [Google Scholar] [CrossRef]
- Gutierrez Fernandez, D.; Briers, Y. Developments and opportunities of bacteriophage lytic proteins for therapeutics against gram-negative pathogens. In Bacterial Viruses: Exploitation for Biocontrol and Therapeutics; Caister Academic Press: Norwich, UK, 2020; pp. 537–586. [Google Scholar]
- Ghose, C.; Euler, C.W. Gram-negative bacterial lysins. Antibiotics 2020, 9, 74. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, X.; Wang, L.; Li, G.; Cong, C.; Li, R.; Cui, H.; Murtaza, B.; Xu, Y. The endolysin of the Acinetobacter baumannii phage vB_AbaP_D2 shows broad antibacterial activity. Microb. Biotechnol. 2021, 14, 403–418. [Google Scholar] [CrossRef]
- Wang, F.; Ji, X.; Li, Q.; Zhang, G.; Peng, J.; Hai, J.; Zhang, Y.; Ci, B.; Li, H.; Xiong, Y. TSPphg Lysin from the extremophilic thermus bacteriophage TSP4 as a potential antimicrobial agent against both gram-negative and gram-positive pathogenic bacteria. Viruses 2020, 12, 192. [Google Scholar] [CrossRef]
- Antonova, N.P.; Vasina, D.V.; Rubalsky, E.O.; Fursov, M.V.; Savinova, A.S.; Grigoriev, I.V.; Usachev, E.V.; Shevlyagina, N.V.; Zhukhovitsky, V.G.; Balabanyan, V.U. Modulation of endolysin LysECD7 bactericidal activity by different peptide tag fusion. Biomolecules 2020, 10, 440. [Google Scholar] [CrossRef]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef]
- Walmagh, M.; Briers, Y.; Dos Santos, S.B.; Azeredo, J.; Lavigne, R. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201ϕ2-1 and PVP-SE1. PLoS ONE 2012, 7, e36991. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J.; Wendlinger, G.; Scherer, S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: A new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 1995, 16, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Payne, K.M.; Hatfull, G.F. Mycobacteriophage endolysins: Diverse and modular enzymes with multiple catalytic activities. PLoS ONE 2012, 7, e34052. [Google Scholar] [CrossRef] [PubMed]
- Borysowski, J.; Weber-Dąbrowska, B.; Górski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Jarábková, V.; Tišáková, L.; Godány, A. Phage endolysin: A way to understand a binding function of C-terminal domains a mini review. Nova Biotechnol. Chim. 2015, 14, 117–134. [Google Scholar] [CrossRef]
- Fenton, M.; McAuliffe, O.; O’Mahony, J.; Coffey, A. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs 2010, 1, 9–16. [Google Scholar] [CrossRef]
- Briers, Y.; Lavigne, R.; Volckaert, G.; Hertveldt, K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods 2007, 70, 531–533. [Google Scholar] [CrossRef]
- Fowler, V.G., Jr.; Das, A.F.; Lipka-Diamond, J.; Schuch, R.; Pomerantz, R.; Jáuregui-Peredo, L.; Bressler, A.; Evans, D.; Moran, G.J.; Rupp, M.E.; et al. Exebacase for patients with Staphylococcus aureus bloodstream infection and endocarditis. J. Clin. Investig. 2020, 130, 3750–3760. [Google Scholar] [CrossRef]
- Paul, V.D.; Rajagopalan, S.S.; Sundarrajan, S.; George, S.E.; Asrani, J.Y.; Pillai, R.; Chikkamadaiah, R.; Durgaiah, M.; Sriram, B.; Padmanabhan, S. A novel bacteriophage Tail-Associated Muralytic Enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein. BMC Microbiol. 2011, 11, 226. [Google Scholar] [CrossRef]
