Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland)
Abstract
1. Introduction
2. Results
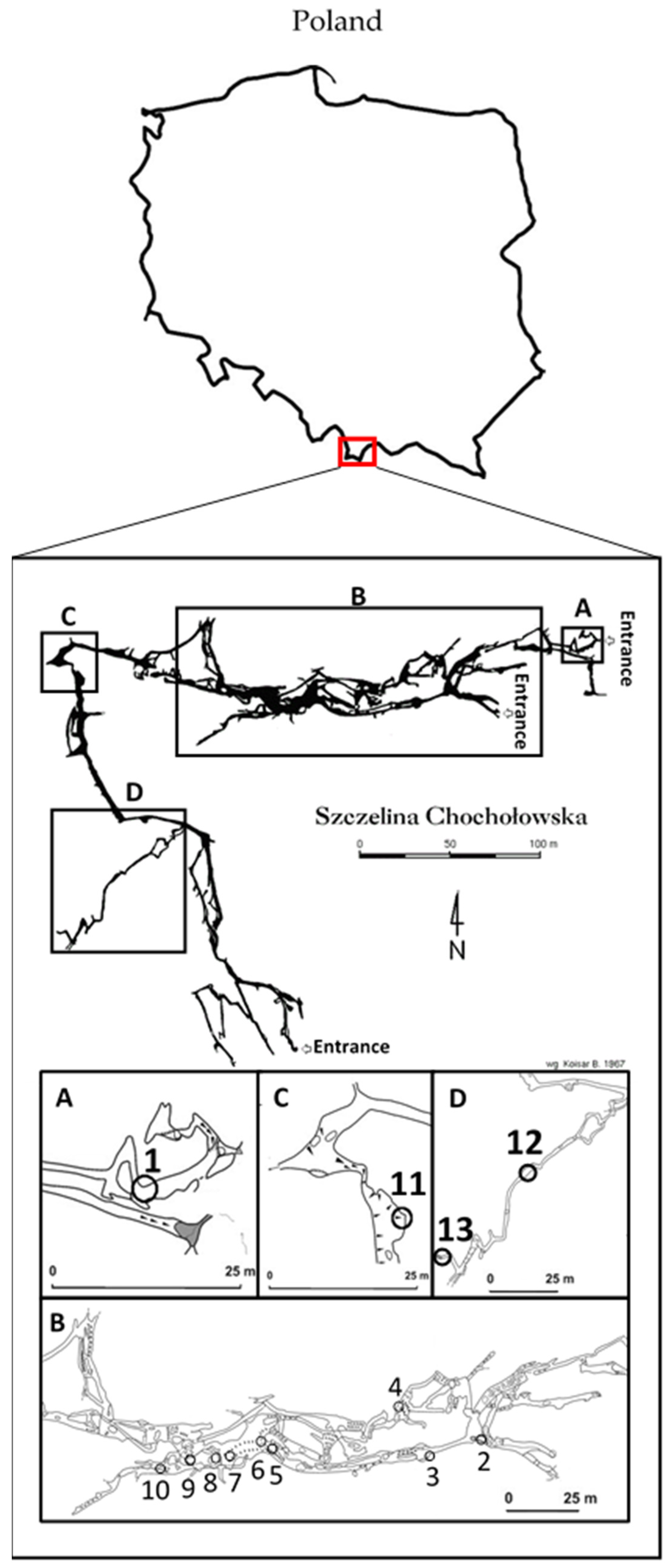
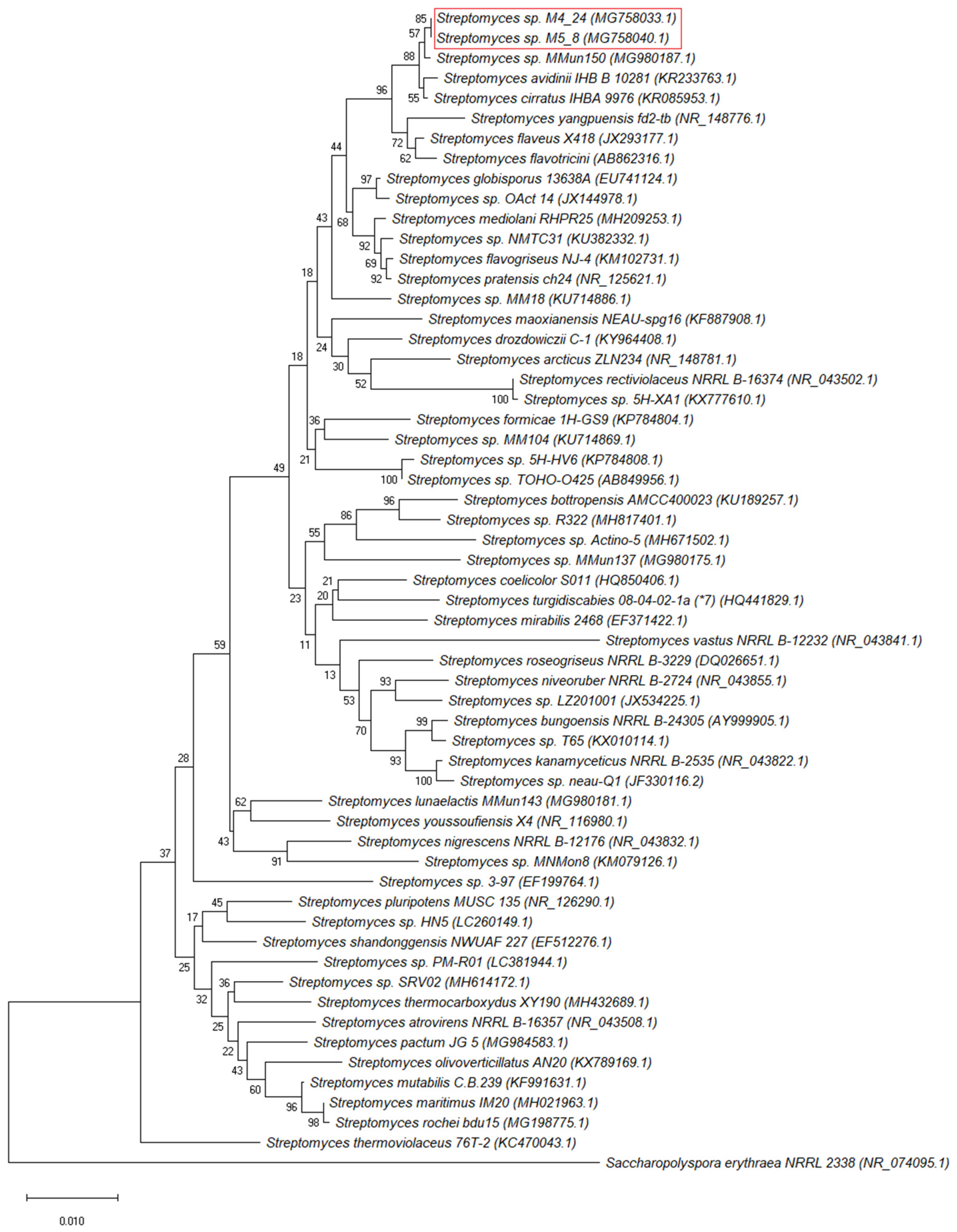
2.1. Isolation and Identification of Bacteria from the Szczelina Chochołowska Cave
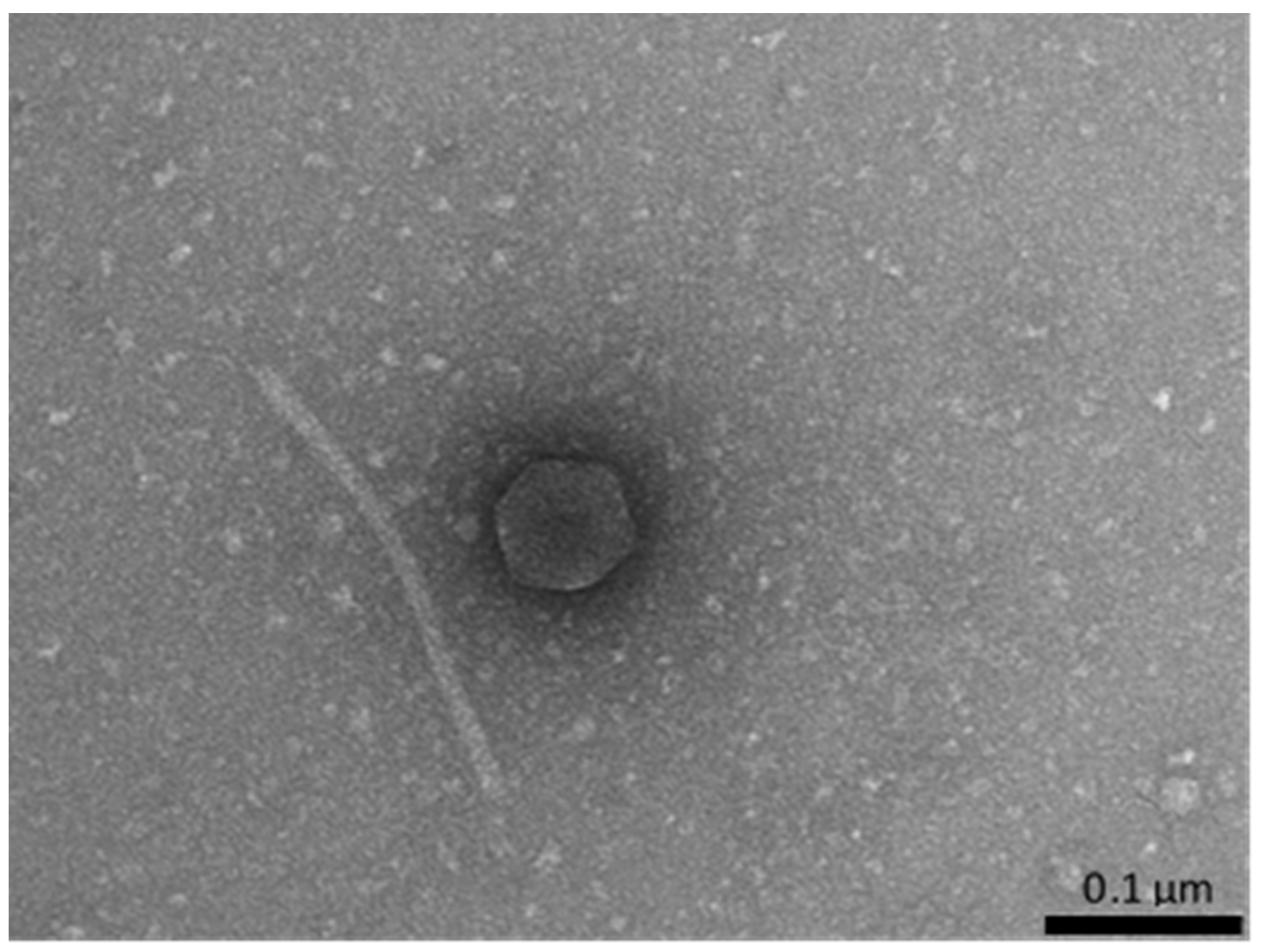
2.2. Prophage Induction and Determination of the Presence of Phage Virions
2.3. Preliminary Screening for Production of Antibacterial Compounds by the Isolates and Selection of Strains for Further Investigation
2.4. Antibacterial and Antifungal Activities of Streptomyces M4_24 and M5_8 Strains
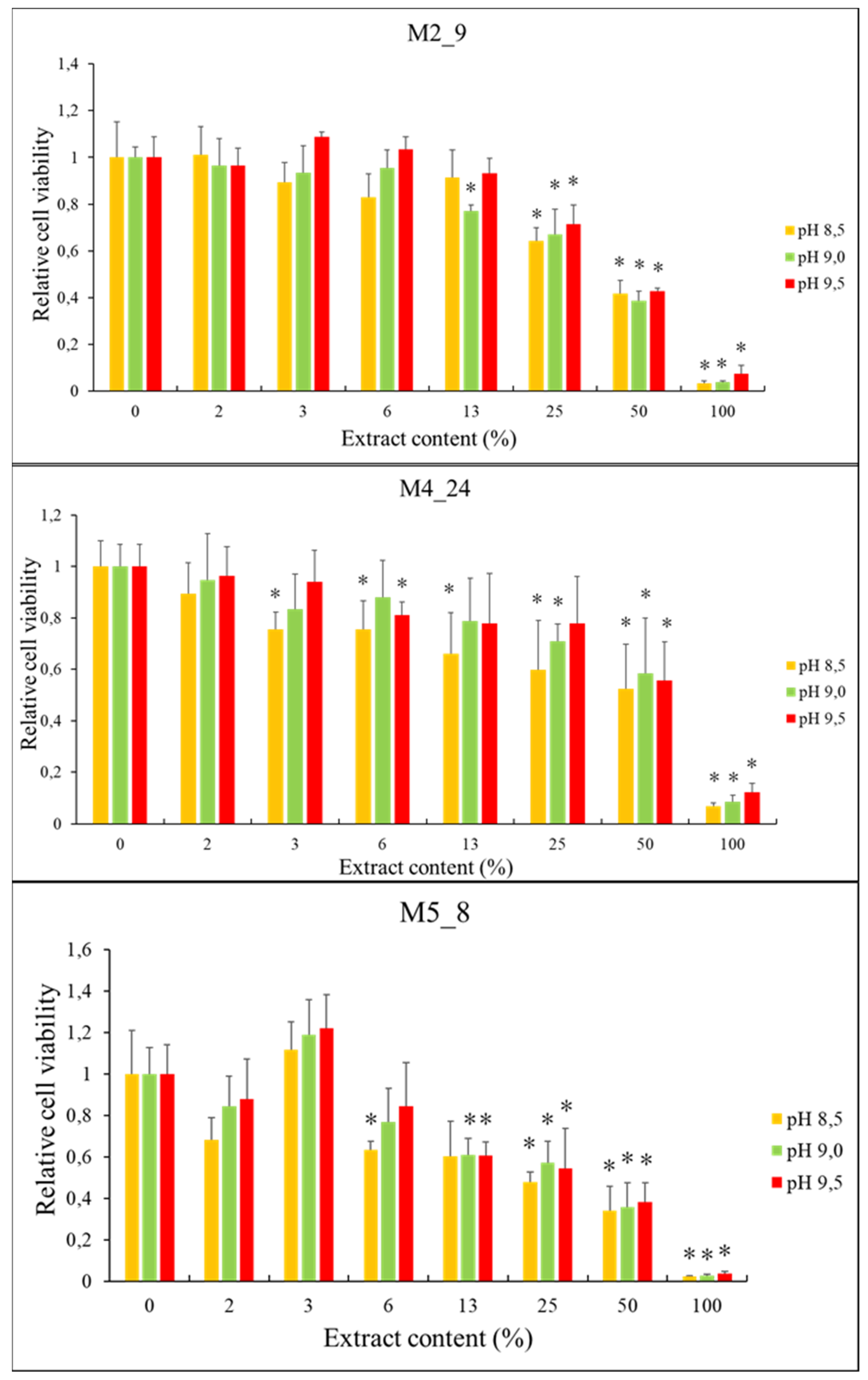
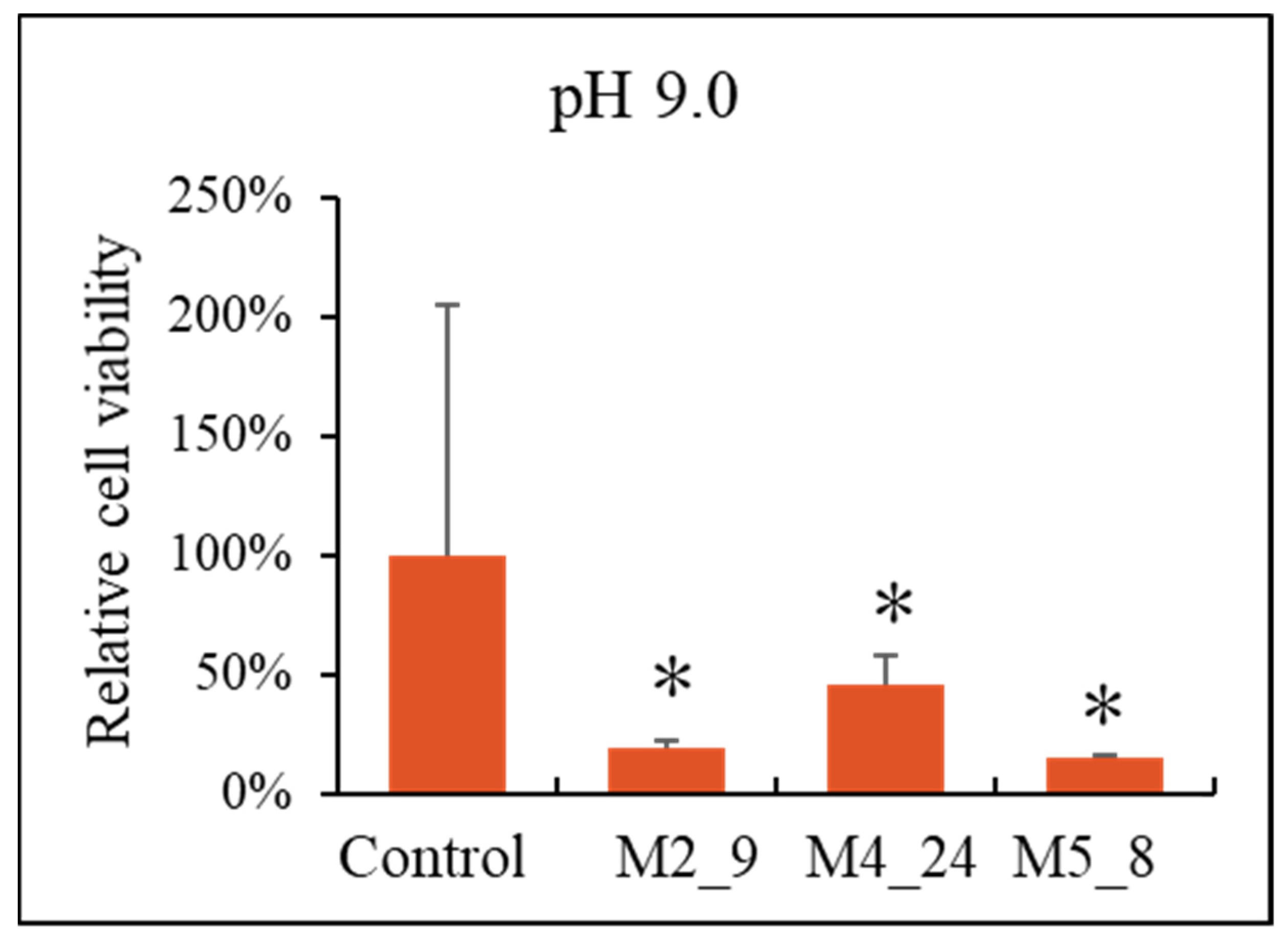
2.5. Anticancer Activities of Streptomyces M2_9, M4_24 and M5_8 Isolates
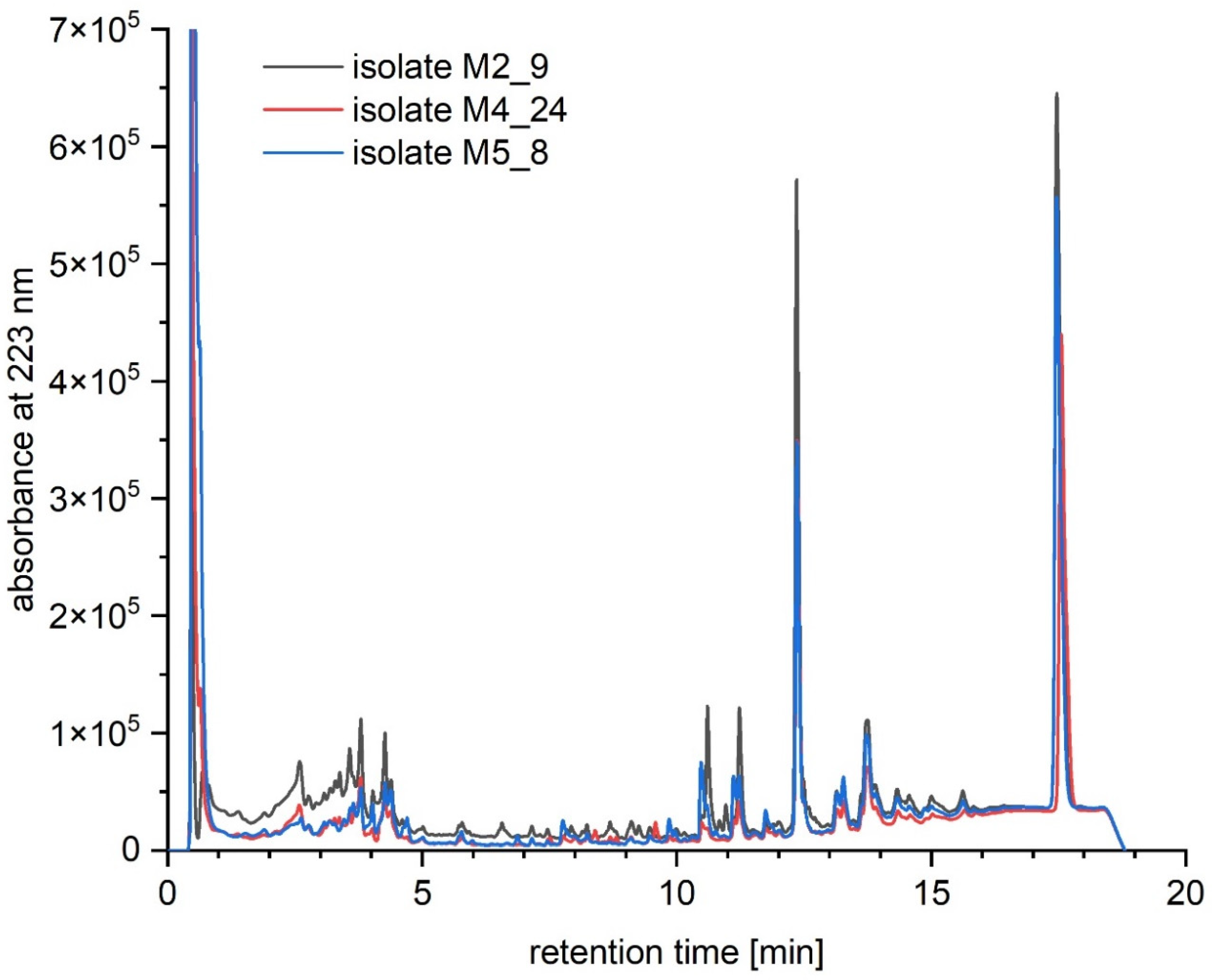
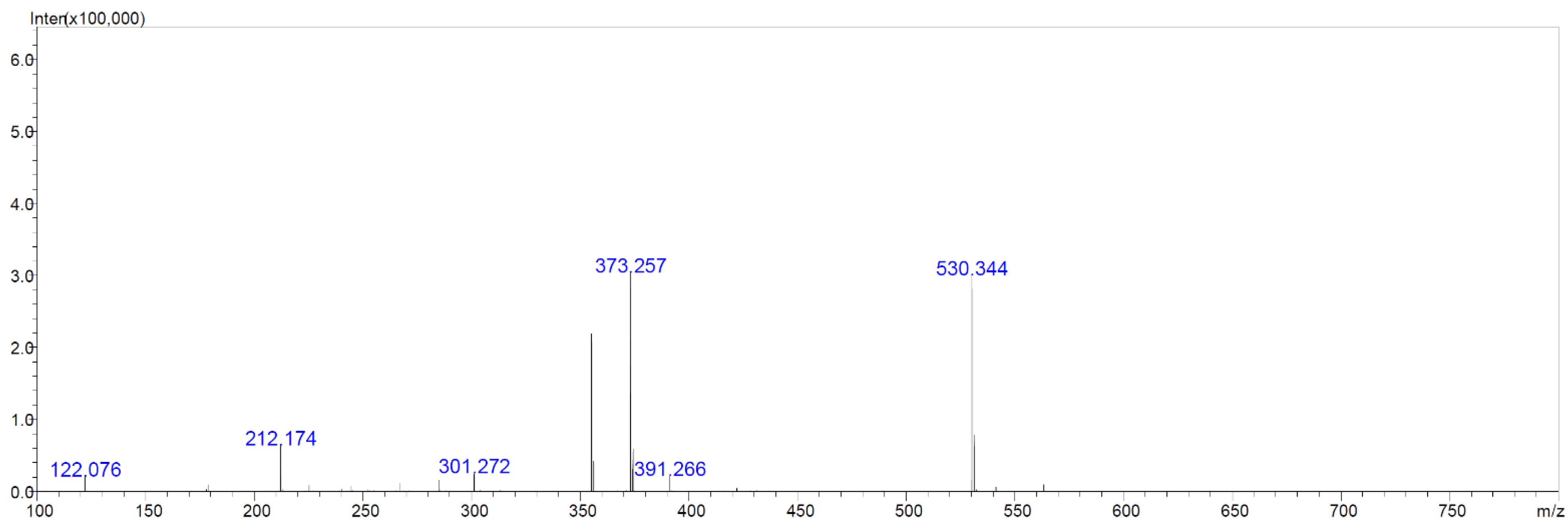
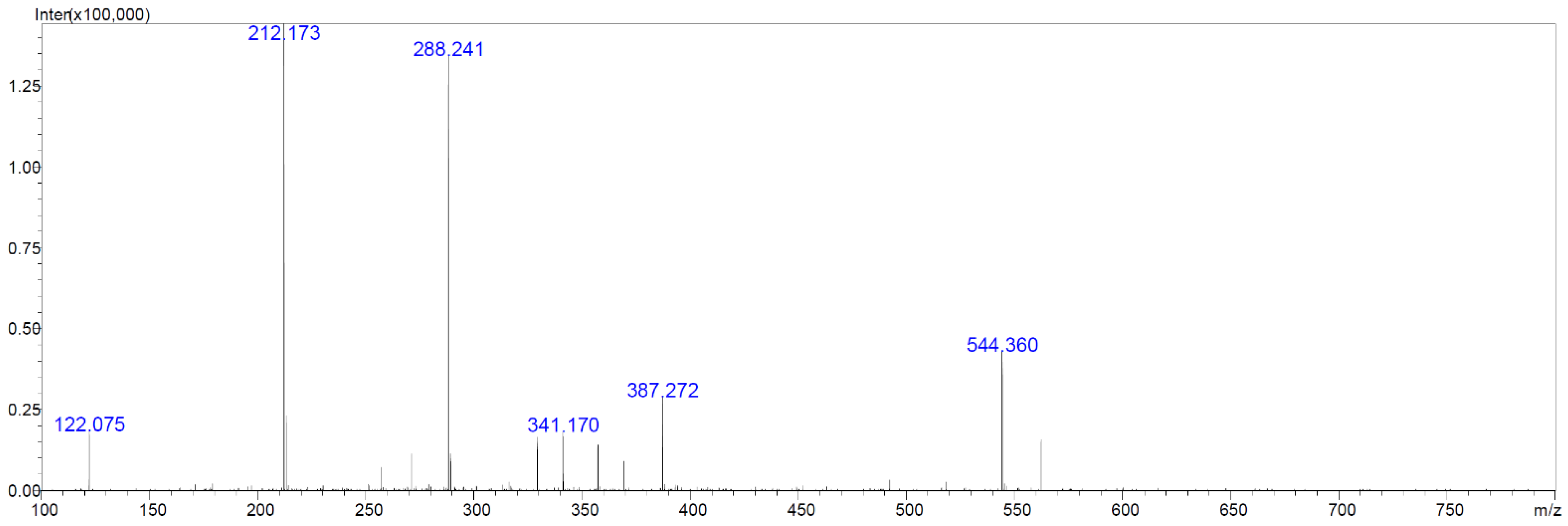
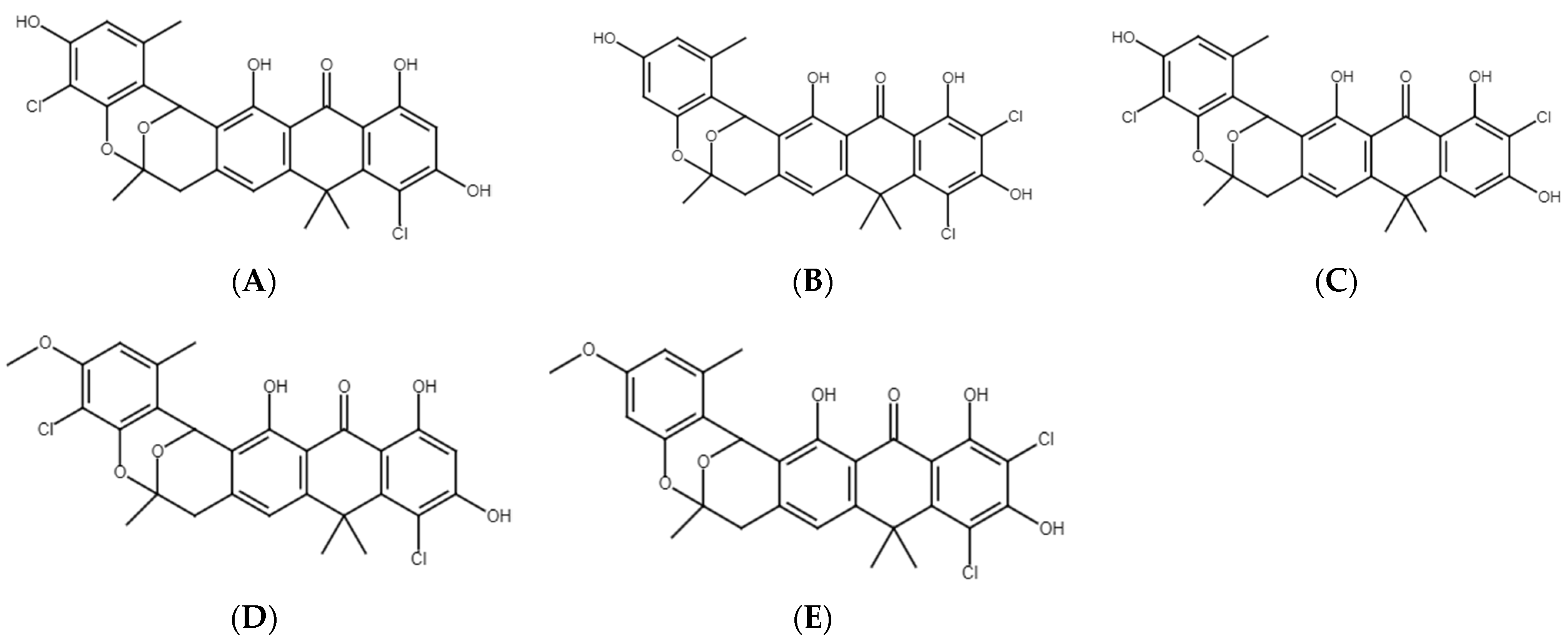
2.6. Chemical Analyses of Extracts from Cultures of Streptomyces M2_9, M4_24 and M5_8 Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Cave Description and Sampling
4.3. Bacterial Growth Conditions
4.4. Prophage Induction and Electron Microscopy of Bacteriophage Virions
4.5. Streak-Test
4.6. Molecular Identification of Isolates and Phylogenetic Analyses
4.7. Preparation of Extracts from Cultures of Isolates
4.8. Cancer and Non-Transformed Cell Lines and Cell Cultures
4.9. Estimation of Cells’ Viability
4.10. Chemical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morehead, M.S.; Scarbrough, C. Emergence of Global Antibiotic Resistance. Prim. Care 2018, 45, 467–484. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Bragg, R.R.; Meyburgh, C.M.; Lee, J.-Y.; Coetzee, M. Potential Treatment Options in a Post-Antibiotic Era. Adv. Exp. Med. Biol. 2018, 1052, 51–61. [Google Scholar] [CrossRef]
- Cyske, Z.; Jaroszewicz, W.; Żabińska, M.; Lorenc, P.; Sochocka, M.; Bielańska, P.; Grabowski, Ł.; Gaffke, L.; Pierzynowska, K.; Węgrzyn, G. Unexplored Potential: Biologically Active Compounds Produced by Microorganisms from Hard-to-Reach Environments and Their Applications. Acta Biochim. Pol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Rangseekaew, P.; Pathom-aree, W. Cave Actinobacteria as Producers of Bioactive Metabolites. Front. Microbiol. 2019, 10, 387. [Google Scholar] [CrossRef]
- Yücel, S.; Yamaç, M. Selection of Streptomyces Isolates from Turkish Karstic Caves against Antibiotic Resistant Microorganisms. Pak. J. Pharm. Sci. 2010, 23, 1–6. [Google Scholar] [PubMed]
- Jiang, Z.; Guo, L.; Chen, C.; Liu, S.; Zhang, L.; Dai, S.; He, Q.; You, X.; Hu, X.; Tuo, L.; et al. Xiakemycin A, a Novel Pyranonaphthoquinone Antibiotic, Produced by the Streptomyces Sp. CC8-201 from the Soil of a Karst Cave. J. Antibiot. 2015, 68, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Klusaite, A.; Vickackaite, V.; Vaitkeviciene, B.; Karnickaite, R.; Bukelskis, D.; Kieraite-Aleksandrova, I.; Kuisiene, N. Characterization of Antimicrobial Activity of Culturable Bacteria Isolated from Krubera-Voronja Cave. Int. J. Speleol. 2016, 45, 275–287. [Google Scholar] [CrossRef]
