Characterization of blaKPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China
Abstract
1. Introduction
2. Results and Discussion
2.1. Case Report
2.2. General Features of P. aeruginosa R31
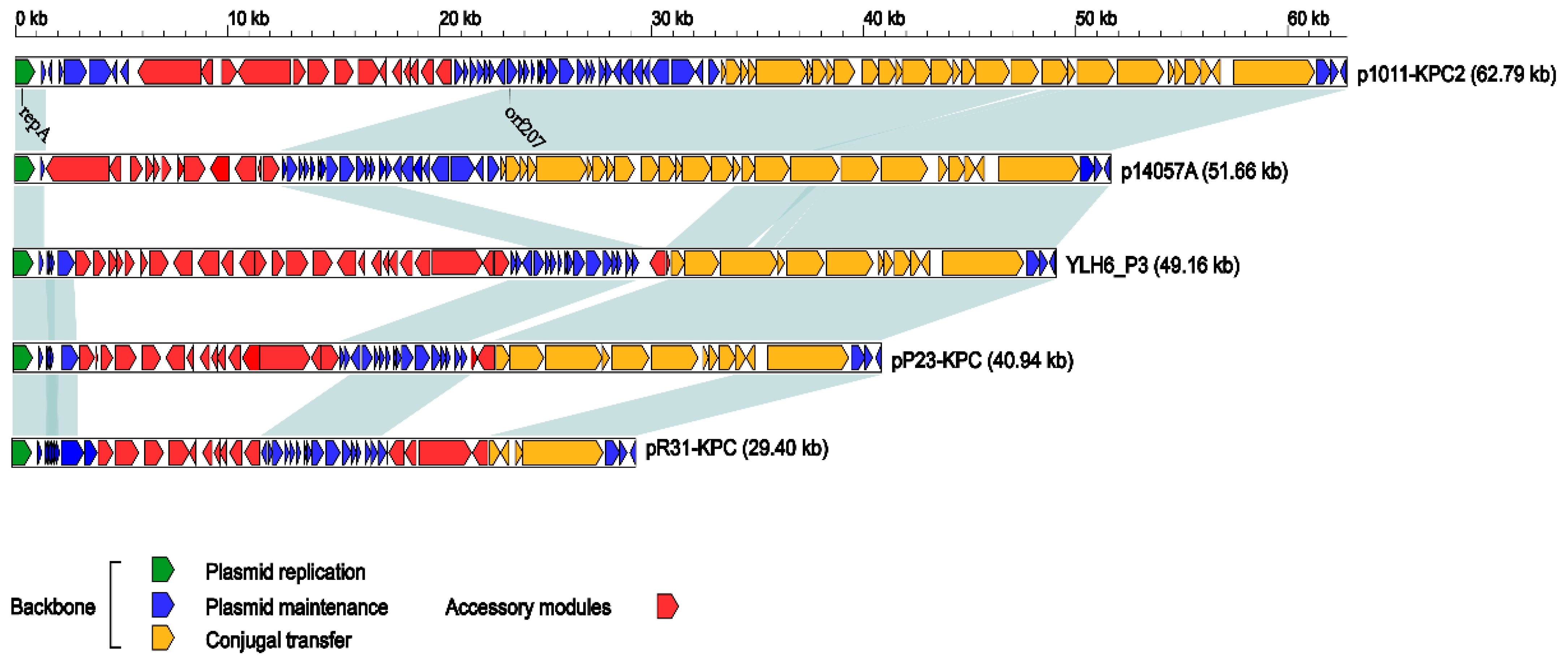
2.3. Overview of pR31-KPC
2.4. The Backbone of pR31-KPC
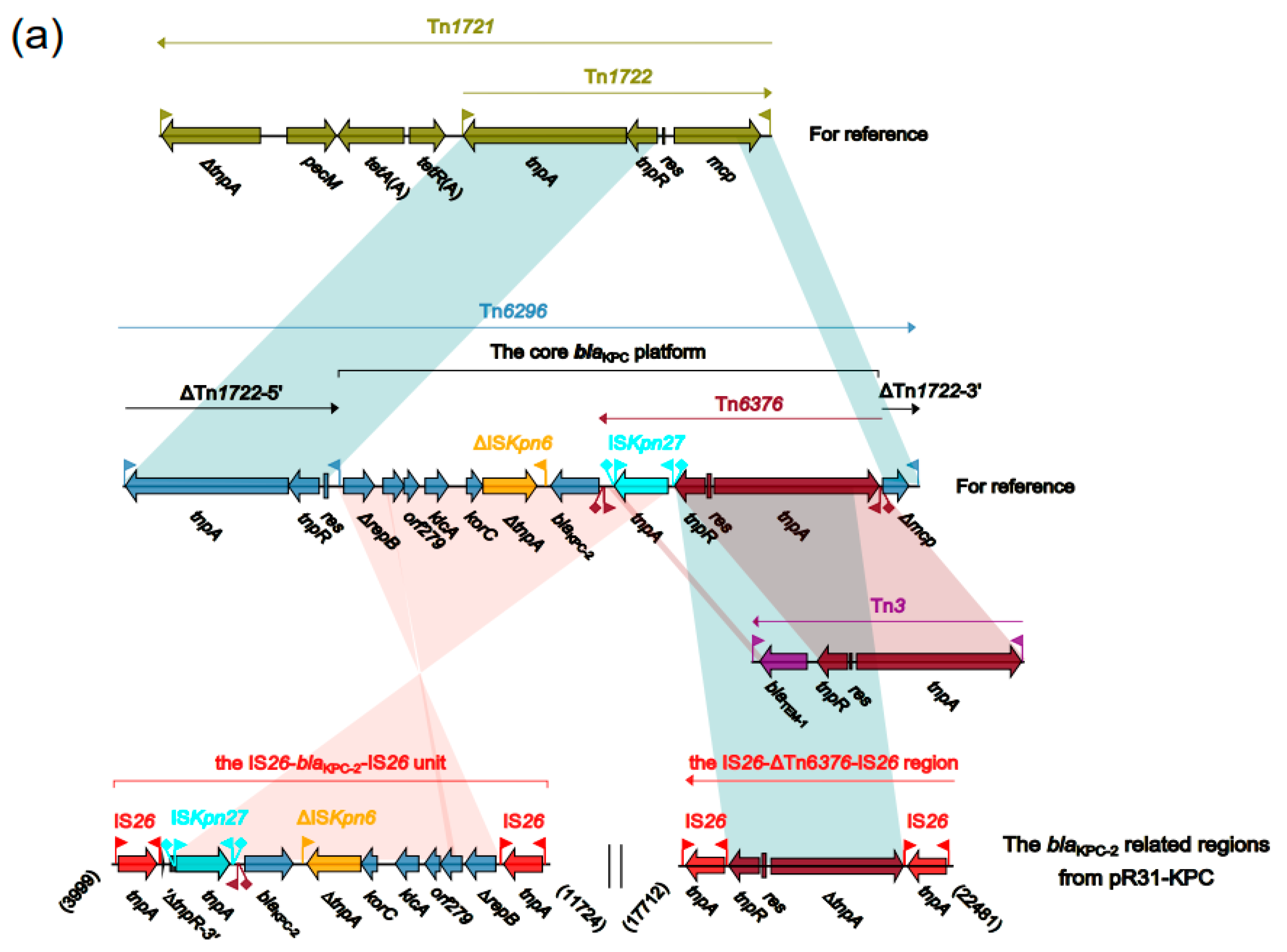
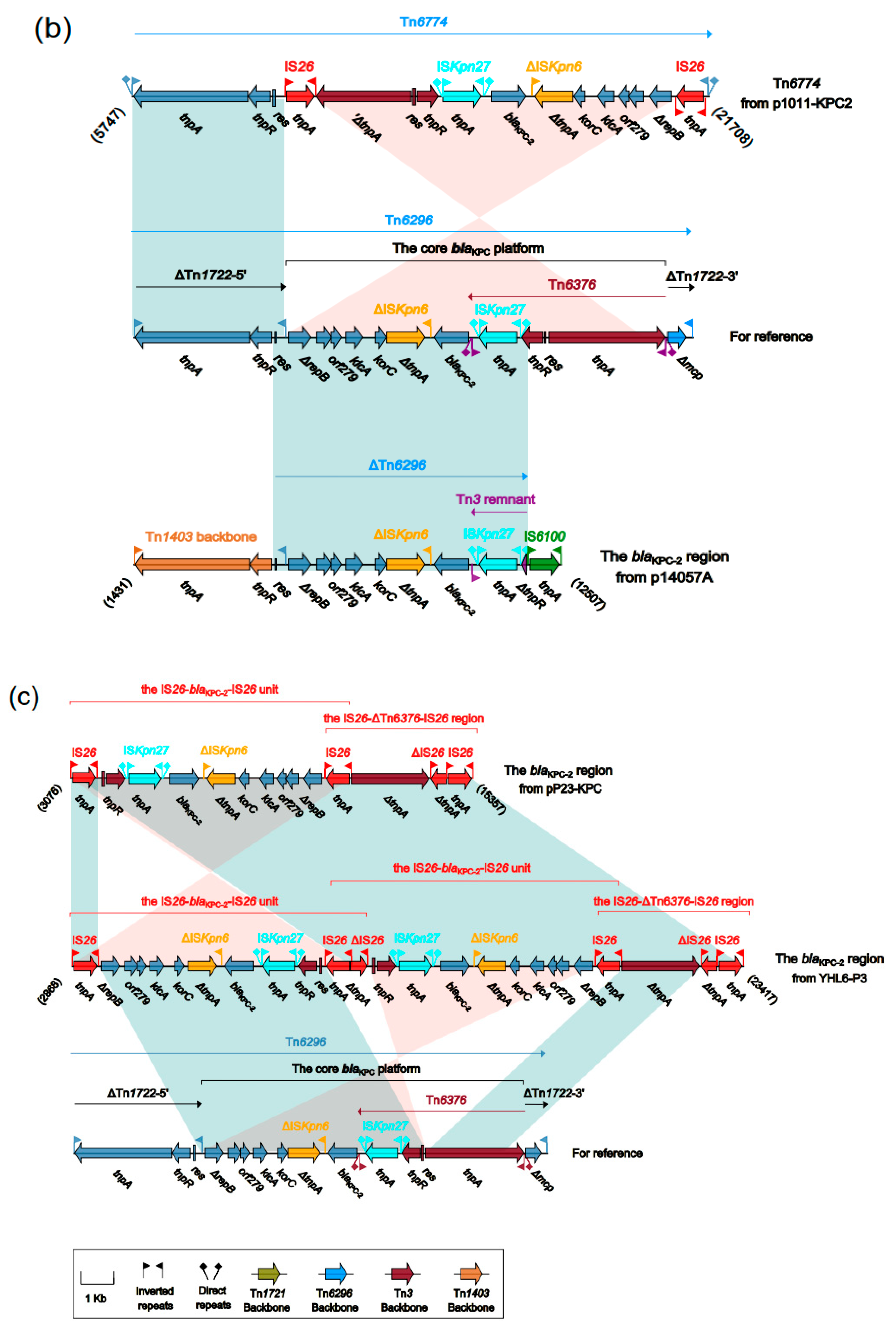
2.5. The Accessory Regions of pR31-KPC
2.6. Genomic Characterization of P. aeruginosa Genomes
3. Materials and Methods
3.1. Ethics Statement
3.2. Identification of Bacterial Strains
3.3. Determination of Minimum Inhibitory Concentration (MIC)
3.4. Detection of Carbapenemase Activity and Screening of Responsible Genes
3.5. Conjugation Experiments
3.6. Determination of R31 Genome Sequences
3.7. Sequence Annotation and Comparison
3.8. Whole Genome Phylogeny and Genetic Background Analysis
3.9. Nucleotide Sequence Accession Number
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.W.; Khan, A.U. Updates on the Pathogenicity Status of Pseudomonas Aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 100640. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Kung, V.L.; Ozer, E.A.; Hauser, A.R. The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2010, 74, 621–641. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Villegas, M.V.; Correa, A.; Quinn, J.P.; Nordmann, P. Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 2011, 55, 5350–5353. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Weldhagen, G.F.; Naas, T.; De Champs, C.; Dove, M.G.; Nordmann, P. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 2001, 45, 2598–2603. [Google Scholar] [CrossRef] [PubMed]
