Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Filters, Inclusion and Exclusion Criteria
2.3. Data Extraction
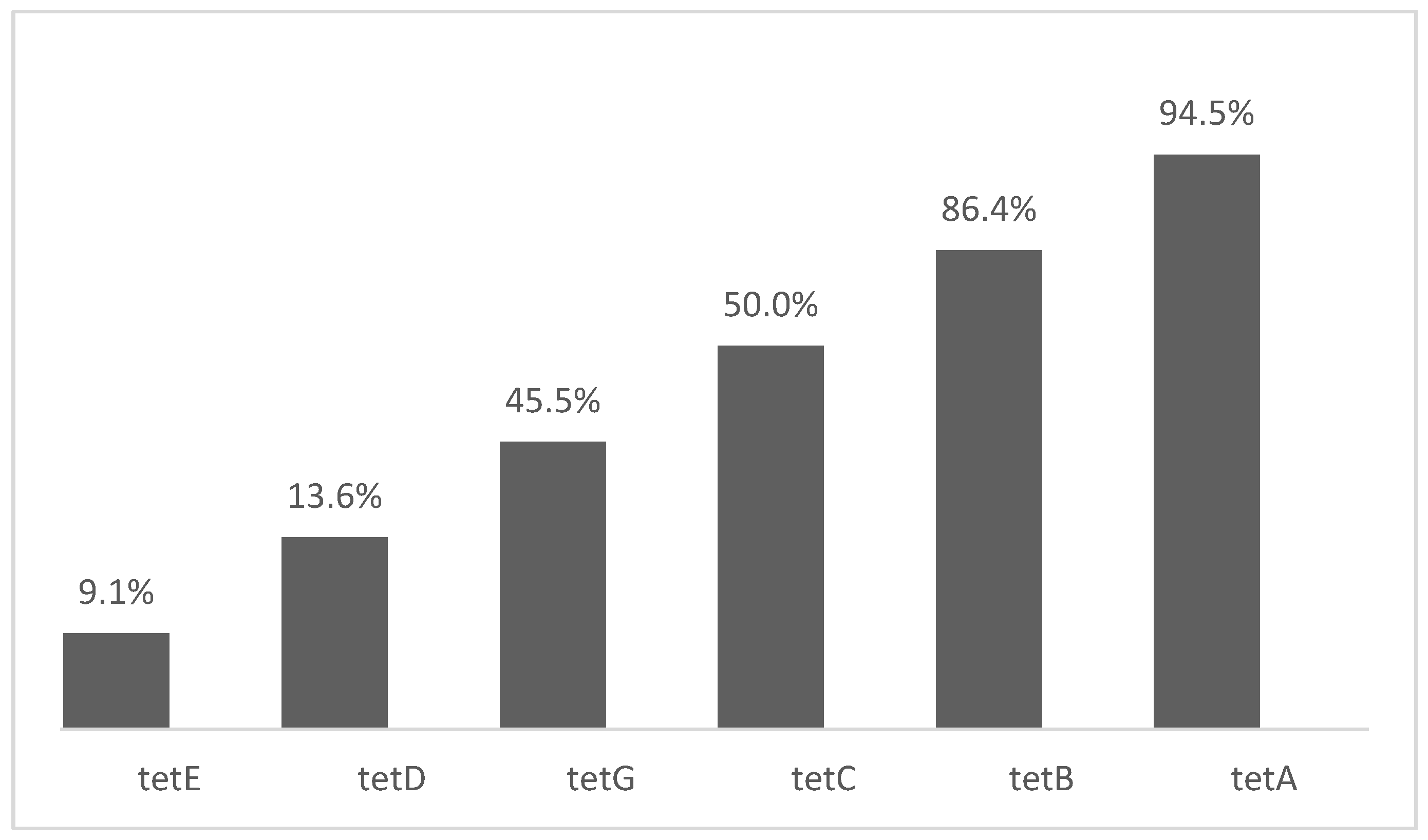
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Adesoji, A.T.; Ogunjobi, A.A.; Olatoye, I.O.; Douglas, D.R. Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 2–8. [Google Scholar] [CrossRef]
- Marosevic, D.; Kaevska, M.; Jaglic, Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage—A review. Ann. Agric. Environ. Med. 2017, 24, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Sheykhsaran, E.; Baghi, H.B.; Soroush, M.H.; Ghotaslou, R. An overview of tetracyclines and related resistance mechanisms. Rev. Med. Microbiol. 2019, 30, 69–75. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Schwarz, S. Tetracycline and chloramphenicol resistance mechanisms. In Antimicrobial Drug Resistance; Meyers, D., Sobel, J., Ouellette, M., Kaye, K., Marchaim, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 231–242. [Google Scholar]
- Hussain, T.; Jamal, M.; Nighat, F.; Andleeb, S. Broad spectrum antibiotics and resistance in nontarget bacteria: An example from tetracycline. J. Pure Appl. Microbiol. 2014, 8, 2667–2671. [Google Scholar]
- Maka, L.; Mackiw, E.; Sciezynska, H.; Modzelewska, M.; Popowska, M. Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland. Foodborne Pathog. Dis. 2015, 12, 383–389. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Cortés, P.; Barbé, J.; Erill, I. Origin of the mobile di-hydro-pteroate synthase gene determining sulfonamide resistance in clinical isolates. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Sköld, O.E.; Swedberg, G. Sulfonamides and Trimethoprim. In Antimicrobial Drug Resistance; Mayers, D., Sobel, J., Ouellette, M., Kaye, K., Marchaim, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 348–358. [Google Scholar]
- Xu, F.; Min, F.; Wang, J.; Luo, Y.; Huang, S.; Chen, M.; Wu, R.; Zhang, Y. Development and evaluation of a Luminex xTAG assay for sulfonamide resistance genes in Escherichia coli and Salmonella isolates. Mol. Cell Probes. 2019, 49, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Nunes, O.C.; Manaia, C.M.; Kolvenbach, B.A.; Corvini, P.F. Living with sulfonamides: A diverse range of mechanisms observed in bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 10389–10408. [Google Scholar] [CrossRef]
- Christian, A.; Vivian, E.B.; Crystal, N.Z.; Frank, B.O. Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance. In Antimicrobial Resistance—A Global Threat; Kumar, Y., Ed.; IntechOpen: London, UK, 2018; pp. 33–51. [Google Scholar]
- Lees, P.; Pelligand, L.; Giraud, E.; Toutain, P.L. A history of antimicrobial drugs in animals: Evolution and revolution. J. Vet. Pharmacol. Therap. 2021, 44, 137–171. [Google Scholar] [CrossRef]
- Ben, W.W.; Wang, J.; Pan, X.; Qiang, Z.M. Dissemination of antibiotic resistance genes and their potential removal by on-farm treatment processes in nine swine feedlots in Shandong Province, China. Chemosphere 2017, 167, 262–268. [Google Scholar] [CrossRef]
- Card, R.; Vaughan, K.; Bagnall, M.; Spiropoulos, J.; Cooley, W.; Strickland, T.; Rob, D.; Anjum, M.F. Virulence characterization of Salmonella enterica isolates of differing antimicrobial resistance recovered from UK livestock and imported meat samples. Front. Microbiol. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Klümper, U.; Shi, L.; Ye, L.; Li, M. From pig breeding environment to subsequently produced pork: Comparative analysis of antibiotic resistance genes and bacterial community composition. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Yuan, J.; Ni, M.; Liu, M.; Zheng, Y.; Gu, Z. Occurrence of antibiotics and antibiotic resistance genes in a typical estuary aquaculture region of Hangzhou Bay, China. Mar. Pollut. Bull. 2019, 138, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, X.; Jiao, S.; Zhang, J.; Ye, B.; Gao, S. Sulfonamide resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS ONE 2014, 9, e112626. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front. Microbiol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Achermann, S.; Bianco, V.; Mansfeldt, C.B.; Vogler, B.; Kolvenbach, B.A.; Corvini, P.F.X.; Kathrin, F.K. Biotransformation of sulfonamide antibiotics in activated sludge: The formation of pterinconjugates leads to sustained risk. Environ. Sci. Technol. 2018, 52, 6265–6274. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Mao, Y.; Li, B.; Yang, C.; Zhang, T. Aerobic degradation of sulfadiazine by Arthrobacter spp.: Kinetics, pathways, and genomic characterization. Environ. Sci. Technol. 2016, 50, 9566–9575. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, S. Overview of sulfonamide biodegradation and the relevant pathways and microorganisms. Sci. Total. Environ. 2018, 640, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 1–24. [Google Scholar] [CrossRef]
- Roberts, M.C.; Schwarz, S. Tetracycline and phenicol resistance genes and mechanisms: Importance for agriculture, the environment, and humans. J. Environ. Qual. 2016, 45, 576–592. [Google Scholar] [CrossRef]
- Xu, X.; Biswas, S.; Gu, G.; Elbediwi, M.; Li, Y.; Yue, M. Characterization of multidrug resistance patterns of emerging Salmonella enterica serovar Rissen along the food chain in China. Antibiotics 2020, 9, 660. [Google Scholar] [CrossRef]
- Dessie, H.K.; Bae, D.H.; Lee, Y.J. Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea. Poult. Sci. 2013, 92, 3036–3043. [Google Scholar] [CrossRef] [PubMed]
- Mąka, Ł.; Popowska, M. Antimicrobial resistance of Salmonella spp. isolated from food. Rocz. Panstw. Zakl. Hig. 2016, 67, 343–358. [Google Scholar]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jové, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Peruzy, M.F.; Capuano, F.; Proroga, Y.T.R.; Cristiano, D.N.; Carullo, M.R.; Murru, N. Antimicrobial susceptibility testing for Salmonella serovars isolated from food samples: Five-year monitoring (2015–2019). Antibiotics 2020, 9, 365. [Google Scholar] [CrossRef]