- Adam, D.; Maciejewska, M.; Naômé, A.; Martinet, L.; Coppieters, W.; Karim, L.; Baurain, D.; Rigali, S. Isolation, Characterization, and Antibacterial Activity of Hard-to-Culture Actinobacteria from Cave Moonmilk Deposits. Antibiotics 2018, 7, 28. [Google Scholar] [CrossRef]
- Doull, J.L.; Singh, A.K.; Hoare, M.; Ayer, S.W. Conditions for the Production of Jadomycin B by Streptomyces Venezuelae ISP5230: Effects of Heat Shock, Ethanol Treatment and Phage Infection. J. Ind. Microbiol. 1994, 13, 120–125. [Google Scholar] [CrossRef]
- Lu, N.; Sun, Y.; Wang, Q.; Qiu, Y.; Chen, Z.; Wen, Y.; Wang, S.; Song, Y. Cloning and Characterization of Endolysin and Holin from Streptomyces Avermitilis Bacteriophage PhiSASD1 as Potential Novel Antibiotic Candidates. Int. J. Biol. Macromol. 2020, 147, 980–989. [Google Scholar] [CrossRef]
- Herath, K.B.; Jayasuriya, H.; Guan, Z.; Schulman, M.; Ruby, C.; Sharma, N.; MacNaul, K.; Menke, J.G.; Kodali, S.; Galgoci, A.; et al. Anthrabenzoxocinones from Streptomyces Sp. as Liver X Receptor Ligands and Antibacterial Agents. J. Nat. Prod. 2005, 68, 1437–1440. [Google Scholar] [CrossRef]
- Herold, K.; Gollmick, F.A.; Groth, I.; Roth, M.; Menzel, K.-D.; Möllmann, U.; Gräfe, U.; Hertweck, C. Cervimycin A-D: A Polyketide Glycoside Complex from a Cave Bacterium Can Defeat Vancomycin Resistance. Chemistry 2005, 11, 5523–5530. [Google Scholar] [CrossRef]
- Nakaew, N.; Pathom-aree, W.; Lumyong, S. First Record of the Isolation, Identification and Biological Activity of a New Strain of Spirillospora Albida from Thai Cave Soil. Actinomycetologica 2009, 23, 1–7. [Google Scholar] [CrossRef]
- Nakaew, N.; Pathom-aree, W.; Lumyong, S. Generic Diversity of Rare Actinomycetes from Thai Cave Soils and Their Possible Use as New Bioactive Compounds. Actinomycetologica 2009, 23, 21–26. [Google Scholar] [CrossRef]
- Rajput, Y.; Biswas, J.; Rai, V. Potentiality Test in Antimicrobial Activity and Antibiotic Sensitivity of Subterranean Streptomyces Strains Isolated from Kotumsar Cave of India. Int. J. Biol. Chem. 2012, 6, 53–60. [Google Scholar] [CrossRef][Green Version]
- Stankovic, N.; Radulovic, V.; Petkovic, M.; Vuckovic, I.; Jadranin, M.; Vasiljevic, B.; Nikodinovic-Runic, J. Streptomyces Sp. JS520 Produces Exceptionally High Quantities of Undecylprodigiosin with Antibacterial, Antioxidative, and UV-Protective Properties. Appl. Microbiol. Biotechnol. 2012, 96, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Tomova, I.; Lazarkevich, I.; Tomova, A.; Kambourova, M.; Vasileva-Tonkova, E. Diversity and Biosynthetic Potential of Culturable Aerobic Heterotrophic Bacteria Isolated from Magura Cave, Bulgaria. IJS 2013, 42, 65–76. [Google Scholar] [CrossRef]
