Antimicrobial Susceptibility and Frequency of bla and qnr Genes in Salmonella enterica Isolated from Slaughtered Pigs
Abstract
1. Introduction
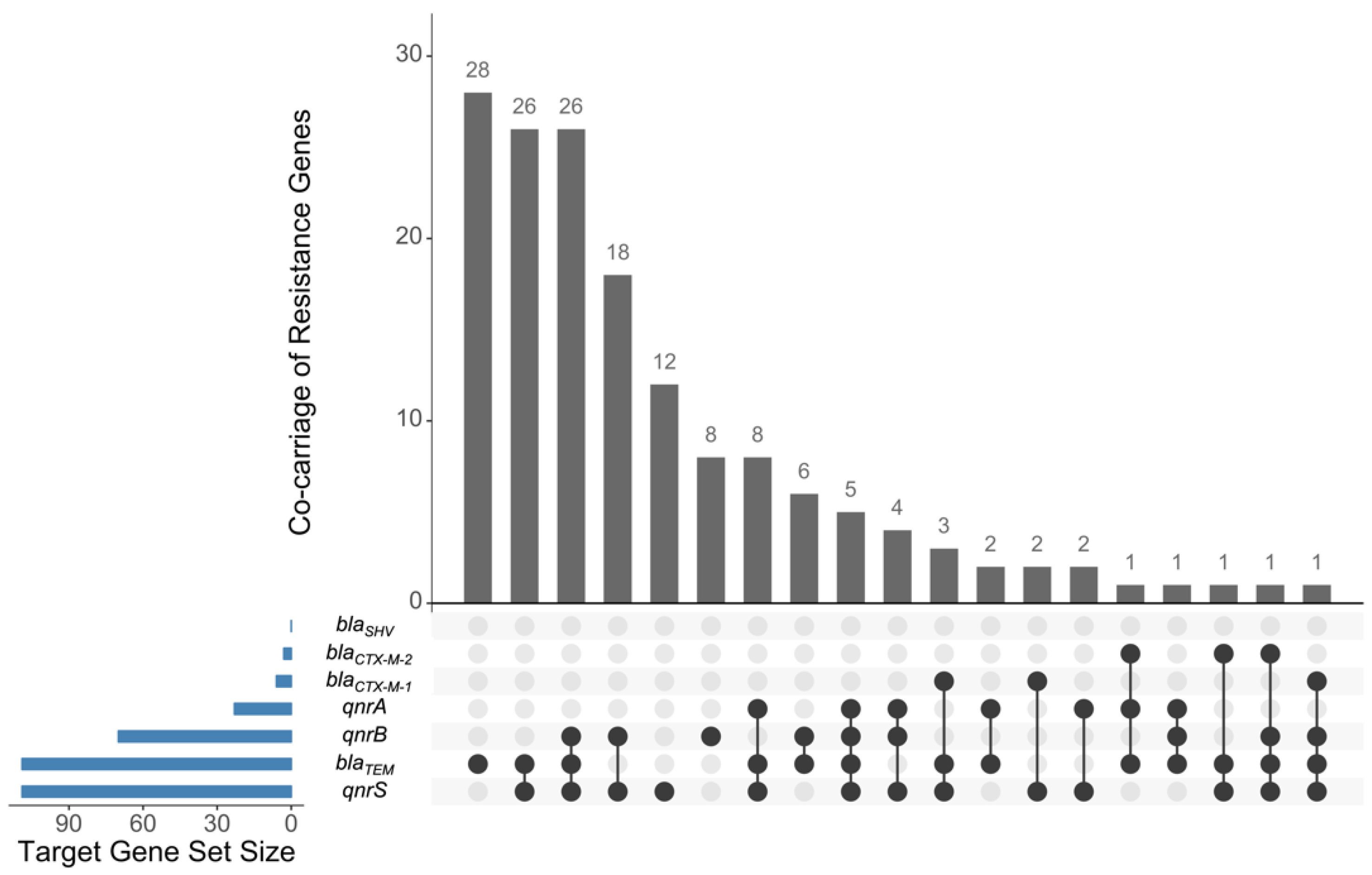
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Bacterial Isolation and Identification
4.3. Molecular Characterization of S. enterica
4.4. Bacterial Storage and Recovery
4.5. Antimicrobial Susceptibility Testing
4.6. Detection of bla and qnr Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Azanza, M.P.V.; Membrebe, B.N.Q.; Sanchez, R.G.R.; Estilo, E.E.C.; Dollete, U.G.M.; Feliciano, R.J.; Garcia, N.K.A. Foodborne disease outbreaks in the Philippines (2005–2018). Philipp. J. Sci. 2019, 148, 317–336. [Google Scholar]
- Azanza, M.P.V. Philippine foodborne-disease outbreaks (1995–2004). J. Food Saf. 2006, 26, 92–102. [Google Scholar] [CrossRef]
- Britto, C.D.; Wong, V.K.; Dougan, G.; Pollard, A.J. A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLoS Negl. Trop. Dis. 2018, 12, e0006779. [Google Scholar] [CrossRef] [PubMed]
- Gut, A.M.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Salmonella infection–Prevention and treatment by antibiotics and probiotic yeasts: A review. Microbiology 2018, 164, 1327–1344. [Google Scholar] [CrossRef]
- Monstein, H.-J.; Östholm-Balkhed, Å.; Nilsson, M.V.; Nilsson, M.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41, S120–S126. [Google Scholar] [CrossRef] [PubMed]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Tomaso, H.; Neubauer, H.; Hafez, H.M.; El-Adawy, H. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017, 9, 57. [Google Scholar] [CrossRef]
- Penha Filho, R.A.C.; Ferreira, J.C.; Kanashiro, A.M.I.; Berchieri Junior, A.; da Costa Darini, A.L. Emergent multidrug-resistant nontyphoidal Salmonella serovars isolated from poultry in Brazil coharboring blaCTX-M-2 and qnrB or blaCMY-2 in large plasmids. Diagn. Microbiol. Infect. Dis. 2019, 95, 93–98. [Google Scholar] [CrossRef]
- Cheung, T.K.M.; Chu, Y.W.; Chu, M.Y.; Ma, C.H.; Yung, R.W.H.; Kam, K.M. Plasmid-mediated resistance to ciprofloxacin and cefotaxime in clinical isolates of Salmonella enterica serotype Enteritidis in Hong Kong. J. Antimicrob. Chemother. 2005, 56, 586–589. [Google Scholar] [CrossRef]
- Gutierrez, B.; Herrera-Leon, S.; Escudero, J.A.; Hidalgo, L.; Gonzalez-Sanz, R.; Arroyo, M.; San Millan, Á.; Echeita, M.A.; Gonzalez-Zorn, B. Novel genetic environment of qnrB2 associated with TEM-1 and SHV-12 on pB1004, an IncHI2 plasmid, in Salmonella Bredeney BB1047 from Spain. J. Antimicrob. Chemother. 2009, 64, 1334–1336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nhung, N.T.; Cuong, N.V.; Thwaites, G.; Carrique-Mas, J. Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia: A review. Antibiotics 2016, 5, 37. [Google Scholar] [CrossRef]