- Senda, K.; Arakawa, Y.; Nakashima, K.; Ito, H.; Ichiyama, S.; Shimokata, K.; Kato, N.; Ohta, M. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 1996, 40, 349–353. [Google Scholar] [CrossRef]
- Lauretti, L.; Riccio, M.L.; Mazzariol, A.; Cornaglia, G.; Amicosante, G.; Fontana, R.; Rossolini, G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999, 43, 1584–1590. [Google Scholar] [CrossRef]
- Murphy, T.A.; Simm, A.M.; Toleman, M.A.; Jones, R.N.; Walsh, T.R. Biochemical characterization of the acquired metallo-beta-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2003, 47, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Jovcic, B.; Lepsanovic, Z.; Suljagic, V.; Rackov, G.; Begovic, J.; Topisirovic, L.; Kojic, M. Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 2011, 55, 3929–3931. [Google Scholar] [CrossRef] [PubMed]
- Leiros, H.K.; Borra, P.S.; Brandsdal, B.O.; Edvardsen, K.S.; Spencer, J.; Walsh, T.R.; Samuelsen, O. Crystal structure of the mobile metallo-beta-lactamase AIM-1 from Pseudomonas aeruginosa: Insights into antibiotic binding and the role of Gln157. Antimicrob. Agents Chemother. 2012, 56, 4341–4353. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Chung, H.S.; Lee, Y.; Yong, D.; Jeong, S.H.; Lee, K.; Chong, Y. Comparison of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry assay with conventional methods for detection of IMP-6, VIM-2, NDM-1, SIM-1, KPC-1, OXA-23, and OXA-51 carbapenemase-producing Acinetobacter spp., Pseudomonas aeruginosa, and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2013, 77, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Pollini, S.; Maradei, S.; Pecile, P.; Olivo, G.; Luzzaro, F.; Docquier, J.D.; Rossolini, G.M. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 2013, 57, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Pfennigwerth, N.; Lange, F.; Belmar Campos, C.; Hentschke, M.; Gatermann, S.G.; Kaase, M. Genetic and biochemical characterization of HMB-1, a novel subclass B1 metallo-beta-lactamase found in a Pseudomonas aeruginosa clinical isolate. J. Antimicrob. Chemother. 2017, 72, 1068–1073. [Google Scholar] [PubMed]
- Boyd, D.A.; Lisboa, L.F.; Rennie, R.; Zhanel, G.G.; Dingle, T.C.; Mulvey, M.R. Identification of a novel metallo-beta-lactamase, CAM-1, in clinical Pseudomonas aeruginosa isolates from Canada. J. Antimicrob. Chemother. 2019, 74, 1563–1567. [Google Scholar] [CrossRef]
- Sevillano, E.; Gallego, L.; Garcia-Lobo, J.M. First detection of the OXA-40 carbapenemase in P. aeruginosa isolates, located on a plasmid also found in A. baumannii. Pathol. Biol. 2009, 57, 493–495. [Google Scholar] [CrossRef]
- Borah, V.V.; Saikia, K.K.; Hazarika, N.K. First report on the detection of OXA-48 beta-lactamase gene in Escherichia coli and Pseudomonas aeruginosa co-infection isolated from a patient in a Tertiary Care Hospital in Assam. Indian J. Med. Microbiol. 2016, 34, 252–253. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Bogaerts, P.; Girlich, D.; Huang, T.D.; Dortet, L.; Glupczynski, Y.; Naas, T. Molecular Characterization of OXA-198 Carbapenemase-Producing Pseudomonas aeruginosa Clinical Isolates. Antimicrob. Agents Chemother. 2018, 62, e02496-17. [Google Scholar] [CrossRef]