- Cheeptham, N.; Sadoway, T.; Rule, D.; Watson, K.; Moote, P.; Soliman, L.; Azad, N.; Donkor, K.; Horne, D. Cure from the Cave: Volcanic Cave Actinomycetes and Their Potential in Drug Discovery. Int. J. Speleol. 2013, 42, 35–47. [Google Scholar] [CrossRef]
- Riquelme, C.; Enes Dapkevicius, M.d.L.; Miller, A.Z.; Charlop-Powers, Z.; Brady, S.; Mason, C.; Cheeptham, N. Biotechnological Potential of Actinobacteria from Canadian and Azorean Volcanic Caves. Appl. Microbiol. Biotechnol. 2017, 101, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, M.; Adam, D.; Martinet, L.; Naômé, A.; Całusińska, M.; Delfosse, P.; Carnol, M.; Barton, H.A.; Hayette, M.-P.; Smargiasso, N.; et al. A Phenotypic and Genotypic Analysis of the Antimicrobial Potential of Cultivable Streptomyces Isolated from Cave Moonmilk Deposits. Front. Microbiol. 2016, 7, 1455. [Google Scholar] [CrossRef]
- Belyagoubi, L.; Belyagoubi-Benhammou, N.; Jurado, V.; Dupont, J.; Lacoste, S.; Djebbah, F.; Ounadjela, F.; Benaissa, S.; Habi, S.; Abdelouahid, D.; et al. Antimicrobial Activities of Culturable Microorganisms (Actinomycetes and Fungi) Isolated from Chaabe Cave, Algeria. IJS 2018, 47, 189–199. [Google Scholar] [CrossRef]
- Ambrožič Avguštin, J.; Petrič, P.; Pašić, L. Screening the Cultivable Cave Microbial Mats for the Production of Antimicrobial Compounds and Antibiotic Resistance. IJS 2019, 48, 295–303. [Google Scholar] [CrossRef]
- Paun, V.I.; Lavin, P.; Chifiriuc, M.C.; Purcarea, C. First Report on Antibiotic Resistance and Antimicrobial Activity of Bacterial Isolates from 13,000-Year Old Cave Ice Core. Sci. Rep. 2021, 11, 514. [Google Scholar] [CrossRef]
- Nakaew, N.; Sungthong, R.; Yokota, A.; Lumyong, S. Nonomuraea Monospora Sp. Nov., an Actinomycete Isolated from Cave Soil in Thailand, and Emended Description of the Genus Nonomuraea. Int. J. Syst. Evol. Microbiol. 2012, 62, 3007–3012. [Google Scholar] [CrossRef] [PubMed]
- Derewacz, D.K.; McNees, C.R.; Scalmani, G.; Covington, C.L.; Shanmugam, G.; Marnett, L.J.; Polavarapu, P.L.; Bachmann, B.O. Structure and Stereochemical Determination of Hypogeamicins from a Cave-Derived Actinomycete. J. Nat. Prod. 2014, 77, 1759–1763. [Google Scholar] [CrossRef]
- Jiang, L.; Pu, H.; Xiang, J.; Su, M.; Yan, X.; Yang, D.; Zhu, X.; Shen, B.; Duan, Y.; Huang, Y. Huanglongmycin A-C, Cytotoxic Polyketides Biosynthesized by a Putative Type II Polyketide Synthase From Streptomyces Sp. CB09001. Front. Chem. 2018, 6, 254. [Google Scholar] [CrossRef]
- Kojiri, K.; Nakajima, S.; Fuse, A.; Suzuki, H.; Suda, H. BE-24566B, a New Antibiotic Produced by Streptomyces Violaceusniger. J. Antibiot. 1995, 48, 1506–1508. [Google Scholar] [CrossRef]