- Saravanan, S.R.; Paul, V.D.; George, S.; Sundarrajan, S.; Kumar, N.; Hebbur, M.; Kumar, N.; Veena, A.; Maheshwari, U.; Appaiah, C.B. Properties and mutation studies of a bacteriophage-derived chimeric recombinant staphylolytic protein P128: Comparison to recombinant lysostaphin. Bacteriophage 2013, 3, e26564. [Google Scholar] [CrossRef]
- Nair, S.; Poonacha, N.; Desai, S.; Hiremath, D.; Tuppad, D.; Mohan, T.; Chikkamadaiah, R.; Durgaiah, M.; Kumar, S.; Channabasappa, S. Restoration of sensitivity of a diverse set of drug-resistant Staphylococcus clinical strains by bactericidal protein P128. J. Med. Microbiol. 2018, 67, 296–307. [Google Scholar] [CrossRef]
- Nair, S.; Desai, S.; Poonacha, N.; Vipra, A.; Sharma, U. Antibiofilm activity and synergistic inhibition of Staphylococcus aureus biofilms by bactericidal protein P128 in combination with antibiotics. Antimicrob. Agents Chemother. 2016, 60, 7280–7289. [Google Scholar] [CrossRef]
- Poonacha, N.; Nair, S.; Desai, S.; Tuppad, D.; Hiremath, D.; Mohan, T.; Vipra, A.; Sharma, U. Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob. Agents Chemother. 2017, 61, e00457-17. [Google Scholar] [CrossRef]
- Vipra, A.A.; Desai, S.N.; Roy, P.; Patil, R.; Raj, J.M.; Narasimhaswamy, N.; Paul, V.D.; Chikkamadaiah, R.; Sriram, B. Antistaphylococcal activity of bacteriophage derived chimeric protein P128. BMC Microbiol. 2012, 12, 41. [Google Scholar] [CrossRef]
- Drilling, A.J.; Cooksley, C.; Chan, C.; Wormald, P.J.; Vreugde, S. Fighting sinus-derived Staphylococcus aureus biofilms in vitro with a bacteriophage-derived muralytic enzyme. Int. Forum Allergy Rhinol. 2016, 6, 349–355. [Google Scholar] [CrossRef]
- George, S.E.; Chikkamadaiah, R.; Durgaiah, M.; Joshi, A.A.; Thankappan, U.P.; Madhusudhana, S.N.; Sriram, B. Biochemical characterization and evaluation of cytotoxicity of antistaphylococcal chimeric protein P128. BMC Res. Notes 2012, 5, 280. [Google Scholar] [CrossRef]
- Climo, M.W.; Ehlert, K.; Archer, G.L. Mechanism and suppression of lysostaphin resistance in oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1431–1437. [Google Scholar] [CrossRef]
- Sriram, B.; Chikkamadaiah, S.; Durgaiah, M.; Hariharan, S.; Jayaraman, R.; Kumar, S.; Maheshwari, U.; Nandish, P. Pharmacokinetics and efficacy of ectolysin P128 in a mouse model of systemic Methicillin Resistant Staphylococcus aureus (MRSA) infection. In Proceedings of the ASM Microbe, New Orleans, LA, USA, 1–5 June 2017. [Google Scholar]
- Channabasappa, S.; Chikkamadaiah, R.; Durgaiah, M.; Kumar, S.; Ramesh, K.; Sreekanthan, A.; Sriram, B. Efficacy of chimeric ectolysin P128 in drug-resistant Staphylococcus aureus bacteraemia in mice. J. Antimicrob. Chemother. 2018, 73, 3398–3404. [Google Scholar] [CrossRef]
- Channabasappa, S.; Durgaiah, M.; Chikkamadaiah, R.; Kumar, S.; Joshi, A.; Sriram, B. Efficacy of novel antistaphylococcal ectolysin P128 in a rat model of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2018, 62, e01358-17. [Google Scholar] [CrossRef]