- Perreten, V.; Boerlin, P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef]
- Razavi, M.; Marathe, N.P.; Gillings, M.R.; Flach, C.F.; Kristiansson, E.; Joakim Larsson, D.G. Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 2017, 5, 1–12. [Google Scholar] [CrossRef]
- Guerra, B.; Junker, E.; Helmuth, R. Incidence of the recently described sulfonamide resistance gene sul3 among German Salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 2004, 48, 2712–2715. [Google Scholar] [CrossRef][Green Version]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Ministério da Saúde do Brasil. Surto de Doenças Transmitidas por Alimentos no Brasil. Informe, 2018. Available online: http://portalarquivos2.saude.gov.br/images/pdf/2019/maio/17/Apresentacao-Surtos-DTA-Maio-2019.pdf (accessed on 9 January 2021).
- Hoffmann, S.; Maculloch, B.; Batz, M. Economic Burden of Major Foodborne Illnesses Acquired in the United States, EIB-140; US Department of Agriculture, Economic Research Service: Washington, DC, USA, 2015; pp. 543–616.
- EFSA-ECDC. European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 1–178. [Google Scholar]
- Deng, W.; Quan, Y.; Yang, S.; Guo, L.; Zhang, X.; Liu, S.; Chen, S.; Zhou, K.; He, L.; Li, B.; et al. Antibiotic resistance in Salmonella from retail foods of animal origin and its association with disinfectant and heavy metal resistance. Microb. Drug Resist. 2018, 24, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Iwu, C.J.; Iweriebor, B.C.; Obi, L.C.; Basson, A.K.; Okoh, A.I. Multidrug-Resistant Salmonella isolates from swine in the Eastern Cape Province, South Africa. J. Food. Prot. 2016, 79, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, M.B.; Tarning, J.; Cho, T.Z.A.; Anal, A.K. Antibacterial activities and possible modes of action of Acacia nilotica (L.) Del. against multidrug-resistant Escherichia coli and Salmonella. Molecules 2017, 22, 47. [Google Scholar] [CrossRef] [PubMed]
- Vital, P.G.; Caballes, M.B.D.; Rivera, W.L. Antimicrobial resistance in Escherichia coli and Salmonella spp. isolates from fresh produce and the impact to food safety. J. Environ. Sci. Health 2017, 52, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Zishiri, O.T.; Mkhize, N.; Mukaratirwa, S. Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil. Onderstepoort J. Vet. Res. 2016, 83, 1–11. [Google Scholar] [CrossRef]
- Zhu, Y.; Lai, H.; Zow, L.; Yin, S.; Wang, C.; Han, X.; Xia, X.; Hu, K.; He, L.; Zhou, K.; et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 2017, 259, 43–51. [Google Scholar] [CrossRef]
- Igbinosa, I. Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals. Trop. Anim. Health Prod. 2014, 47, 37–43. [Google Scholar] [CrossRef]
- Aslam, M.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Gensler, G.; Reid-Smith, R.; Boerlin, P. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012, 32, 110–117. [Google Scholar] [CrossRef]
- Dahshan, H.; Chuma, T.; Shahada, F.; Akiba, M.; Fujimoto, H.; Akasaka, K.; Kamimura, Y.; Okamoto, K. Characterization of antibiotic resistance and the emergence of AmpC-producing Salmonella Infantis from pigs. J. Vet. Med. Sci. 2010, 72, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, H.; Tahoun, A.; El-Gohary, A.E.-G.A.; El-Abasy, M.; El-Khayat, F.; Gillespie, T.; Kitade, Y.; Hafez, H.M.; Neubauer, H.; El-Adawy, H. Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 2017, 9, 8. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Hsu, B.M.; Ji, W.T.; Chen, J.S.; Hsu, T.K.; Ji, D.D.; Tseng, S.F.; Chiu, Y.C.; Kao, P.M.; Huang, Y.L. Antibiotic resistance pattern and gene expression of non-typhoid Salmonella in riversheds. Environ. Sci. Pollut. Res. 2015, 22, 7843–7850. [Google Scholar] [CrossRef] [PubMed]
- Khoshbakht, R.; Derakhshandeh, A.; Jelviz, L.; Azhdari, F. Tetracycline resistance genes in Salmonella enterica serovars with animal and human origin. Int. J. Enteric Pathog. 2018, 6, 60–64. [Google Scholar] [CrossRef]
- Kozak, G.K.; Pearl, D.L.; Parkman, J.; Reid-Smith, R.J.; Deckert, A.; Boerlin, P. Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at abattoirs in Ontario and Quebec, Canada. Appl. Environ. Microb. 2009, 75, 5999–6001. [Google Scholar] [CrossRef]
- Lapierre, L.; San Martín, B.; Araya-Jordán, C.; Borie, C. Comparison of integron-linked antibiotic resistance genes in strains of Salmonella spp. isolated from swine in Chile in 2005 and 2008. Can. J. Microbiol. 2010, 56, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Lopes, G.V.; Pissetti, C.; da Cruz, P.P.D.; da Silva, L.E.; Cardoso, M. Resistance phenotypes and genotypes of Salmonella enterica subsp. enterica isolates from feed, pigs, and carcasses in Brazil. J. Food Prot. 2015, 78, 407–413. [Google Scholar]
- Márquez, F.M.L.; Burgos, M.J.G.; Pulido, R.P.; Gálvez, A.; López, R.L. Biocide tolerance and antibiotic resistance in Salmonella isolates from hen eggshells. Foodborne Pathog. Dis. 2017, 14, 89–95. [Google Scholar] [CrossRef]
- Mthembu, T.P.; Zishiri, O.T.; El Zowalaty, M.E. Molecular detection of multidrug-resistant Salmonella isolated from livestock production systems in South Africa. Infect. Drug Resist. 2019, 12, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Soyer, Y.; Richards, J.; Hoelzer, K.; Warnick, L.D.; Fortes, E.; McDonough, P.; Dumas, N.B.; Grohn, Y.T.; Wiedmann, M. Antimicrobial drug resistance patterns among cattle-and human-associated Salmonella strains. J. Food. Prot. 2013, 76, 1676–1688. [Google Scholar] [CrossRef]
- Tajbakhsh, M.; Hendriksen, R.S.; Nochi, Z.; Zali, M.R.; Aarestrup, F.M.; Garcia-Migura, L. Antimicrobial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008. Folia Microbiol. 2012, 57, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Thai, T.H.; Lan, N.T.; Hirai, T.; Yamaguchi, R. Antimicrobial resistance in Salmonella serovars isolated from meat shops at the markets in North Vietnam. Foodborne Pathog. Dis. 2012, 9, 986–991. [Google Scholar] [CrossRef]