- Lü, Y.; Yue, C.; Shao, M.; Qian, S.; Liu, N.; Bao, Y.; Wang, M.; Liu, M.; Li, X.; Wang, Y.; et al. Molecular Genetic Characterization of an Anthrabenzoxocinones Gene Cluster in Streptomyces Sp. FJS31-2 for the Biosynthesis of BE-24566B and Zunyimycin Ale. Molecules 2016, 21, 711. [Google Scholar] [CrossRef] [PubMed]
- Lü, Y.; Shao, M.; Wang, Y.; Qian, S.; Wang, M.; Wang, Y.; Li, X.; Bao, Y.; Deng, C.; Yue, C.; et al. Zunyimycins B and C, New Chloroanthrabenzoxocinones Antibiotics against Methicillin-Resistant Staphylococcus Aureus and Enterococci from Streptomyces Sp. FJS31-2. Molecules 2017, 22, 251. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Yan, X.; Zhang, H.; Yu, M.; Shen, G.; Zhou, L.; Deng, Z.; Lei, C.; Qu, X. Expanding the Bioactive Chemical Space of Anthrabenzoxocinones through Engineering the Highly Promiscuous Biosynthetic Modification Steps. ACS Chem. Biol. 2018, 13, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lew, C.I.; Neuhaus, G.F.; Adpressa, D.A.; Zakharov, L.N.; Kaweesa, E.N.; Plitzko, B.; Loesgen, S. Biodiversity, Bioactivity, and Metabolites of High Desert Derived Oregonian Soil Bacteria. Chem. Biodivers. 2021, 18, e2100046. [Google Scholar] [CrossRef] [PubMed]
- Morshed, M.T.; Lacey, E.; Vuong, D.; Lacey, A.E.; Lean, S.S.; Moggach, S.A.; Karuso, P.; Chooi, Y.-H.; Booth, T.J.; Piggott, A.M. Chlorinated Metabolites from Streptomyces Sp. Highlight the Role of Biosynthetic Mosaics and Superclusters in the Evolution of Chemical Diversity. Org. Biomol. Chem. 2021, 19, 6147–6159. [Google Scholar] [CrossRef]
- Łubowska, N.; Grygorcewicz, B.; Kosznik-Kwaśnicka, K.; Zauszkiewicz-Pawlak, A.; Węgrzyn, A.; Dołęgowska, B.; Piechowicz, L. Characterization of the Three New Kayviruses and Their Lytic Activity Against Multidrug-Resistant Staphylococcus Aureus. Microorganisms 2019, 7, 471. [Google Scholar] [CrossRef]
- Van der Linde, K.; Lim, B.T.; Rondeel, J.M.M.; Antonissen, L.P.M.T.; de Jong, G.M.T. Improved Bacteriological Surveillance of Haemodialysis Fluids: A Comparison between Tryptic Soy Agar and Reasoner’s 2A Media. Nephrol. Dial. Transplant. 1999, 14, 2433–2437. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory: New York, NY, USA, 2001. [Google Scholar]
- Kosznik-Kwaśnicka, K.; Ciemińska, K.; Grabski, M.; Grabowski, Ł.; Górniak, M.; Jurczak-Kurek, A.; Węgrzyn, G.; Węgrzyn, A. Characteristics of a Series of Three Bacteriophages Infecting Salmonella Enterica Strains. Int. J. Mol. Sci. 2020, 21, 6152. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. ISBN 978-1-4832-3211-9. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Kim, T.; Yoo, K.H.; Kang, K. The T47D Cell Line Is an Ideal Experimental Model to Elucidate the Progesterone-Specific Effects of a Luminal A Subtype of Breast Cancer. Biochem. Biophys. Res. Commun. 2017, 486, 752–758. [Google Scholar] [CrossRef]