- Son, J.; Kim, E.B.; Lee, S.; Jun, S.; Yoon, S.-J.; Kang, S.; Choi, Y. Characterization of Staphylococcus aureus derived from bovine mastitis and isolation of two lytic bacteriophages. J. Gen. Appl. Microbiol. 2010, 56, 347–353. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jung, G.M.; Son, J.-S.; Yoon, S.J.; Choi, Y.-J.; Kang, S.H. Comparison of the Antibacterial Properties of Phage Endolysins SAL-1 and LysK. Antimicrob. Agents Chemother. 2011, 55, 1764–1767. [Google Scholar] [CrossRef]
- Schuch, R.; Lee, H.M.; Schneider, B.C.; Sauve, K.L.; Law, C.; Khan, B.K.; Rotolo, J.A.; Horiuchi, Y.; Couto, D.E.; Raz, A. Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus–induced murine bacteremia. J. Infect. Dis. 2014, 209, 1469–1478. [Google Scholar] [CrossRef]
- Schuch, R.; Khan, B.K.; Raz, A.; Rotolo, J.A.; Wittekind, M. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob. Agents Chemother. 2017, 61, e02666-16. [Google Scholar] [CrossRef]
- Sauve, K.; Jandourek, A.; Cassino, C.; Schuch, R. Lysin CF-301 demonstrates in vitro synergy with conventional antibiotics against Staphylococcus aureus. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2017; p. 370. [Google Scholar]
- Oh, J.T.; Cassino, C.; Schuch, R. Postantibiotic and sub-MIC effects of exebacase (lysin CF-301) enhance antimicrobial activity against Staphylococcus aureus. Antimicrob. Agents Chemother. 2019, 63, e02616-18. [Google Scholar] [CrossRef]
- Indiani, C.; Sauve, K.; Raz, A.; Abdelhady, W.; Xiong, Y.Q.; Cassino, C.; Bayer, A.S.; Schuch, R. The antistaphylococcal lysin, CF-301, activates key host factors in human blood to potentiate methicillin-resistant Staphylococcus aureus bacteriolysis. Antimicrob. Agents Chemother. 2019, 63, e02291-18. [Google Scholar] [CrossRef]
- Asempa, T.E.; Abdelraouf, K.; Carabeo, T.; Schuch, R.; Nicolau, D.P. Synergistic activity of exebacase (CF-301) in addition to daptomycin against Staphylococcus aureus in a neutropenic murine thigh infection model. Antimicrob. Agents Chemother. 2019, 64, e02176-19. [Google Scholar] [CrossRef]
- Anastasiou, D.; Jandourek, A.; Traczewski, M.; Cassino, C.; Schuch, R. 1342. Comparison of lysin CF-301 (Exebacase) activity against S. aureus isolates. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2018; p. 410. [Google Scholar]
- Anastasiou, D.; Cassino, C.; Schuch, R. 711. Exebacase (Lysin CF-301) activity against Staphylococcus aureus (S. aureus) isolates from bacteremic patients enrolled in a phase 2 study (CF-301-102). In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2019. [Google Scholar]
- Hendricks, A.J.; Mills, B.W.; Shi, V.Y. Skin bacterial transplant in atopic dermatitis: Knowns, unknowns and emerging trends. J. Dermatol. Sci. 2019, 95, 56–61. [Google Scholar] [CrossRef]
- Herpers, B.; Badoux, P.; Pietersma, F.; Eichenseher, F.; Loessner, M. Specific lysis of methicillin susceptible and resistant Staphylococcus aureus by the endolysin Staphefekt SA. 100 TM. In Proceedings of the 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Barcelona, Spain, 10–13 May 2014. [Google Scholar]