- Vuthy, Y.; Lay, K.S.; Seiha, H.; Kerleguer, A.; Aidara-Kane, A. Antibiotic susceptibility and molecular characterization of resistance genes among Escherichia coli and among Salmonella subsp. in chicken food chains. Asian Pac. J. Trop. Biomed. 2017, 7, 670–674. [Google Scholar] [CrossRef]
- Zhu, A.; Zhi, W.; Qiu, Y.; Wei, L.; Tian, J.; Pan, Z.; Kang, X.; Gu, W.; Duan, L. Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China. Food Control. 2019, 98, 474–480. [Google Scholar] [CrossRef]
- Li, S.; Zhou, Y.; Miao, Z. Prevalence and antibiotic resistance of non-typhoidal Salmonella isolated from raw chicken carcasses of commercial broilers and spent hens in Tai’an, China. Front. Microbiol. 2017, 8, 2106. [Google Scholar] [CrossRef]
- Li, Y.C.; Pan, Z.M.; Kang, X.L.; Geng, S.Z.; Liu, Z.Y.; Cai, Y.Q.; Jiao, X.A. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu province, eastern China. J. Food Prot. 2014, 77, 236–245. [Google Scholar] [CrossRef]
- Dallal, M.M.S.; Doyle, M.P.; Rezadehbashi, M.; Dabiri, H.; Sanaei, M.; Modarresi, S. Prevalence and antimicrobial resistance profiles of Salmonella serotypes, Campylobacter and Yersinia spp. isolated from retail chicken and beef, Tehran, Iran. Food Control. 2010, 21, 388–392. [Google Scholar] [CrossRef]
- Romero-Barrios, P.; Deckert, A.; Parmley, E.J.; Leclair, D. Antimicrobial resistance profiles of Escherichia coli and Salmonella isolates in Canadian broiler chickens and their products. Foodborne Pathog. Dis. 2020, 17, 672–678. [Google Scholar] [CrossRef]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, X.; Jiang, Z.; Qi, Y.; Ed-Dra, A.; Yue, M. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan, China. Front. Cell Infect. Microbiol. 2020, 10, 497. [Google Scholar] [CrossRef]
- Moe, A.Z.; Paulsen, P.; Pichpol, D.; Fries, R.; Irsigler, H.; Baumann, M.P.O.; Oo, K.N. Prevalence and antimicrobial resistance of Salmonella isolates from chicken carcasses in retail markets in Yangon, Myanmar. J. Food. Prot. 2017, 80, 947–951. [Google Scholar] [CrossRef]
- Terentjeva, M.; Avsejenko, J.; Streikisa, M.; Utinane, A.; Kovalenko, K.; Berzins, A. Prevalence and antimicrobial resistance of Salmonella in meat and meat products in Latvia. Ann. Agric. Environ. Med. 2017, 24, 317–321. [Google Scholar] [CrossRef]
- Sodagari, H.R.; Mashak, Z.; Ghadimianazar, A. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J. Infect. Dev. Ctries. 2015, 9, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.B.; Xiong, L.G.; Tan, M.F.; Li, H.Q.; Yan, H.; Zhang, L.; Yin, D.F.; Kang, Z.F.; Wei, Q.P.; Luo, L.G. Prevalence and antimicrobial resistance of Salmonella in pork, chicken, and duck from retail markets of China. Foodborne Pathog. Dis. 2019, 16, 339–345. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Potter, L.; Vaz, C.S.; Pereira, D.I.; Sangioni, L.A.; Vargas, Á.C.; de Avila Botton, S. Antimicrobial resistance in nontyphoidal Salmonella isolated from human and poultry-related samples in Brazil: 20-Year Meta-Analysis. Foodborne Pathog. Dis. 2017, 14, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Vaez, H.; Ghanbari, F.; Sahebkar, A.; Khademi, F. Antibiotic resistance profiles of Salmonella serotypes isolated from animals in Iran: A meta-analysis. Iran. J. Vet. Res. 2020, 21, 188–197. [Google Scholar]
- Zhang, C.M.; Xu, L.M.; Mou, X.; Xu, H.; Liu, J.; Miao, Y.H.; Xiaochang, C.; Wang, X.L. Characterization and evolution of antibiotic resistance of Salmonella in municipal wastewater treatment plants. J. Environ. Manag. 2019, 251, 109547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.M.; Du, C.; Xu, H.; Miao, Y.H.; Cheng, Y.Y.; Tang, H.; Zhou, J.H.; Wang, X.C. Occurrence of tetracycline-resistant fecal coliforms and their resistance genes in an urban river impacted by municipal wastewater treatment plant discharges. J. Environ. Sci. Health A 2015, 50, 744–749. [Google Scholar] [CrossRef]
- Mattiello, S.P.; Drescher, G.; Barth, V.C.; Ferreira, C.A.; Oliveira, S.D. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie Van Leeuwenhoek. 2015, 108, 1227–1238. [Google Scholar] [CrossRef]
- Sanchez-Maldonado, A.F.; Aslam, M.; Service, C.; Narváez-Bravo, C.; Avery, B.P.; Johnson, R.; Jones, T.H. Prevalence and antimicrobial resistance of Salmonella isolated from two pork processing plants in Alberta, Canada. Int. J. Food Microbiol. 2017, 241, 49–59. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The tetracycline resistome. Cell Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef]
- Ma, S.; Lei, C.; Kong, L.; Jiang, W.; Liu, B.; Men, S.; Yang, Y.; Cheng, G.; Chen, Y.; Wang, H. Prevalence, antimicrobial resistance, and relatedness of Salmonella isolated from chickens and pigs on farms, abattoirs, and markets in Sichuan Province, China. Foodborne Pathog. Dis. 2017, 14, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; Coque, T.M.; Cantón, R.; Sousa, J.C.; Peixe, L. Commensal Enterobacteriaceae as reservoirs of extended-spectrum beta-lactamases, integrons, and sul genes in Portugal. Front. Microbiol. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, C.G.; Zhong, X.H. Survey on sulfonamide antibiotic-resistant genotype and phenotype of avian Escherichia coli in North China. Poult. Sci. 2012, 91, 884–887. [Google Scholar] [CrossRef]
- Deekshit, V.K.; Kumar, B.K.; Rai, P.; Srikumar, S.; Karunasagar, I. Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in south-west coast of India. J. Appl. Microbiol. 2012, 112, 1113–1122. [Google Scholar] [CrossRef]
- Roberts, M.C.; Kuchmiy, E.; Miranda, C.D. The tetracycline resistant tet gene identified in three new genera of bacteria isolated in 1999 from Chilean salmon farms. J. Antimicrob. Chemother. 2015, 70, 619–620. [Google Scholar] [CrossRef]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef]