- Pierzynowska, K.; Gaffke, L.; Hać, A.; Mantej, J.; Niedziałek, N.; Brokowska, J.; Węgrzyn, G. Correction of Huntington’s Disease Phenotype by Genistein-Induced Autophagy in the Cellular Model. Neuro Mol. Med. 2018, 20, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
| Isolate 1 | Strongest 16S rDNA Sequence Match (BLASTN) | |||
|---|---|---|---|---|
| Organism | Accession No. 2 | Bits | % | |
| JHARAB1_N | Arthrobacter sp. strain VTT E-052904 | EF093123 | 2691 | 99.9 |
| JHARN2 | Rhodococcus sp. strain UFZ-B528 | AF235012 | 2667 | 99.9 |
| JSZCO2 | Microbacterium sp. strain JSZCO2 | KU643207 | 2705 | 100 |
| JSZCZL7 | Nocardia sp. strain JSZCL7 | KU643201 | 2219 | 99.9 |
| M1_4 | Nocardia sp. strain OAct 132 | JX047071 | 2671 | 99.9 |
| M1_7 | Arthrobacter sp. strain 3S-5 | KM434250 | 2670 | 99.9 |
| M1_9 | Tomitella biformata strain AHU 1821 | NR_112905 | 2575 | 98.9 |
| M2_1 | Arthrobacter sp. (uncultured clone) | KJ650689 | 2671 | 100 |
| M2_11 | Frigoribacterium sp. strain FB3 | AM933497 | 2657 | 100 |
| M2_15 | Rhodococcus jialingiae strain djl-6-2 16S | NR_115708 | 2675 | 99.9 |
| M2_4 | Arthrobacter sp. strainRKS6-4 | GQ477171 | 2670 | 99.9 |
| M2_9 | Streptomyces sp. strain MM56 | KU714908 | 2684 | 100 |
| M3_10 | Streptomyces sp. strain MM56 | KU714908 | 2679 | 99.9 |
| M3_8 | Arthrobacter sp. strain 3S-5 | KM434250 | 2647 | 99.7 |
| M3_9 | Arthrobacter sp. strain MNPB6 | FM213396 | 2555 | 98.3 |
| M4_18 | Rhodococcus maanshanensis strain GMC121 | AB741451 | 2465 | 97.6 |
| M4_21 | Arthrobacter sp. strain EM0174 | HM165266 | 2559 | 98.5 |
| M4_24 | Streptomyces sp. strain MM56 | KU714908 | 2679 | 99.9 |
| M4_9 | Nocardiopsis umidischolae strain NBRC 100349 | NR_112746 | 2690 | 100 |
| M5_2 | Nocardia sp. strain OAct 132 | JX047071 | 2644 | 99.6 |
| M5_6 | Nocardia sp. strain OAct 132 | JX047071 | 2633 | 99.4 |
| M5_8 | Streptomyces sp. strain MM56 | KU714908 | 2694 | 100 |
| M5_9 | Streptomyces sp. strain MM56 | KU714908 | 2675 | 99.9 |
| W2_1 | Microbacterium phyllosphaerae IHBB 11136 | KR085857 | 2686 | 100 |
| Bacterial or Fungal Strain | Source |
|---|---|
| Staphylococcus aureus MRSA 200 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA ATCC 6538 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 108 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 271 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 203 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 122 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 116 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 115 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 342 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 352 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 44 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 298 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 199 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 343 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 297 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 202 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 124 | Medical University of Gdańsk |