- Totté, J.; de Wit, J.; Pardo, L.; Schuren, F.; van Doorn, M.; Pasmans, S. Targeted anti-staphylococcal therapy with endolysins in atopic dermatitis and the effect on steroid use, disease severity and the microbiome: Study protocol for a randomized controlled trial (MAAS trial). Trials 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Lysin therapy for Staphylococcus aureus and other bacterial pathogens. In Staphylococcus aureus; Springer: Cham, Switzerland, 2015; pp. 529–540. [Google Scholar]
- Lysando. Available online: www.lysando.com (accessed on 2 December 2021).
- Zipfel, C.; Robatzek, S. Pathogen-associated molecular pattern-triggered immunity: Veni, vidi…? Plant Physiol. 2010, 154, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef] [PubMed]
- Agu, R.U.; Ugwoke, M.I.; Armand, M.; Kinget, R.; Verbeke, N. The lung as a route for systemic delivery of therapeutic proteins and peptides. Respir. Res. 2001, 2, 1–12. [Google Scholar]
- Portilla, S.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Encapsulation of the antistaphylococcal endolysin LysRODI in pH-sensitive liposomes. Antibiotics 2020, 9, 242. [Google Scholar] [CrossRef]
- Hathaway, H.; Ajuebor, J.; Stephens, L.; Coffey, A.; Potter, U.; Sutton, J.M.; Jenkins, A.T.A. Thermally triggered release of the bacteriophage endolysin CHAPK and the bacteriocin lysostaphin for the control of methicillin resistant Staphylococcus aureus (MRSA). J. Control. Release 2017, 245, 108–115. [Google Scholar] [CrossRef]
- Heselpoth, R.D.; Euler, C.W.; Schuch, R.; Fischetti, V.A. Lysocins: Bioengineered antimicrobials that deliver lysins across the outer membrane of Gram-negative bacteria. Antimicrob. Agents Chemother. 2019, 63, e00342-19. [Google Scholar] [CrossRef]
- Ozsoy, Y.; Gungor, S.; Cevher, E. Nasal delivery of high molecular weight drugs. Molecules 2009, 14, 3754–3779. [Google Scholar] [CrossRef]
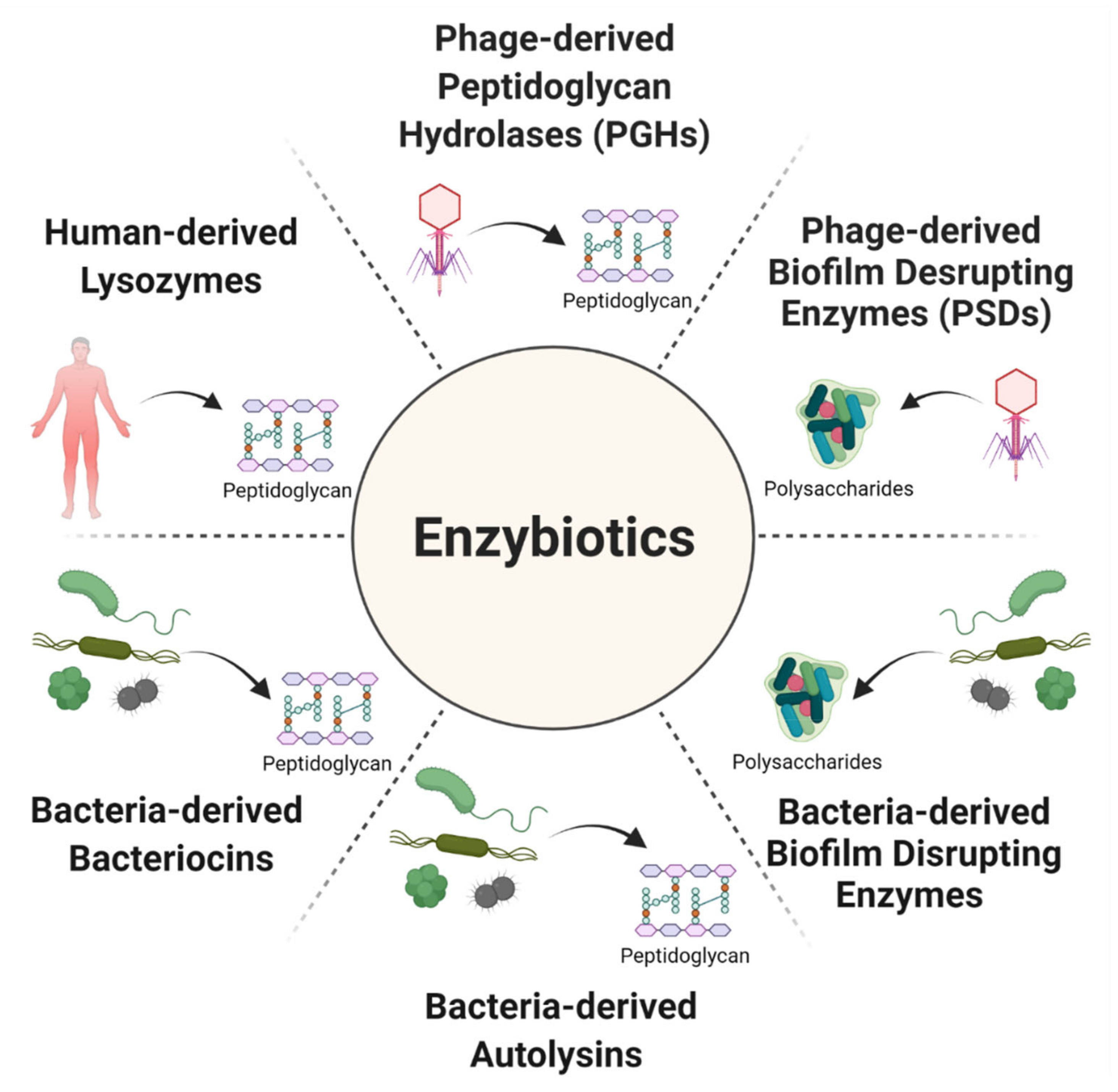

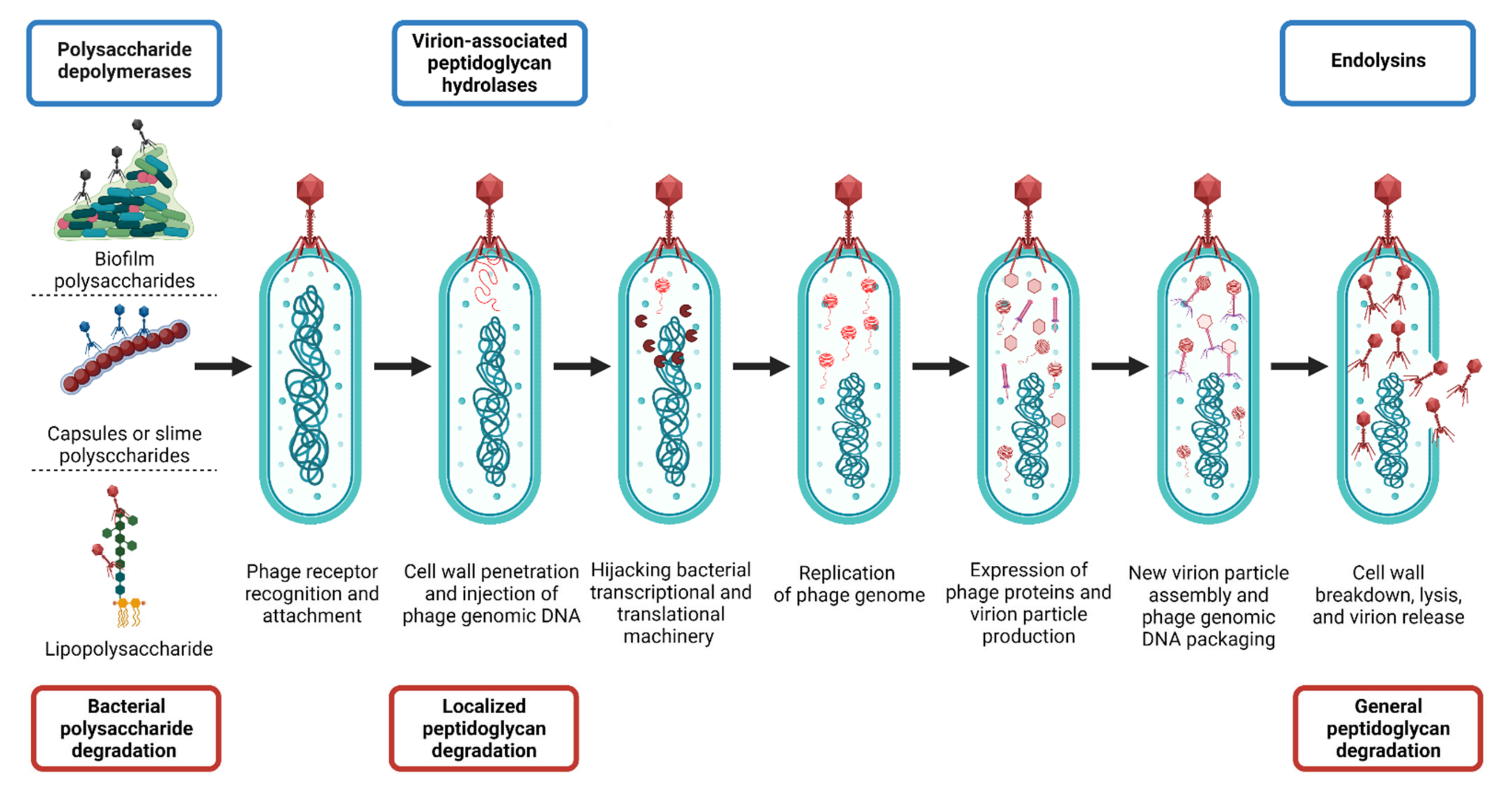
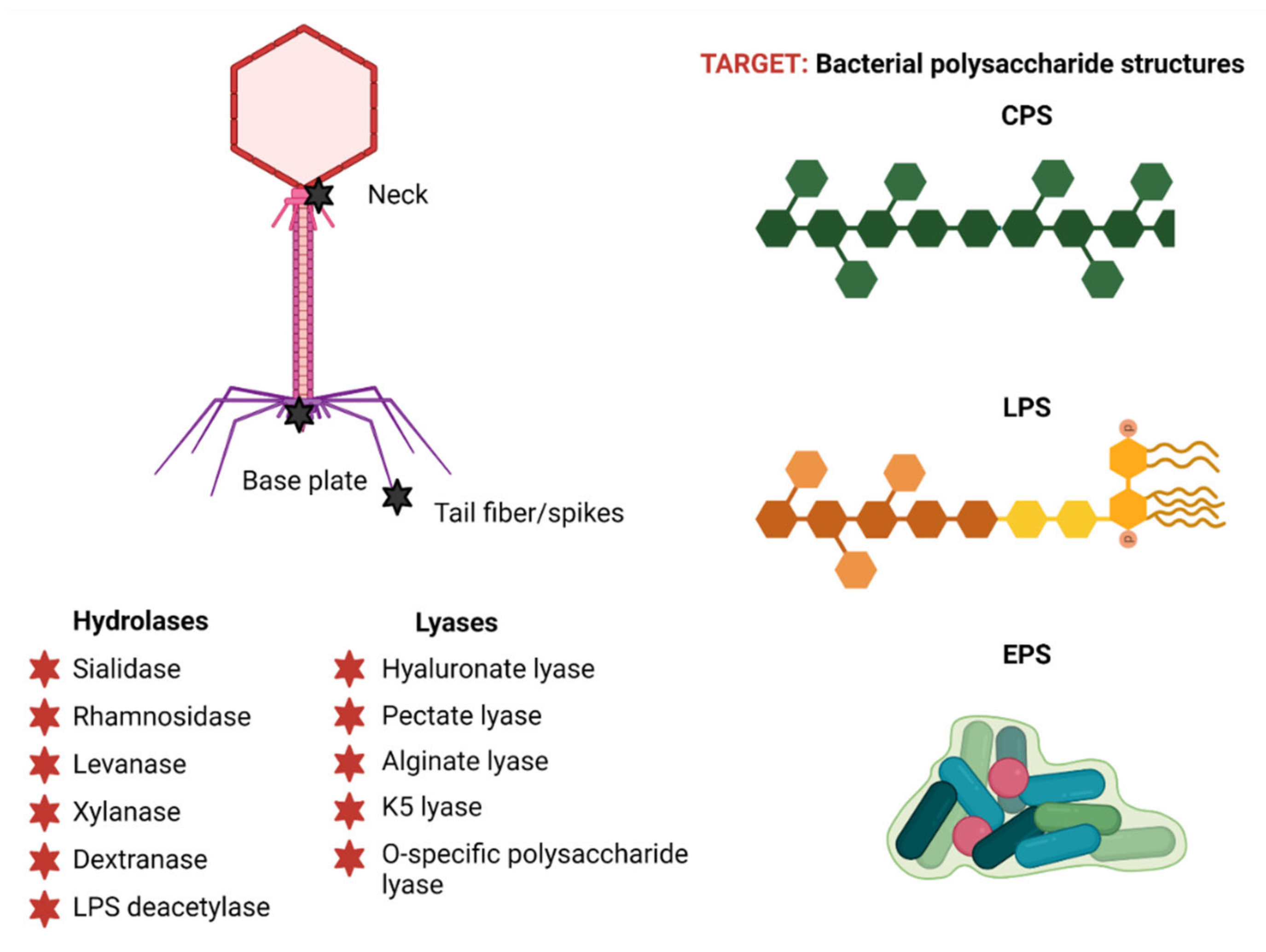

| Enzyme | Pathogen | Animal | Infection | Inoculum | Dosing | Results | Ref. |
|---|---|---|---|---|---|---|---|
| P22 tailspike protein (P22sTsp) recognizing LPS | Salmonella enterica serovar Typhimurium | Leghorn chicks | Intestinal colonization | Oral gavaging, 300 µL PBS containing 104 to 107 CFU | Oral gavaging, 300 µL in 10% BSA containing 30 mg; 3 doses: 1st 1 h post-infection, 2nd and 3rd dose given in 24 h intervals | 100-fold reduction of Salmonella colonization in the gut as well as reduced liver and spleen penetration; Salmonella motility was impaired | [150] |
| Dep-ORF8 targeting capsular serogroup A | Pasteurella multocida capsular serogroup A | BALB/c mouse model | Systemic infection | IP injection of 100 µL containing 80 CFU | 3 treatment groups: IP injection of 100 µL containing 36 µg at 6 h (group 1), 12 h post-infection (group 2), and 12 h post-infection, and then once daily for 5 days (group 3) | Treatment: group 1 showed ~70%, and 50% survival within 3 and 5 days, respectively; group 2 showed 70%, 50%, and ~35% survival within 3, 5, and 12 days, respectively; group 3 showed ~80%, 70% survival within 4 and 6 days, respectively; control group 100% mortality within 5 days | [151] |