- Ljubojević, D.; Pelić, M.; Puvača, N.; Milanov, D. Resistance to tetracycline in Escherichia coli isolates from poultry meat: Epidemiology, policy and perspective. World’s Poult. Sci. J. 2017, 73, 409–417. [Google Scholar] [CrossRef]
- Adesiji, Y.O.; Deekshit, V.K.; Karunasagar, I. Antimicrobial-resistant genes associated with Salmonella spp. isolated from human, poultry, and seafood sources. Food Sci. Nutr. 2014, 2, 436–442. [Google Scholar] [CrossRef]
- Lorenz, T.C. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. 2012, 63, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 2–11. [Google Scholar] [CrossRef]
- Śpibida, M.; Krawczyk, B.; Olszewski, M.; Kur, J. Modified DNA polymerases for PCR troubleshooting. J. Appl. Genet. 2017, 58, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Helke, K.L.; McCrackin, M.A.; Galloway, A.M.; Poole, A.Z.; Salgado, C.D.; Marriott, B.P. Effects of antimicrobial use in agricultural animals on drug-resistant foodborne salmonellosis in humans: A systematic literature review. Crit. Rev. Food. Sci. Nutr. 2017, 11, 472–488. [Google Scholar] [CrossRef] [PubMed]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Ren, X.; Li, M.; Xu, C.; Cui, K.; Feng, Z.; Fu, Y.; Zhang, J.; Liao, M. Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 2016, 144, 2989–2999. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Ha, J.S.; Kim, B.Y.; Lee, D.H.; Park, J.K.; Youn, H.N.; Hong, Y.H.; Lee, S.B.; Lee, J.B.; Park, S.Y.; et al. Prevalence and characterization of Salmonella species in entire steps of a single integrated broiler supply chain in Korea. Poul. Sci. 2014, 93, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Nakao, J.H.; Pringle, J.; Jones, R.W.; Nix, B.E.; Borders, J.; Heseltine, G.; Gomez, T.M.; McCluskey, B.; Roney, C.S.; Brinson, D.; et al. ‘One Health’ investigation: Outbreak of human Salmonella Braenderup infections traced to a mail-order hatchery—United States, 2012–2013. Epidemiol. Infect. 2015, 143, 2178–2186. [Google Scholar] [CrossRef]
- Paine, S.; Thornley, C.; Wilson, M.; Dufour, M.; Sexton, K.; Miller, J. An outbreak of multiple serotypes of Salmonella in New Zealand linked to consumption of contaminated tahini imported from Turkey. Foodborne Pathog. Dis. 2014, 11, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Yang, X.; Tan, H.; Ke, B.; He, D.; Wang, H. Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the Southern coastal area of China. Int. J. Food Microbiol. 2018, 266, 317–323. [Google Scholar] [CrossRef]
- Miller, A.J.; Twomey, D.F.; Davies, R.H.; Teale, C.J.; Williamson, S.M.; Reichel, R.; Featherstone, C.A.; Cook, A.J.; Snow, L.C.; Armstrong, J.D. Salmonella serovars and antimicrobial resistance patterns on a sample of high seroprevalence pig farms in England and Wales (2003–2008). Zoonoses Public Health 2011, 58, 549–559. [Google Scholar] [CrossRef]
- Bier, D.; Kich, J.D.; Duarte, S.C.; Silva, M.R.; Valsoni, L.M.; Ramos, C.A.N.; Rodrigues, D.P.; Araújo, F.R. Survey of Salmonella spp. in beef meat for export at slaughterhouses in Brazil. Pesq. Vet. Bras. 2018, 38, 2037–2043. [Google Scholar] [CrossRef]
- Castro-Vargas, R.E.; Herrera-Sánchez, M.P.; Rodríguez-Hernández, R.; Rondón-Barragán, I.S. Antibiotic resistance in Salmonella spp. isolated from poultry: A global overview. Vet. World. 2020, 13, 2070–2084. [Google Scholar] [PubMed]
- McDermott, P.F.; Zhao, S.; Tate, H. Antimicrobial resistance in nontyphoidal Salmonella. Microbiol. Spectrum. 2018, 6, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Brasil, Lista de Substâncias Proibidas e Legislação Correspondente. 2017. Available online: http://www.agricultura.gov.br/assuntos/insumos-agropecuarios/insumos-pecuarios/arquivos-de-insumos-pecuarios/Substnciasproibidas.pdf (accessed on 9 January 2021).
- Eng, S.K.; Pusparajah, P.; Mutalib, N.-A.S.; Ser, H.; Chan, K.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar]
- Fair, R.J.; Tor, Y. Perspectives in medicinal chemistry antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar]
- Jajere, S.M. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World. 2019, 12, 504–521. [Google Scholar] [CrossRef]
- Bengtsson, B.; Greko, C. Antibiotic resistance–consequences for animal health, welfare, and food production. Upsala J. Med. Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Paudyal, N.; Li, X.; Fang, W.; Yue, M. Multiple food animal- borne route in transmission of antibiotic-resistant Salmonella Newport to humans. Front. Microbiol. 2018, 9, 23. [Google Scholar] [CrossRef]

| Studies | Authors | Title | Year | Genes Searched | Reference |
|---|---|---|---|---|---|
| 1 | Aslam et al. | Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada | 2012 | tetA, tetB, tetC, sul1, sul2, sul3 | [45] |
| 2 | Dahshan et al. | Characterization of antibiotic resistance and the emergence of AmpC-producing Salmonella infantis from pigs | 2010 | tetA, tetB, tetG, sul1 | [46] |
| 3 | Deng et al. | Antibiotic resistance in Salmonella from retail foods of animal origin and its association with disinfectant and heavy metal resistance | 2017 | tetA, tetB, tetC, tetG, sul1, sul2, sul3 | [38] |
| 4 | Dessie et al. | Characterization of integrons and their cassettes in Escherichia coli and Salmonella isolates from poultry in Korea | 2013 | tetA, tetB, tetC, tetD, tetE, tetG, sul1, sul2 | [27] |
| 5 | El-Sharkawy et al. | Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt | 2017 | tetA, tetB, tetC, sul1, sul2, sul3 | [47] |
| 6 | Hsu et al. | Antibiotic resistance pattern and gene expression of non-typhoid Salmonella in river sheds | 2014 | tetA, tetB, sul1 | [48] |
| 7 | Igbinosa | Prevalence and detection of antibiotic-resistant determinant in Salmonella isolated from food-producing animals | 2014 | tetC | [44] |
| 8 | Iwu et al. | Multidrug-resistant Salmonella isolates from swine in the Eastern Cape Province, South Africa | 2016 | tetA | [39] |