| Staphylococcus aureus MRSA 149 | Medical University of Gdańsk |
| Salmonella enterica Virchow 41 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Enteritidis 64 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Kentucky 1368 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Heidelberg 16 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Cholerasuis 1439 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Typhimurium 12 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Typhimurium 13 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Agona 1408 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Thompson 39 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Gallinarum 74 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Hadar 1784 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Cholerasuis 39 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Infantis 155 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Bovismorbificans 300 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Seftenberg 87 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Newport 50 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Newport 51 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Cholerasuis 37 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Dubin 65 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Saindpaul 435 | National Salmonella Center, Gdańsk, Poland |
| Salmonella enterica Enteritidis 1392 | National Salmonella Center, Gdańsk, Poland |
| Escherichia coli STEC 35 | University of Gdańsk collection |
| Escherichia coli STEC 36 | University of Gdańsk collection |
| Escherichia coli STEC 37 | University of Gdańsk collection |
| Escherichia coli STEC 38 | University of Gdańsk collection |
| Escherichia coli STEC 39 | University of Gdańsk collection |
| Pseudomonas aeruginosa 02113 | University of Gdańsk collection |
| Pseudomonas aeruginosa 02109 | University of Gdańsk collection |
| Pseudomons aeruginosa 02108 | University of Gdańsk collection |
| Pseudomonas aeruginosa RA743 | University of Gdańsk collection |
| Bacillus subtilis 3610 | University of Gdańsk collection |
| Bacillus subtilis wt168 | University of Gdańsk collection |
| Bacillus megaterium | University of Gdańsk collection |
| Bacillus cereus | University of Gdańsk collection |
| Candida parapsilosis D2 | Bruss Laboratories, Gdynia, Poland |
| Candida glabrata D3 | Bruss Laboratories, Gdynia, Poland |