| gp49, O-specific polysaccharide lyase | Pseudomonas aeruginosa | Galleria mellonella (Wax moth larvae) | Hemocoel infection | Injection into the last pro-leg of 10 CFU | Pretreatment: 1h incubation of bacteria with 50 µg/mL; treatment: 5 or 50 µg/mL was injected 15 min post-infection | Pretreatment: 24 h post injection, 50% of larvae survived (~30% more than in the control); 35% larvae also survived to the end of the experiment (>72 h); treatment: 24 h post-treatment, the larvae survival rate was at least 20% higher compared to the control, independent of gp49 concentration; 20% of larvae survived up to 72 h with treatment, while 100% of control group died 48 h post injection | [105] |
| depoKP36 targeting KP36 capsule | Klebsiella pneumoniae | G. mellonella (Wax moth larvae) | Hemocoel infection | Injection into the last pro-leg of 10 µL containing 107 CFU | Pretreatment: bacteria were pretreated with depoKP36 (280 µg/mL) for 2 h before infection; treatment: depoKP36 was administered 5 min post-infection | Pretreatment: 77% of larvae were saved within 24 h, and 47% and 43% after 48h and 72 h, respectively; treatment: survival increased up to 40%, 30%, and 20% after 24 h, 48 h, and 72 h post-treatment, respectively; 100% of untreated larvae died | [132] |
| Dp42 targeting capsular polysaccharide type KN1 | K. pneumoniae | BALB/c mouse model | Systemic infection | IP injection of 2 × 107 CFU | Prevention: IP injection of 200 µL containing 0.25 mg/mL 6 h before bacterial infection; pretreatment: 0.25 mg/mL for 30 min; treatment: IP injection of 200 mL containing ~50 mg 30 min post-infection | Prevention: 100% survival within 96 h post-infection, while 100% of control group died within 9 h; pretreatment: 1 mouse died (12.5%) 54 h post-infection, while 100% of control group died within 12 h; treatment: identical to pretreatment results. | [152] |
| K64dep targeting K64 capsular type polysaccharides | K. pneumoniae | BALB/cByl mouse model CP treated, 200 mg/kg IP injections in 2 days intervals | Systemic infection | IP injection of 6 × 106 CFU | IP injection with 150 μg, 37.5 μg, or 18.75 μg at 1 h, 8 h, and 24 h post-infection | 100% survival with 18.75 µg dose applied 1 h post-infection; in control group, 100% mortality was observed; 150 µg dose applied 8 h post-infection had no effect; no K64dep-related toxicity was observed as well as no changes in liver, kidney, and spleen histopathology; treatment sensitizes carbapenem-resistant K64 to serum killing in vitro as well as increased its susceptibility to neutrophil killing (~40% improved killing) | [153] |