| 9 | Khoshbakht et al. | Tetracycline resistance genes in Salmonella enterica serovars with animal and human origin | 2018 | tetA, tetB, tetC, tetG | [49] |
| 10 | Kozak et al. | Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at Abattoirs in Ontario and Québec, Canada | 2009 | sul1, sul2, sul3 | [50] |
| 11 | Lapierre et al. | Comparison of integron-linked antibiotic resistance genes in strains of Salmonella spp. isolated from swine in Chile in 2005 and 2008 | 2010 | tetA, tetB, tetG | [51] |
| 12 | Lopes et al. | Resistance phenotypes and genotypes of Salmonella enterica subsp. enterica isolates from feed, pigs, and carcasses in Brazil | 2015 | tetA, tetB, sul1, sul2, sul3 | [52] |
| 13 | Maka et al. | Resistance to sulfonamides and dissemination of sul genes among Salmonella spp. isolated from food in Poland | 2015 | sul1, sul2, sul3 | [7] |
| 14 | Marquéz et al. | Biocide tolerance and antibiotic resistance in Salmonella isolates from hen eggshells | 2017 | tetA, tetB, tetC, tetD, tetE, tetG, sul1 | [53] |
| 15 | Mthembu et al. | Molecular detection of multidrug-resistant Salmonella isolated from livestock production systems in South Africa | 2019 | tetA, tetC, sul2 | [54] |
| 16 | Sadiq et al. | Antibacterial activities and possible modes of action of Acacia nilotica (L.) Del. against multidrug-resistant Escherichia coli and Salmonella | 2017 | tetA, tetB | [40] |
| 17 | Soyer et al. | Antimicrobial drug resistance patterns among cattle-and human-associated Salmonella strains | 2013 | tetA, tetB, tetG, sul1, sul2 | [55] |
| 18 | Tajbakhsh et al. | Antimicrobial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008 | 2012 | tetA, tetB, tetC, tetD, tetG, sul1 | [56] |
| 19 | Thai et al. | Antimicrobial resistance in Salmonella serovars isolated from meat shops at markets in North Vietnam. | 2012 | tetA, tetB, tetG, sul1 | [57] |
| 20 | Vital et al. | Antimicrobial resistance in Escherichia coli and Salmonella spp. isolates from fresh produce and the impact to food safety. | 2017 | tetA, tetB, tetC | [41] |
| 21 | Vuthy et al. | Antibiotic susceptibility and molecular characterization of resistance genes among Escherichia coli and among Salmonella subsp. in chicken food chains. | 2017 | tetA, tetB, sul1, sul2 | [58] |
| 22 | Xu et al. | Development and evaluation of a Luminex xTAG assay for sulfonamide resistance genes in Escherichia coli and Salmonella isolates | 2019 | sul1, sul2, sul3, sul4 | [10] |
| 23 | Zhu et al. | Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China | 2017 | tetA, tetB, tetC, tetG, sul1, sul2, sul3 | [43] |
| 24 | Zhu et al. | Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China | 2019 | tetA, tetB, sul1, sul2 | [59] |
| 25 | Zishiri et al. | Prevalence of virulence and antimicrobial resistance genes in Salmonella spp. isolated from commercial chickens and human clinical isolates from South Africa and Brazil | 2016 | tetA, tetB, sul1, sul2 | [42] |
| Studies | No. of Salmonella Isolates | Tetracycline-Resistant Isolates n (%) | Isolates with tet Genes n (%) | Sulfonamide-Resistant Isolates n (%) | Isolates with sul Genes n (%) |
|---|---|---|---|---|---|
| Aslam et al. 2012 [45] | 110 | 54 (49.0%) | 45 (40.9%) | 9 (8.0%) | 9 (8.0%) |
| Dahshan et al. 2010 [46] | 44 | 44 (100%) | 10 (22.7%) | 44 (100%) | 8 (18.2%) |
| Deng et al. 2017 [38] | 152 | 123 (80.9%) | 123 (80.9%) | 98 (64.5%) | 60 (39.5%) |
| Dessie et al. 2013 [27] | 33 | 23 (69.7%) | 8 (24.2%) | 31 (93.9%) | 26 (78.8%) |
| El-Sharkawy et al. 2017 [47] | 67 | 61 (91.0%) | 58 (86.6%) | 3 (5.2%) | 58 (86.6%) |
| Hsu et al. 2014 [48] | 54 | 18 (33.3%) | 14 (26.0%) | 20 (37.0%) | 16 (29.6%) |
| Igbinosa 2015 [44] | 150 | 73 (48.7%) | 0 | 99 (66.0%) | * |
| Iwu et al. 2016 [39] | 48 | 48 (100%) | 30 (61.0%) | 36 (75.0%) | * |
| Khoshbakht et al. 2018 [49] | 60 | 60 (100%) | 6 (10.0%) | * | * |
| Kozak et al. 2009 [50] | 234 | * | * | * | 210 (89.7%) |
| Lapierre et al. 2010 [51] | 69 | 65 (94.2%) | 49 (71.0%) | 19 (27.5%) | * |
| Lopes et al. 2015 [52] | 225 | 122 (54.5%) | 73 (32.5%) | 89 (39.6%) | 65 (28.9%) |
| Maka et al. 2015 [7] | 84 | * | * | 84 (100%) | 76 (90.5%) |
| Marquéz et al. 2017 [53] | 39 | 19 (47.6%) | 6 (14.3%) | 15 (38.1%) | 4 (9.5%) |
| Mthembu et al. 2019 [54] | 106 | 67 (63.0%) | 25 (26.0%) | 41 (38.0%) | 22 (21.0%) |
| Sadiq et al. 2017 [40] | 4 | 3 (75.0%) | 3 (75.0%) | * | * |
| Soyer et al. 2013 [55] | 336 | 296 (88.0%) | 44 (13.1%) | 282 (84.0%) | 49 (14.6%) |
| Tajbakhsh et al. 2012 [56] | 71 | 18 (25.0%) | 34 (48.0%) | 21 (30.0%) | 23 (32.0%) |
| Thai et al. 2012 [57] | 97 | 47 (48.5%) | 40 (41.2%) | 55 (56.7%) | 52 (53.6%) |
| Vital et al. 2017 [41] | 24 | 16 (66.7%) | 21 (87.5%) | * | * |
| Vuthy et al. 2017 [58] | 181 | 157 (86.7%) | 117 (64.6%) | 156 (86.2%) | 78 (43.1%) |
| Xu et al. 2019 [10] | 18 | * | * | 13 (72.2%) | 14 (77.8%) |
| Zhu et al. 2017 [43] | 189 | 98 (51.9%) | 84 (44.4%) | 91 (48.1%) | 89 (47.1%) |
| Zhu et al. 2019 [59] | 155 | 143 (92.0%) | 32 (20.6%) | 81 (52.2%) | 29 (18.7%) |
| Zishiri et al. 2016 [42] | 146 | 136 (93.0%) | 128 (87.7%) | 123 (84.0%) | 125 (85.6%) |
| Studies | Salmonella Isolates (n) | tet and sul Genes in Salmonella Isolates n (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| tetA | tetB | tetC | tetD | tetE | tetG | sul1 | sul2 | sul3 | sul4 | ||
| Aslam et al. 2012 [45] | 45 tet 9 sul | 31 (68.7%) | 14 (31.2%) | 0% | * | * | * | 5 (55.6%) | 3 (33.3%) | 1 (11.2%) | * |
| Dahshan et al. 2010 [46] | 10 tet 10 sul | 6 (60.0%) | 2 (20.0%) | * | * | * | 2 (20.0%) | 8 (80.0%) | * | * | * |
| Deng et al. 2017 [38] | 123 tet 60 sul | 54 (44.7%) | 11 (9.0%) | 42 (34.1%) | * | * | 27 (21.9%) | 20 (33.3%) | 20 (33.3%) | 20 (33.3%) | * |
| Dessie et al. 2013 [27] | 33 tet 33 sul | 8 (24.2%) | 0% | 0% | 0% | 0% | 0% | 0% | 26 (78.8%) | * | * |