| Candida tropicalis D4 | Bruss Laboratories, Gdynia, Poland |
| Candida dubliniensis D5 | Bruss Laboratories, Gdynia, Poland |
| Candida albicans D6 | Bruss Laboratories, Gdynia, Poland |
| Candida albicans D7 | Medical University of Gdańsk |
| Candida albicans D8 | Medical University of Gdańsk |
| Candida albicans D9 | University Clinical Centre in Gdańsk |
| Candida albicans E1 | University Clinical Centre in Gdańsk |
| Candida guilliermondii E2 | University Clinical Centre in Gdańsk |
| Candida guilliermondii E3 | University Clinical Centre in Gdańsk |
| Candida albicans E4 | University Clinical Centre in Gdańsk |
| Candida albicans E5 | University Clinical Centre in Gdańsk |
| Candida glabrata E6 | University Clinical Centre in Gdańsk |
| Candida glabrata E7 | University Clinical Centre in Gdańsk |
| Candida sp. E8 | University Clinical Centre in Gdańsk |
| Candida sp. E9 | University Clinical Centre in Gdańsk |
| Bacteruial Strain | Growth Inhibition Zone (mm) 1 | |
|---|---|---|
| M4_24 | M5_8 | |
| S. aureus MRSA 200 | 5.0 ± 1.0 | 5.5 ± 1.5 |
| S. aureus MRSA ATCC 6538 | 7.5 ± 0.3 | 6.5 ± 0.5 |
| S. aureus MRSA 108 | 5.0 ± 2.0 | 6.5 ± 0.5 |
| S. aureus MRSA 271 | 7.5 ± 1.5 | 7.5 ± 0.5 |
| S. aureus MRSA 203 | 6.5 ± 1.5 | 7.0 ± 1.0 |
| S. aureus MRSA 122 | 4.5 ± 0.5 | 4.75 ± 1.25 |
| S. aureus MRSA 116 | 5.0 ± 1.0 | 5.5 ± 0.5 |
| S. aureus MRSA 115 | 6.75 ± 0.75 | 5.75 ± 0.75 |
| S. aureus MRSA 342 | 0.0 | 0.0 |
| S. aureus MRSA 352 | 6.0 ± 1.0 | 5.25 ± 1.75 |
| S. aureus MRSA 44 | 12.0 ± 3.0 | 0.0 |
| S. aureus MRSA 298 | 4.0 ± 0.0 | 0.0 |
| S. aureus MRSA 199 | 8.0 ± 2.0 | 0.0 |
| S. aureus MRSA 343 | 8.0 ± 1.0 | 0.0 |
| S. aureus MRSA 297 | 7.25 ± 1.25 | 6.75 ± 0.25 |
| S. aureus MRSA 202 | 8.25 ± 1.25 | 3.5 ± 0.5 |
| S. aureus MRSA 124 | 3.0 ± 0 | 3.0 ± 0.0 |
| S. aureus MRSA 149 | 6.0 ± 0.0 | 6.0 ± 0.0 |
| S. enterica Virchow 41 | 0.0 | 0.0 |
| S. enterica Enteritidis 64 | 6.0 ± 1.0 | 0.0 |
| S. enterica Kentucky 1368 | 5.5 ± 0.5 | 8.0 ± 1.0 |
| S. enterica Heidelberg 16 | 6.0 ± 1.0 | 5.0 ± 1.0 |
| S. enterica Cholerasuis 1439 | 11.5 ± 0.5 | 0.0 |
| S. enterica Typhimurium 12 | 7.5 ± 1.5 | 0.0 |
| S. enterica Typhimurium 13 | 6.5 ± 0.5 | 0.0 |
| S. enterica Agona 1408 | 0.0 | 0.0 |
| S. enterica Thompson 39 | 0.0 | 0.0 |
| S. enterica Gallinarum 74 | 5.5 ± 0.5 | 0.0 |
| S. enterica Hadar 1784 | 0.0 | 0.0 |
| S. enterica Cholerasuis 39 | 6.5 ± 1.5 | 5.0 ± 2.0 |
| S. enterica Infantis 155 | 6.5 ± 0.5 | 8.0 ± 10 |
| S. enterica Bovismorbificans 300 | 6.25 ± 0.25 | 0.0 |
| S. enterica Seftenberg 87 | 5.0 ± 1.0 | 4.5 ± 0.5 |
| S. enterica Newport 50 | 5.5 ± 0.5 | 5.0 ± 1.0 |
| S. enterica Newport 51 | 5.5 ± 0.5 | 5.0 ± 1.0 |
| S. enterica Cholerasuis 37 | 4.5 ± 0.5 | 6.5 ± 0.5 |
| S. enterica Dubin 65 | 6.5 ± 0.5 | 4.5 ± 0.5 |