| Endosialidase E (endoE) | Escherichia coli producing K1 antigen | Neonatal rats | Intestinal colonization and E. coli-related bacteremia | Oral administration of 20 μL containing 2 to 6 × 106 CFU | IP injection of 20 µg on days 1, 2, 3, 4, and 5 post-infection | No direct effect on E. coli viability but pathogen is sensitized to complement system killing; single dose on day 1 of endoE prevents the death of infected pups and E. coli invasion of the bloodstream; 80–100% survival in comparison to 0–10% survival in untreated control | [133] |
| Endosialidase E (endoE) | E. coli producing K1 antigen | Neonatal rats | Intestinal colonization and E. coli-related bacteremia | Oral administration of 20 μL containing 2 to 6 × 106 CFU | IP injection of 0.125–20 µg range on days 1 post-infection | Minimal dose of 0.25 µg prevented death of at least 80% of rats; treatment sensitizes E. coli to serum killing in vitro, and improved macrophage ingestion of E. coli | [134] |
| Dep6, O91-specific polysaccharide depolymerase | Shiga toxin-producing E. coli | BALB/c mouse thigh model | Systemic infection | Injection near the right thigh of 100 μL containing 2.4 × 108 CFU | Dose: 100 μL containing 0.3 μg/μL; toxicity: IP injection; prophylactic: delivery 3 h prior to infection; simultaneous treatment: delivery at the same time as bacterial inoculum; delayed treatment: delivery 3 h post-infection | Toxicity analysis: no pathological changes in liver, kidney, or small intestine observed; pretreatment: 100% survival; simultaneous treatment: 83% survival; delayed treatment: 33% survival; significant reduction in the levels of proinflammatory cytokines was observed at 24 h post-infection | [138] |
| Capsule depolymerases active against three different capsule types: K1, K5, and K30 | E. coli | NIH Swiss Mouse thigh model | Systemic infection | Injection into thigh of 100 µL containing 1 to 4 × 108 CFU | Injection of 100 µL PBS containing 0, 2, 5, or 20 µg doses, 30 min post-infection; different depolymerases tested | Toxicity: no toxicity observed; treatment: control group did not survive, whereas most mice were rescued by treatment with 20 µg dose per mouse; effective doses of K1F and K1H enzymes were between 2 µg (both partially rescuing) and 5 µg (both rescuing 100% mice) per mouse; for K5, the effective dose was between 2 and 20 µg per mouse; K30 gp41 rescued mice at the higher dose tested (20 µg per mouse); a mixture of K30 gp41 and K30 gp42 yielded the same survival outcome as K30 gp41 alone | [122] |
| ϕAB6 targeting capsular polysaccharide | Acinetobacter baumannii | Zebrafish | Systemic infection | Injection through cloaca of 1 to 4 × 107 CFU | Injection through cloaca of 20 μL protein (1 μg/μL), 30 min post-infection | Treatment: survival rate was significantly improved (80%) compared with untreated control (10%); toxicity: none observed | [154] |
| Dpo48 capsule depolymerase | A. baumannii | G. mellonella (Wax moth larvae) | Hemocoel infection | Injected into the last pro-leg of 10 µL PBS containing 106 CFU | Pretreatment: 50 µg/mL for 1 h; treatment: Injection of 10 µL PBS containing 5 µg 5 min post-infection | Pretreatment: 100% survival, while, in control group, ~65% and 84% of larvae died within 24 h and 72 h, respectively; treatment: 76% survival, while, in control group, ~65% and 84% of larvae died within 24 h and 72 h, respectively | [155] |