| El-Sharkawy et al. 2017 [47] | 67 tet 67 sul | 55 (82.0%) | 0% | 58 (86.6%) | * | * | * | 34 (50.7%) | 0% | 57 (85.1%) | * |
| Hsu et al. 2014 [48] | 54 tet 54 sul | 13 (24.1%) | 1 (1.9%) | * | * | * | * | 16 (29.6%) | * | * | * |
| Igbinosa 2015 [44] | 73 tet | * | * | 0% | * | * | * | * | * | * | * |
| Iwu et al. 2016 [39] | 48 tet | 30 (61.0%) | * | * | * | * | * | * | * | * | * |
| Khoshbakht et al. 2018 [49] | 60 tet | 6 (10.0%) | 0% | 3 (5.0%) | * | * | 0% | * | * | * | * |
| Kozak et al. 2009 [50] | 234 sul | * | * | * | * | * | * | 180 (76.9%) | 25 (10.7%) | 5 (2.1%) | * |
| Lapierre et al. 2010 [51] | 65 tet | 10 (15.4%) | 39 (60.0%) | * | * | * | 0% | * | * | * | * |
| Lopes et al. 2015 [52] | 91 tet 91 sul | 61 (67.0%) | 30 (32.9%) | * | * | * | * | 47 (51.6%) | 14 (15.4%) | 11 (12.1%) | * |
| Maka et al. 2015 [7] | 84 sul | * | * | * | * | * | * | 37 (44.0%) | 39 (46.4%) | 0 | * |
| Marquéz et al. 2017 [53] | 39 tet 39 sul | 4 (9.5%) | 0% | 2 (4.8%) | 0% | 0% | 0% | 4 (9.5%) | * | * | * |
| Mthembu et al. 2019 [54] | 106 tet 106 sul | 9 (8.0%) | * | 19 (18.0%) | * | * | * | 22 (21.0%) | * | * | * |
| Sadiq et al. 2017 [40] | 4 tet | 2 (50.0%) | 3 (75.0%) | * | * | * | * | * | * | * | * |
| Soyer et al. 2013 [55] | 48 tet 48 sul | 36 (75.0%) | 3 (6.3%) | * | * | * | 5 (10.4%) | 23 (47.9%) | 26 (54.2%) | * | * |
| Tajbakhsh et al. 2012 [56] | 71 tet 71 sul | 20 (28.0%) | 10 (14.0%) | 0% | 0% | * | 4 (6.0%) | 23 (32.0%) | * | * | * |
| Thai et al. 2012 [57] | 50 tet 58 sul | 37 (74.0%) | 3 (6.0%) | * | * | * | 13 (26.0%) | 52 (89.7%) | * | * | * |
| Vital et al. 2017 [41] | 24 tet | 21 (87.5%) | 0% | 0% | * | * | * | * | * | * | * |
| Vuthy et al. 2017 [58] | 157 tet 156 sul | 117 (64.6%) | 0% | * | * | * | * | 39 (25.0%) | 38 (24.3%) | * | * |
| Xu et al. 2019 [10] | 18 sul | * | * | * | * | * | * | 10 (55.6%) | 13 (72.2%) | 5 (27.8%) | 1 (5.6%) |
| Zhu et al. 2017 [43] | 98 tet 91 sul | 23 (23.5%) | 49 (50.0%) | 70 (71.4%) | * | * | 0% | 43 (50.0%) | 89 (97.8%) | 43 (50.0%) | * |
| Zhu et al. 2019 [59] | 29 sul 45 tet | 32 (71.1%) | 0% | * | * | * | * | 18 (62.1%) | 18 (62.1%) | * | * |
| Zishiri et al. 2016 [42] | 146 tet 146 sul | 79 (54.1%) | 49 (33.6%) | * | * | * | * | 76 (52.1%) | 74 (50.7%) | * | * |
| Authors | Genes Searched | Primers | PCR Amplification Conditions |
|---|---|---|---|
| Aslam et al. [45] | tetA | F: GGCGGTCTTCTTCATCATGC R: CGGCAGGCAGAGCAAGTAGA | Initial denaturation at 94 °C for 15 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 63 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. |
| tetB | F: CGCCCAGTGCTGTTGTTGTC R: CGCGTTGAGAAGCTGAGGTG | ||
| tetC | F: GCTGTAGGCATAGGCTTGGT R: GCCGGAAGCGAGAAGAATCA | ||
| sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG | Initial denaturation at 95 °C for 15 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 66 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. | |
| sul2 | F: CGGCATCGTCAACATAACCT R: TGTGCGGATGAAGTCAGCTC | ||
| sul3 | F: CAACGGAAGTGGGCGTTGTGGA R: GCTGCACCAATTCGCTGAACG | ||
| Dahshan et al. [46] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Annealing temperature: 64 °C |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| tetG | F: GCTCGGTGGTATCTCTGCTC R: AGCAACAGAATCGGGAACAC | Annealing temperature: 59 °C | |
| sul1 | F: TCGGATCAGACGTCGTGG R: CCAGCCTGCAGTCCGCCT | Annealing temperature: 60 °C | |
| Deng et al. [38] | tetA | F: CTCAGTATTCCAAGCCTTTG R: ACTCCCCTGAGCTTGAGGGG | 30 cycles of denaturation at 94 °C for 1 min, annealing at 60 °C for 45 s, and extension at 72 °C for 90 s, with an additional extension at 72 °C for 5 min. |
| tetB | F: CTAATCTAGACATCATTAATTCC R: TTTGAAGCTAAATCTTCTTTAT | ||
| tetG | F: AGTTTCAGGTGCGCAGC R: CCAATCGCCATGACTAAT | ||
| sul1 | F: CATCATTTTCGGCATCGTC R: TCTTGCGGTTTCTTTCAGC | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 50 s, annealing at 54 °C for 50 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. | |
| sul2 | F: AGATGTGATTGATTTGGGAGC R: TAGTTGTTTCTGGATTAGAGCCT | ||
| sul3 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | ||
| Dessie et al. [27] | tetA | F: GTAATTCTGAGCACTGTCGC R: CTGCCTGGACAACATTGCTT | Initial denaturation at 94 °C for 4 min, followed by 34 cycles of denaturation at 94 °C for 1 min, annealing at 43 °C for 2 min, and extension at 72 °C for 3 min, with an additional extension at 72 °C for 7 min. |
| tetB | F: CTCAGTATTCCAAGCCTTTG R: ACTCCCCTGAGCTTGAGGGG | ||
| tetC | F: CCTCTTGCGGGATATCGTCC R: GGTTGAAGGCTCTCAAGGGC | ||
| tetD | F: GGATATCTCACCGCATCTGC R: CATCCATCCGGAAGTGATAGC | ||
| tetE | F: AAACCACATCCTCCATACGC R: AAATAGGCCACAACCGTCAG | ||
| sul1 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 15 s, annealing at 69 °C for 30 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 7 min. | |
| sul2 | F: CGGCATCGTCAACATAACC R: GTGTGCGGATGAAGTCAG | ||
| El-Sharkawy et al. [47] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 2 min, and extension at 72 °C for 90 s. |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | Same conditions, with the specific annealing temperature: 53 °C | |
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | Same conditions, with the specific annealing temperature: 56 °C | |
| sul1 | F: TCACCGAGGACTCCTTCTTC R: AATATCGGGATAGAGCGCAG | Initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 1 min, specific annealing temperature at 60 °C, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 7 min. | |
| sul2 | F: CGGTCCGGCATCCAGCAATCC R: CGAGAGCCACGACCGCGCC | Same conditions, with the specific annealing temperature: 64 °C | |
| sul3 | F: GAGCAAGATTTTTGGAATCG R: CATCTGCAGCTAACCTAGGGCTTGGA | Same conditions, with the specific annealing temperature: 51 °C | |
| Hsu et al. [48] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Annealing temperature: 55 °C |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| sul1 | F: TCGGATCAGACGTCGTGG R: CCAGCCTGCAGTCCGCCT | Annealing temperature: 60 °C | |