- Phongaran, D.; Khang-Air, S.; Angkititrakul, S. Molecular epidemiology and antimicrobial resistance of Salmonella isolates from broilers and pigs in Thailand. Vet. World 2019, 12, 1311–1318. [Google Scholar] [CrossRef]
- Lalruatdiki, A.; Dutta, T.K.; Roychoudhury, P.; Subudhi, P.K. Extended-spectrum β-lactamases producing multidrug resistance Escherichia coli, Salmonella and Klebsiella pneumoniae in pig population of Assam and Meghalaya, India. Vet. World 2018, 11, 868–873. [Google Scholar] [CrossRef]
- Cameron-Veas, K.; Fraile, L.; Napp, S.; Garrido, V.; Grilló, M.J.; Migura-Garcia, L. Multidrug resistant Salmonella enterica isolated from conventional pig farms using antimicrobial agents in preventative medicine programmes. Vet. J. 2018, 234, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chen, K.; Chan, E.W.-C.; Chen, S. Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sinwat, N.; Angkittitrakul, S.; Coulson, K.F.; Pilapil, F.M.I.R.; Meunsene, D.; Chuanchuen, R. High prevalence and molecular characteristics of multidrug-resistant Salmonella in pigs, pork and humans in Thailand and Laos provinces. J. Med. Microbiol. 2016, 65, 1182–1193. [Google Scholar] [CrossRef] [PubMed]
- Khachatourians, G.G. Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. Can. Med. Assoc. J. 1998, 159, 1129–1136. [Google Scholar]
- Barton, M.D. Impact of antibiotic use in the swine industry. Curr. Opin. Microbiol. 2014, 19, 9–15. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar]
- Kerr, J.R. Antibiotic treatment and susceptibility testing. J. Clin. Pathol. 2005, 58, 786–787. [Google Scholar] [CrossRef]
- Kakkar, M.; Chatterjee, P.; Chauhan, A.S.; Grace, D.; Lindahl, J.; Beeche, A.; Jing, F.; Chotinan, S. Antimicrobial resistance in South East Asia: Time to ask the right questions. Glob. Health Action 2018, 11, 1483637. [Google Scholar] [CrossRef] [PubMed]
- Philippine Statistics Authority. Available online: http://openstat.psa.gov.ph/ (accessed on 16 October 2020).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Calayag, A.M.B.; Paclibare, P.A.P.; Santos, P.D.M.; Bautista, C.A.C.; Rivera, W.L. Molecular characterization and antimicrobial resistance of Salmonella enterica from swine slaughtered in two different types of Philippine abattoir. Food Microbiol. 2017, 65, 16. [Google Scholar] [CrossRef]
- Tu, L.T.P.; Hoang, N.V.M.; Cuong, N.V.; Campbell, J.; Bryant, J.E.; Hoa, N.T.; Kiet, B.T.; Thompson, C.; Duy, D.T.; Phat, V.V.; et al. High levels of contamination and antimicrobial-resistant non-typhoidal Salmonella serovars on pig and poultry farms in the Mekong Delta of Vietnam. Epidemiol. Infect. 2015, 143, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Viana, C.; Sereno, M.J.; Pegoraro, K.; Yamatogi, R.S.; Call, D.R.; dos Santos Bersot, L.; Nero, L.A. Distribution, diversity, virulence genotypes and antibiotic resistance for Salmonella isolated from a Brazilian pork production chain. Int. J. Food Microbiol. 2019, 310, 108310. [Google Scholar] [CrossRef]