| Dpo48 capsule depolymerase | A. baumannii | BALB/c mice model | Systemic infection | IP injection of 107 CFU | IP injection of 200 µL PBS containing 50 µg 2 h post-infection | 100% mice treated survived and appeared healthy for 7 days, while 100% of the untreated control died within 24 h due to peritoneal sepsis; bacterial count in tissue and organs was significantly reduced with treatment 6 h post-infection in comparison to control group | |
| BALB/c mice model, IP injection of CP (300 mg/kg) in 200 µL PBS, 3 days before infection | Systemic infection | IP injection of 107 CFU | IP injection of 200 µL of PBS containing 50 µg 2 h post- infection | 100% of mice treated survived and appeared healthy for 7 days, while 100% of untreated control died within 24 h due to peritoneal sepsis | |||
| K2 capsular depolymerase | A. baumannii capsular type K2 | G. mellonella (Wax moth larvae) | Hemocoel infection | Injection into the last pro-leg of 5.5 µL of 20 mM HEPES containing 106 CFU | Pretreatment: bacteria pretreated with protein for 2 h; treatment: injection of enzyme 30 min post-infection; in both scenarios, a range of protein dosages were used (0.25 g, 0.5 g, and 3 g/larvae) | No toxicity, 100% survival of larvae; pretreatment: untreated control group survival rate was 25%, 20%, and 10% after 24, 48, and 72 h, respectively; in group with pretreatment after 72 h, 53%, 69%, and 88% of larvae survived using 0.25 g, 0.5 g, and 3 g pretreatments; treatment: only 35%, 22%, and <15% larvae survived in untreated control after 24 h, 48 h, 72 h, respectively, while 73%, 40–76%, 56–70% survived with treatment; K2 depolymerase is highly refractory to resistance development | [156] |
| BALB/c mouse model, IP injection of CP (100 mg/kg), 4 and 1 day before infection | Systemic infection | IP injection of 107 CFU | IP injection with 50 µg dose 1 h post-infection | 20 h post-infection control group had to be euthanized, while in a treatment group 90% mice had survived, decreasing to 60% at 42 h post-infection |
| Descriptor | Company | Type | Route | Phase | # | Start | Status | Registry # | Protocol and Observations | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| P128 (StaphTAME) | GangaGen | Ectolysin | IN | I/II | 74 | 2012 | Completed | NCT01746654 | Type: randomized, double-blind, placebo-controlled study; goal: (1) evaluation of safety, tolerability via single or multiple doses (3 doses/day for 5 days) of 0.1 mg, 0.3 mg, and 1mg concentrations of P128, administrated intranasally to healthy individuals; (2) evaluation of safety, tolerability, and efficacy of P128 in chronic kidney disease patients or any patients who are nasal carriers of S. aureus or MRSA strain with single dose or 3 escalating concentrations of P128; Initial results: drug was well tolerated; results: reduction of nasal carriage | NA |