| Igbinosa [44] | tetC | F: GGTTGAAGGCTCTCAAGGGC R: GGTTGAAGGCTCTCAAGGGC | Initial denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 65 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. |
| Iwu et al. [39] | tetA | F: GGCCTCAATTTCCTGACG R: AAGCAGGATGTAGCCTGTGC | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1.5-min, with an additional extension at 72 °C for 5 min. |
| Khoshbakht et al. [49] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Annealing temperature: 50 °C |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | Annealing temperature: 49 °C | |
| tetG | F: GCTCGGTGGTATCTCTGCTC R: AGCAACAGAATCGGGAACAC | ||
| Kozak et al. [50] | sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG | Initial denaturation at 95 °C for 15 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 66 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. |
| sul2 | F: CGGCATCGTCAACATAACCT R: TGTGCGGATGAAGTCAGCTC | ||
| sul3 | F: CAACGGAAGTGGGCGTTGTGGA R: GCTGCACCAATTCGCTGAACG | ||
| Lapierre et al. [51] | tetA | F: GGTTCACTCGAACGACGTCA R: CTGTCCGACAAGTTGCATGA | Annealing temperature: 52 °C |
| tetB | F: CTGGATTACTTATTGCTGGC R: CACCTTGCTGATGACTCTT | ||
| tetG | F: CCGGTCTTATGGGTGCTCTA R: GACTGGCTTCGTTCTTCTGG | Annealing temperature: 56 °C | |
| Lopes et al. [52] | tetA | F: GTAATTCTGAGCACTGT R: CCTGGACAACATTGCTT | Initial denaturation at 94 °C for 4 min, followed by 34 cycles of denaturation at 94 °C for 1 min, annealing at 43 °C for 2 min, and extension at 72 °C for 3 min, with an additional extension at 72 °C for 7 min. |
| tetB | F: ACGTTACTCGATGCCAT R: AGCACTTGTCTCCTGTT | ||
| tetG | F: CTGCTGATCGTGGGTCT R: TTGCGAATGGTCTGCGT | ||
| sul1 | F: ATGGTGACGGTGTTCGGCATTCTGA R: CTAGGCATGATCTAACCCTCGGTCT | Initial denaturation at 94 °C for 2 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 51 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 7 min. | |
| sul2 | F: ACAGTTTCTCCGATGGAGGCC R: CTCGTGTGTGCGGATGAAGTC | Same conditions, with the specific annealing temperature of 64 °C | |
| sul3 | F: GAGCAAGATTTTTGGAATCG R: CATCTGCAGCTAACCTAGGGCTTTGGA | Same conditions, with the specific annealing temperature of 51 °C | |
| Maka et al. [7] | sul1 | F: CGGCGTGGGCTACCTGAACG R: GCCGATCGCGTGAAGTTCCG | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 68 °C for 25 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. |
| sul2 | F: GCGCTCAAGGCAGATGGCAT R: GCGTTTGATACCGGCACCCGT | ||
| sul3 | F: CAGATAAGGCAATTGAGCATGCTCTGC R: AGAATGATTTCCGTGACACTGCAATCATT | ||
| Marquéz et al. [53] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1-5 min. |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | ||
| tetD | F: AAACCATTACGGCATTCTGC R: GACCGGATACACCATCCATC | ||
| tetE | F: AAACCACATCCTCCATACGC R: AAATAGGCCACAACCGTCAG | ||
| tetG | F: GCTCGGTGGTATCTCTGCTC R: AGCAACAGAATCGGGAACAC | ||
| sul1 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | Annealing temperature: 65 °C for 30 s | |
| Mthembu et al. [54] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Initial denaturation at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 8 min. |
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | Same conditions, with the specific annealing temperature: 42 °C | |
| sul2 | F: CGGCATCGTCAACATAACC R: GTGTGCGGATGAAGTCAG | Same conditions, with the specific annealing temperature: 60 °C | |
| Sadiq et al. [40] | tetA | F: GGTTCACTCGAACGACGTCA R: CTGTCCGACAAGTTGCATGA | Initial denaturation at 95 °C for 30 s, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 61.1 °C for 30 s, and extension at 68 °C for 1 min, with an additional extension at 68 °C for 5 min. |
| tetB | F: CCTCAGCTTCTCAACGCGTG R: GCACCTTGCTGATGACTCT | ||
| Soyer et al. [55] | tetA | F: GCGCCTTTCCTTTGGGTTCT R: CCACCCGTTCCACGTTGTTA | |
| tetB | F: CCCAGTGCTGTTGTTGTCAT R: CCACCACCAGCCAATAAAAT | ||
| tetG | F: AGCAGGTCGCTGGACACTAT R: CGCGGTGTTCCACTGAAAAC | Initial denaturation at 95 °C for 10 min, followed by 32 to 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 7 min. | |
| sul1 | F: TCACCGAGGACTCCTTCTTC R: CAGTCCGCCTCAGCAATATC | ||
| sul2 | F: CCTGTTTCGTCCGACACAGA R: GAAGCGCAGCCGCAATTCAT | ||
| Tajbakhsh et al. [56] | tetA | F: GTAATTCTGAGCACTGTCGC R: CTGCCTGGACAACATTGCTT | Annealing temperature: 58 °C |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | Annealing temperature: 60 °C | |
| tetC | F: ATGGTCGTCATCTACCTGCC R: GGTTGAAGGCTCTCAAGGGC | Annealing temperature: 53 °C | |
| tetD | F: AAACCATTACGGCATTCTGC R: GACCGGATACACCATCCATC | Annealing temperature: 60 °C | |
| tetG | F: CAGCTTTCGGATTCTACGG R: GATTGGTGAGGCTCGTTAGC | ||
| Thai et al. [57] | tetA | F: GCTACATCCTGCTTGCCT R: CATAGATCGCCGTGAAGA | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, the corresponding temperature of each primer pair for 30 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 5 min. |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| tetG | F: GCTCGGTGGTATCTCTGC R: AGCAACAGAATCGGGAAC | ||
| sul1 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | ||
| Vital et al. [41] | tetA | F: GTGAAACCCAACATACCCC R: GAAGGCAAGCAGGATGTAG | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50º C for 30 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 10 min. |
| tetB | F: CCTTATCATGCCAGTCTTGC R: ACTGCCGTTTTTTCGCC | ||
| tetC | F: ACTTGGAGCCACTATCGAC R: CTACAATCCATGCCAACCC | ||
| Vuthy et al. [58] | tetA | F: GCTACATCCTGCTTGCCTTC R: CATAGATCGCCGTGAAGAGG | Annealing temperature: 58 °C |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| sul1 | F: GTGACGGTGTTCGGCATTCT R: TTTACAGGAAGGCCAACGGT | ||
| sul2 | F: GGCAGATGTGATCGACCTCG R: ATGCCGGGATCAAGGACAAG | ||
| Xu et al. [10] | sul1 | F: CTAAACATACAAATACACATTTCA R: TGAAGTTCCGCCGCAAGGCTCG | Initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58º C for 30 s, and extension at 72 °C for 15 s, with an additional extension at 72 °C for 8 min. Initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 63º C for 30 s, and extension at 72 °C for 90 s, with an additional extension at 72 °C for 5 min. |