- Nguyen Thi, H.; Pham, T.-T.-T.; Turchi, B.; Fratini, F.; Ebani, V.V.; Cerri, D.; Bertelloni, F. Characterization of Salmonella spp. isolates from swine: Virulence and antimicrobial resistance. Animals 2020, 10, 2418. [Google Scholar] [CrossRef]
- Trongjit, S.; Angkititrakul, S.; Tuttle, R.E.; Poungseree, J.; Padungtod, P.; Chuanchuen, R. Prevalence and antimicrobial resistance in Salmonella enterica isolated from broiler chickens, pigs and meat products in Thailand–Cambodia border provinces. Microbiol. Immunol. 2017, 61, 23–33. [Google Scholar] [CrossRef]
- Li, X.; Yao, Z.; Yuan, L.; Qi, W.; Shuchang, A.; Jichao, C.; Yusheng, C.; Lin, L.; Jiabin, L.; Zhancheng, G. Identification of Klebsiella pneumoniae strains harboring inactive extended-spectrum beta-lactamase antibiotic-resistance genes. Chin. Med. J. 2014, 127, 3051–3057. [Google Scholar]
- Cruz, M.C.; Hedreyda, C.T. Detection of plasmid-borne β-lactamase genes in extended-spectrum β-lactamase (ESBL) and non-ESBL-producing Escherichia coli clinical isolates. Philipp. J. Sci 2017, 146, 167–175. [Google Scholar]
- Temmerman, R.; Garmyn, A.; Antonissen, G.; Vanantwerpen, G.; Vanrobaeys, M.; Haesebrouck, F.; Devreese, M. Evaluation of fluoroquinolone resistance in clinical avian pathogenic Escherichia coli isolates from Flanders (Belgium). Antibiotics 2020, 9, 800. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.A.; Awasthi, S.P.; Hoang, P.H.; Nguyen, P.D.; Jayedul, H.; Hatanaka, N.; Hinenoya, A.; Van Dang, C.; Faruque, S.M.; Yamasaki, S. Prevalence, serovar and antimicrobial resistance of nontyphoidal Salmonella in vegetable, fruit and water samples in Ho Chi Minh City, Vietnam. Foodborne Pathog. Dis. 2021, 18, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.C.S.; Rivera, W.L. Multiplex PCR-based serogrouping and serotyping of Salmonella enterica from tonsil and jejunum with jejunal lymph nodes of slaughtered swine in Metro Manila, Philippines. J. Food Prot. 2015, 78, 873–880. [Google Scholar] [CrossRef]
- Soguilon-Del Rosario, S.A.; Rivera, W.L. Incidence and molecular detection of Salmonella enterica serogroups and spvC virulence gene in raw and processed meats from selected wet markets in Metro Manila, Philippines. Int. J. Philipp. Sci. Tech. 2015, 8, 52–56. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, T.; Lee, M.D.; Hofacre, C.L.; Maier, M.; White, D.G.; Ayers, S.; Wang, L.; Berghaus, R.; Maurer, J.J. Rapid screening of Salmonella enterica serovars Enteritidis, Hadar, Heidelberg and Typhimurium using a serologically-correlative allelotyping PCR targeting the O and H antigen alleles. BMC Microbiol. 2008, 8, 1–8. [Google Scholar] [CrossRef]
- Hirose, K.; Itoh, K.-I.; Nakajima, H.; Kurazono, T.; Yamaguchi, M.; Moriya, K.; Ezaki, T.; Kawamura, Y.; Tamura, K.; Watanabe, H. Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J. Clin. Microbiol. 2002, 40, 633–636. [Google Scholar] [CrossRef]