| N-Rephasin® SAL200 (SAL-1, tonabacase) | iNtRON Biotechnology | Endolysin | IV | I | 36 | 2013 | Completed | NCT01855048 | Type: randomized, double-blind, placebo-controlled study; goal: evaluation of safety, pharmacokinetics, and pharmacodynamics of single intravenous dose of SAL-1 at various concentrations: 0.1 mg/kg, 0.3 mg/kg, 1 mg/kg, 3 mg/kg, 10 mg/kg, administrated to healthy male individuals; results: no severe side effects observed | [157] |
| II | 25 | 2017 | Terminated 1 | NCT03089697 | Type: randomized, double-blind, placebo-controlled study; goal: evaluation of safety and efficacy of SAL-1 (3mg/kg), administrated once a day intravenously to individuals with persistent S. aureus bacteremia; results: serious adverse effects occurred (2/12 patients, 16.67% of test group), including pneumonia (one patient, 8.33% of test group) and respiratory failure (one patient, 8.33% of test group), as well as several other minor adverse events (10/12 patients, 83.33% of the test group), e.g., anemia, chills, back pain, headache, gastrointestinal disorders | NA | ||||
| Lysin CF-301 (PlySs2, exebacase) | ContraFect | Endolysin | IV | I | 20 | 2015 | Completed | NCT02439359 | Type: placebo-controlled, dose-escalating study; goal: evaluation of safety and tolerability of single intravenous dose of CF-301; healthy male and female individuals; results: CF-301 has a safe profile with no side effects observed | [266,267,268,269,270] |
| II | 121 | 2017 | Completed | NCT03163446 | Type: multicenter, randomized, double-blind, placebo-controlled study; goal: evaluation of safety, tolerability, efficacy, and pharmacokinetics of CF-301; study performed in addition to standard-of-care antibacterial therapy; adult individuals with bloodstream infections (bacteremia), including endocarditis | [325] | ||||
| III | 348 | 2019 | Ongoing | NCT04160468 | Type: randomized, double-blind, placebo-controlled study; goal: evaluation of the efficacy and safety of a single dose of Exebacase in addition to standard-of-care antibacterial therapy; adult individuals with bloodstream infections (bacteremia), including endocarditis | NA | ||||
| Staphefekt SA.100 | Micreos | Endolysin | T | I/II 1 | 100 | 2016 | Completed | NCT02840955 | Goal: evaluation on disease severity and skin microbiome; individuals with atopic dermatitis; results: no side effects observed, decrease in bacterial burden | [189] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics 2021, 10, 1497. https://doi.org/10.3390/antibiotics10121497
Danis-Wlodarczyk KM, Wozniak DJ, Abedon ST. Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics. 2021; 10(12):1497. https://doi.org/10.3390/antibiotics10121497
Chicago/Turabian StyleDanis-Wlodarczyk, Katarzyna M., Daniel J. Wozniak, and Stephen T. Abedon. 2021. "Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application" Antibiotics 10, no. 12: 1497. https://doi.org/10.3390/antibiotics10121497
APA StyleDanis-Wlodarczyk, K. M., Wozniak, D. J., & Abedon, S. T. (2021). Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics, 10(12), 1497. https://doi.org/10.3390/antibiotics10121497