| sul2 | F: TACTTAAACATACAAACTTACTCA R: TGCCAAACTCGTCGTTATGC | ||
| sul3 | F: ATCTCAATTACAATAACACACAAA R: CGGGTATGGGCTTCTTTTTAG | ||
| sul4 | F: TACTACTTCTATAACTCACTTAAA R: CGGACCTATTAAGATGGGAAA | ||
| Zhu et al. [43] | tetA tetB | F: GTAATTCTGAGCACTGTCGC R: GAGACGCAATCGAATTCGG F: GAGACGCAATCGAATTCGG R: TTTAGTGGCTATTCTTCCTGCC | Initial denaturation at 95 °C for 10 min, followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 55-70º C for 50 s, and extension at 72 °C for 50 s, with an additional extension at 72 °C for 10 min. |
| tetC | F: CTTGAGAGCCTTCAACCCAG R: ATGGTCGTCATCTACCTGCC | ||
| tetG | F: GCTCGGTGGTATCTCTGCTC R: AGCAACAGAATCGGGAACAC | ||
| sul1 | F: CTTCGATGAGAGCCGGCGGC R: GCAAGGCGGAAACCCGCGCC | ||
| sul2 | F: GCGCTCAAGGCAGATGGCATT R: GCGTTTGATACCGGCACCCGT | ||
| sul3 | F: AGATGTGATTGATTTGGGAGC R: TAGTTGTTTCTGGATTAGAGCCT | ||
| Zhu et al. [59] | tetA | F: TCGCTTGCCGCATTT R: CGCGTATAGCTTGCCG | Initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55º C for 30 s, and extension at 72 °C for 1 min, with an additional extension at 72 °C for 6 min. |
| tetB | F: GACACTCTATCATTGAT R: GACAATATTTAGCAACG | ||
| sul1 | F: TGCAGGCTGGTGGTGGTTA R: CGCGTGGGTGCGGACGT | ||
| sul2 | F: CATTCCCGTCTCGCTCGA R: GCGCGCAGAAAGGATTT | ||
| Zishiri et al. [42] | tetA | F: GCTACATCCTGCTTGCCTT R: CATAGATCGCCGTGAAGAGG | Initial denaturation at 94 °C for 5 min, followed by 34 cycles of denaturation at 94 °C for 25 s, annealing at 55º C for 50 s, and extension at 72 °C for 50 s, with an additional extension at 72 °C for 5 min. |
| tetB | F: TTGGTTAGGGGCAAGTTTTG R: GTAATGGGCCAATAACACCG | ||
| sul1 | F: GCGCGGCGTGGGCTACCT R: GATTTCCGCGACACCGAGACAA | Same conditions, with the specific annealing temperature at 65 °C. | |
| sul2 | F: CGGCATCGTCAACATAACC R: GTGTGCGGATGAAGTCAG |
| Studies | Type of Samples | Salmonella spp. Isolates n (%) |
|---|---|---|
| Aslam et al. 2012 [45] | 564 meat samples (206 chicken, 91 turkey, 134 beef and 133 pork) | 210 isolates (183 strains from chicken; 24 strains from turkey and 3 strains from pork) (37.2%) |
| Dahshan et al. 2010 [46] | 270 pig fecal samples | 44 isolates (16.3%) |
| Deng et al. 2017 [38] | 327 meat samples (137 pork, 91chicken and 99 beef) | 252 isolates (175 strains from pork, 43 strains from chicken and 34 strains from beef) (46.5%) |
| Dessie et al. 2013 [27] | Chicken fecal samples | 33 isolates |
| El-Sharkawy et al. 2017 [47] | 615 samples collected from intestine, liver, and gall bladder from chickens | 67 isolates (10.9%) |
| Hsu et al. 2014 [48] | 236 water samples from river sheds | 54 isolates (22.9%) |
| Igbinosa 2015 [44] | Cow and goat fecal samples | 250 isolates (182 strains from cow feces and 68 strains from goat feces) |
| Iwu et al. 2016 [39] | 500 adult pig fecal samples | 48 isolates (9.6%) |
| Khoshbakht et al. 2018 [49] | Human and poultry samples | 60 isolates |
| Kozak et al. 2009 [50] | 938 chicken and swine meat samples | 234 isolates (13 strains from chicken and 221 strains from swine) (24.9%) |
| Lapierre et al. 2010 [51] | 580 healthy swine samples (290 fecal samples and 290 lymph node samples) | 65 isolates (11.2%) |
| Lopes et al. 2015 [52] | 1771 samples from pig feces and carcasses | 225 isolates (12.7%) |
| Maka et al. 2015 [7] | Retail meat samples (poultry, pork, and beef) | 84 isolates |
| Marquéz et al. 2017 [53] | 120 hen eggshells | 39 isolates (32.5%) |
| Mthembu et al. 2019 [54] | 361 fecal samples (cattle, sheep, goats, pigs, ducks, and chickens) | 106 isolates (29.4%) |
| Sadiq et al. 2017 [40] | Beef, poultry, and human samples | 4 isolates (2 strains from human clinical samples; 1 strain from poultry and 1 strain from beef) |
| Soyer et al. 2013 [55] | Human and bovine samples | 336 isolates (178 isolates from human and 158 isolates from bovine) |
| Tajbakhsh et al. 2012 [56] | 1.120 samples of humans with diarrhea symptoms | 71 isolates (6.4%) |
| Thai et al. 2012 [57] | 245 pork and chicken meat shops samples (116 carcass, 84 table surfaces and 45 sewage effluent) | 97 isolates (51 strains from carcass; 30 strains from table surfaces and 16 strains from sewage effluent) (39.6%) |
| Vital et al. 2017 [41] | 410 fresh vegetables samples | 24 isolates (5.85%) |
| Vuthy et al. 2017 [58] | 762 chicken samples (80 feces, 82 chicken caeca, 440 chicken neck skins, 80 rinse water and 80 chopping boards samples selected inside chicken slaughter) | 181 isolates (23.4%) |
| Xu et al. 2019 [10] | Agricultural samples | 18 isolates |
| Zhu et al. 2017 [43] | 627 broiler chicken samples | 189 isolates (30.1%) |
| Zhu et al. 2019 [59] | 324 pork meat samples | 155 isolates (47.8%) |
| Zishiri et al. 2016 [42] | 200 chicken samples | 102 isolates (51.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavelquesi, S.L.S.; de Oliveira Ferreira, A.C.A.; Rodrigues, A.R.M.; de Souza Silva, C.M.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. https://doi.org/10.3390/antibiotics10111314
Pavelquesi SLS, de Oliveira Ferreira ACA, Rodrigues ARM, de Souza Silva CM, Orsi DC, da Silva ICR. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics. 2021; 10(11):1314. https://doi.org/10.3390/antibiotics10111314
Chicago/Turabian StylePavelquesi, Sabrina Lunara Santos, Ana Carolina Almeida de Oliveira Ferreira, Angeislenie Ricelle Magalhães Rodrigues, Calliandra Maria de Souza Silva, Daniela Castilho Orsi, and Izabel Cristina Rodrigues da Silva. 2021. "Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review" Antibiotics 10, no. 11: 1314. https://doi.org/10.3390/antibiotics10111314
APA StylePavelquesi, S. L. S., de Oliveira Ferreira, A. C. A., Rodrigues, A. R. M., de Souza Silva, C. M., Orsi, D. C., & da Silva, I. C. R. (2021). Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics, 10(11), 1314. https://doi.org/10.3390/antibiotics10111314