- Vanegas, R.A.; Joys, T.M. Molecular analyses of the phase-2 antigen complex 1, 2 of Salmonella spp. J. Bacteriol. 1995, 177, 3863–3864. [Google Scholar] [CrossRef] [PubMed]
- Herrera-León, S.; McQuiston, J.R.; Usera, M.A.; Fields, P.I.; Garaizar, J.; Echeita, M.A. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 2004, 42, 2581–2586. [Google Scholar] [CrossRef]
- Agron, P.G.; Walker, R.L.; Kinde, H.; Sawyer, S.J.; Hayes, D.C.; Wollard, J.; Andersen, G.L. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 2001, 67, 4984–4991. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Hujer, K.M.; Hujer, A.M.; Yeiser, B.; Bonomo, M.D.; Rice, L.B.; Bonomo, R.A.; the International Klebsiella Study Group. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 2003, 47, 3554–3560. [Google Scholar] [CrossRef]
- Boyd, D.A.; Tyler, S.; Christianson, S.; McGeer, A.; Muller, M.P.; Willey, B.M.; Bryce, E.; Gardam, M.; Nordmann, P.; Mulvey, M.R.; et al. Complete nucleotide sequence of a 92-Kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 2004, 48, 3758–3764. [Google Scholar] [CrossRef]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2006, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Weill, F.-X.; Poirel, L.; Fabre, L.; Soussy, C.-J.; Nordmann, P. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 2007, 59, 751–754. [Google Scholar] [CrossRef]
| Serogroup | First-Phase Flagellar (H1) Antigens | Sdf I | Other | ||||
|---|---|---|---|---|---|---|---|
| H:d | H: e,h | H:g | H:i | H:r | |||
| O:3,10 (n = 69) | 39 | 23 | 7 | ||||
| O:4 (n = 38) | 1 | 2 | 25 | 1 | 9 | ||
| O:7 (n = 54) | 2 | 27 | 9 | 10 | 6 | ||
| O:8 (n = 3) | 2 | 1 | |||||
| O:9 (n = 3) | 3 | ||||||
| Other (n = 11) | |||||||
| Class | Antimicrobial | % Non-Susceptibility |
|---|---|---|
| Penicillin | Ampicillin | 71.9% (±8.8) |
| Penicillin/β-lactamase inhibitor | Amoxicillin/clavulanic acid | 10.1% (±5.9) |
| Antipseudomonal penicillin/β-lactamase inhibitor | Piperacillin/tazobactam | 0.6% (±1.5) |
| Extended-spectrum cephalosporin | Ceftazidime | 8.4% (+5.4) |
| Ceftriaxone | 7.9% (±5.3) | |
| Cefepime | 0.0% (±0) | |
| Carbapenem | Ertapenem | 0.0% (±0) |
| Imipenem | 1.7% (±2.5) | |
| Meropenem | 0.0% (±0) | |
| Fluoroquinolone | Ciprofloxacin | 15.7% (±7.1) |
| Folate pathway inhibitor | Trimethoprim/sulfamethoxazole | 70.8% (±8.9) |
| Number of S. enterica Isolates | Multidrug Resistance Pattern 1 |
|---|---|
| 1 | Pen, Pen/BI, APen/BI, ESC, Flu |
| 14 | Pen, Pen/BI, FPI, ESC |
| 1 | Pen, Pen/BI, FPI, Car |
| 1 | Pen, Pen/BI, Car, Flu |
| 7 | Pen, FPI, Flu |
| Target | Nucleotide Sequence (5′-3′) | Amplicon Length (bp) | Reference |
|---|---|---|---|
| O:4 | F: GGCTTCCGGCTTTATTGG R: TCTCTTATCTGTTCGCCTGTTG | 561 | [38] |
| O:9 | F: GAGGAAGGGAAATGAAGCTTTT R: TAGCAAACTGTCTCCCACCATAC | 615 | [39] |