| S. enterica Saindpaul 435 | 3.0 ± 1.0 | 0.0 |
| S. enterica Enteritidis 1392 | 9.75 ± 0.25 | 4 ± 0 |
| Enterococcus sp. | 10.5 ± 0.5 | 8.0 ± 1.0 |
| E. coli 35 | 8.5 ± 1.5 | 6.5 ± 0.5 |
| E. coli 36 | 10.5 ± 1.5 | 7.0 ± 1.0 |
| E. coli 37 | 10.0 ± 1.0 | 7.0 ± 0.5 |
| E. coli 38 | 11.0 ± 0.5 | 6.5 ± 0.5 |
| E. coli 39 | 8.5 ± 0.5 | 8.5 ± 0.5 |
| B. subtilis 3610 | 7.0 ± 1.0 | 0.0 |
| B. subtilis wt168 | 8.0 ± 1.0 | 0.0 |
| B. megaterium | 8.0 ± 1.0 | 0.0 |
| B. cereus | 7.0 ± 2.0 | 0.0 |
| P. aeruginosa 02113 | 8.0 ± 2.0 | 6.0 ± 1.0 |
| P. aeruginosa 02109 | 6.5 ± 0.5 | 5.0 ± 0.5 |
| P. aeruginosa 02108 | 10.0 ± 1.0 | 5.0 ± 0.0 |
| P. aeruginosa RA743 | 8.0 ± 1.0 | 7.0 ± 2.0 |
| Fungal Strain | Growth Inhibition Zone (mm) 1 | |
|---|---|---|
| M4_24 | M5_8 | |
| Candida parapsilosis D2 | 0.0 | 0.0 |
| Candida glabrata D3 | 10.3 ± 2.1 | 18.7 ± 3.5 |
| Candida tropicalis D4 | 0.0 | 0.0 |
| Candida dubliniensis D5 | 5.7 ± 1.5 | 0.0 |
| Candida albicans D6 | 3.3 ± 1.2 | 0.0 |
| Candida albicans D7 | 3.7 ± 0.6 | 3.0 ± 1.0 |
| Candida albicans D8 | 0.0 | 0.0 |
| Candida albicans D9 | 4.3 ± 1.5 | 2.3 |
| Candida albicans E1 | 3.3 ± 2.3 | 0.0 |
| Candida guilliermondii E2 | 6.0 ± 2.6 | 0.0 |
| Candida guilliermondii E3 | 6.3 ± 0.6 | 0.0 |
| Candida albicans E4 | 3.3 ± 0.6 | 0.0 |
| Candida albicans E5 | 0.0 | 0.0 |
| Candida glabrata E6 | 12.0 ± 4.4 | 5.7 ± 2.5 |
| Candida glabrata E7 | 6.0 ± 2.0 | 2.0 ± 3.5 |
| Candida sp. E8 | 3.7 ± 0.6 | 3.7 ± 1.2 |
| Candida sp. E9 | 3.3 ± 1.2 | 3.3 ± 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaroszewicz, W.; Bielańska, P.; Lubomska, D.; Kosznik-Kwaśnicka, K.; Golec, P.; Grabowski, Ł.; Wieczerzak, E.; Dróżdż, W.; Gaffke, L.; Pierzynowska, K.; et al. Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland). Antibiotics 2021, 10, 1212. https://doi.org/10.3390/antibiotics10101212
Jaroszewicz W, Bielańska P, Lubomska D, Kosznik-Kwaśnicka K, Golec P, Grabowski Ł, Wieczerzak E, Dróżdż W, Gaffke L, Pierzynowska K, et al. Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland). Antibiotics. 2021; 10(10):1212. https://doi.org/10.3390/antibiotics10101212
Chicago/Turabian StyleJaroszewicz, Weronika, Patrycja Bielańska, Daria Lubomska, Katarzyna Kosznik-Kwaśnicka, Piotr Golec, Łukasz Grabowski, Ewa Wieczerzak, Weronika Dróżdż, Lidia Gaffke, Karolina Pierzynowska, and et al. 2021. "Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland)" Antibiotics 10, no. 10: 1212. https://doi.org/10.3390/antibiotics10101212
APA StyleJaroszewicz, W., Bielańska, P., Lubomska, D., Kosznik-Kwaśnicka, K., Golec, P., Grabowski, Ł., Wieczerzak, E., Dróżdż, W., Gaffke, L., Pierzynowska, K., Węgrzyn, G., & Węgrzyn, A. (2021). Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland). Antibiotics, 10(10), 1212. https://doi.org/10.3390/antibiotics10101212