| O:2, O:9 | F: CTTGCTATGGAAGACATAACGAACC R: CGTCTCCATCAAAAGCTCCATAGA | 258 | [39] |
| O:6,7 | F: ATTTGCCCAGTTCGGTTTG R: CCATAACCGACTTCCATTTCC | 341 | [38] |
| O:8 | F: CGTCCTATAACCGAGCCAAC R: R: CTGCTTTATCCCTCTCACCG | 397 | [38] |
| O:3, 10 | F: GATAGCAACGTTCGGAAATTC R: CCCAATAGCAATAAACCAAGC | 281 | [38] |
| Sense60 | F: GCAGATCAACTCTCAGACCCTGGG | [40] | |
| H:r | R: AAGTGACTTTTCCATCGGCTG | 275 | [41] |
| H:i | R: ATAGCCATCTTTACCAGTTCC | 250 | [41] |
| H:e,h | R: AACGAAAGCGTAGCAGACAAG | 200 | [41] |
| H:b | R: CGCACCAGTCYWACCTAAGGCGG | 150 | [41] |
| H:d | F: CCCGAAAGAAACTGCTGTAACCG R: TGGATATCAGTATTGCTCTGGGC | 100 | [41] |
| G complex alleles (H:g) | F: GTGATCTGAAATCCAGCTTCAAG R: AAGTTTCGCACTCTCGTTTTTGG | 500 | [41] |
| Sdf I | F: TGTGTTTTATCTGATGCAAGAGG R: CGTTCTTCTGGTACTTACGATGAC | 333 | [42] |
| Target Gene | Nucleotide Sequence (5′-3′) | Amplicon Length (bp) | Reference |
|---|---|---|---|
| blaSHV | F: ATGCGTTATATTCGCCTGTG | 747 | [43] |
| R: TGCTTTGTTATTCGGGCCAA | |||
| blaTEM | F: TCGCCGCATACACTATTCTCAGAAT GA | 445 | [5] |
| R: ACGCTCACCGGCTCCAGATTTAT | |||
| blaCTX-M | F: ATGTGCAGYACCAGTAARGTKATGG C | 593 | [44] |
| R: TGGGTRAARTARGTSACCAGAAYCA GCGG | |||
| blaCTX-M-1 | F: AAAAATCACTGCGCCAGTTC | 415 | [45] |
| R: AGCTTATTCATCGCCACGTT | |||
| blaCTX-M-2 | F: CGATATCGTTGGTGGTRCCAT | 404 | [6] |
| R: CGTTAACGGCACGATGAC | |||
| blaCTX-M-9 | F: CAAAGAGAGTGCAACGGATG | 205 | [45] |
| R: ATTGGAAAGCGTTCATCACC | |||
| blaCTX-M-8/25 | F: AACCCACGATGTGGGTAGC | [45] | |
| blaCTX-M-8 | R: TCGCGTTAAGCGGATGATGC | 666 | |
| blaCTX-M-25 | R: GCACGATGACATTCGGG | 327 | |
| qnrA | F: AGAGGATTTCTCACGCCAGG | 580 | [46] |
| R: TGCCAGGCACAGATCTTGAC | |||
| qnrB | F: GGAATAGAAATTCGCCACTG | 264 | [47] |
| R: TTTGCTGTTCGCCAGTCGAA | |||
| qnrS | F: GCAAGTTCATTGAACAGGGT | 428 | [46] |
| R: TCTAAACCGTCGAGTTCGGCG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calayag, A.M.B.; Widmer, K.W.; Rivera, W.L. Antimicrobial Susceptibility and Frequency of bla and qnr Genes in Salmonella enterica Isolated from Slaughtered Pigs. Antibiotics 2021, 10, 1442. https://doi.org/10.3390/antibiotics10121442
Calayag AMB, Widmer KW, Rivera WL. Antimicrobial Susceptibility and Frequency of bla and qnr Genes in Salmonella enterica Isolated from Slaughtered Pigs. Antibiotics. 2021; 10(12):1442. https://doi.org/10.3390/antibiotics10121442
Chicago/Turabian StyleCalayag, Alyzza Marie B., Kenneth W. Widmer, and Windell L. Rivera. 2021. "Antimicrobial Susceptibility and Frequency of bla and qnr Genes in Salmonella enterica Isolated from Slaughtered Pigs" Antibiotics 10, no. 12: 1442. https://doi.org/10.3390/antibiotics10121442
APA StyleCalayag, A. M. B., Widmer, K. W., & Rivera, W. L. (2021). Antimicrobial Susceptibility and Frequency of bla and qnr Genes in Salmonella enterica Isolated from Slaughtered Pigs. Antibiotics, 10(12), 1442. https://doi.org/10.3390/antibiotics10121442