- Yoon, E.J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Naas, T.; Cuzon, G.; Villegas, M.V.; Lartigue, M.F.; Quinn, J.P.; Nordmann, P. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob. Agents Chemother. 2008, 52, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Akpaka, P.E.; Swanston, W.H.; Ihemere, H.N.; Correa, A.; Torres, J.A.; Tafur, J.D.; Montealegre, M.C.; Quinn, J.P.; Villegas, M.V. Emergence of KPC-producing Pseudomonas aeruginosa in Trinidad and Tobago. J. Clin. Microbiol. 2009, 47, 2670–2671. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Robledo, I.E.; Aquino, E.E.; Vázquez, G.J. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 2011, 55, 2968–2970. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Wei, Z.; Jiang, Y.; Shen, P.; Yu, Y.; Li, L. Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J. Antimicrob. Chemother. 2011, 66, 1184–1186. [Google Scholar] [CrossRef]
- Jacome, P.R.; Alves, L.R.; Cabral, A.B.; Lopes, A.C.; Maciel, M.A. First report of KPC-producing Pseudomonas aeruginosa in Brazil. Antimicrob. Agents Chemother. 2012, 56, 4990. [Google Scholar] [CrossRef]
- Hagemann, J.B.; Pfennigwerth, N.; Gatermann, S.G.; von Baum, H.; Essig, A. KPC-2 carbapenemase-producing Pseudomonas aeruginosa reaching Germany. J. Antimicrob. Chemother. 2018, 73, 1812–1814. [Google Scholar] [CrossRef]
- Naas, T.; Bonnin, R.A.; Cuzon, G.; Villegas, M.V.; Nordmann, P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013, 68, 1757–1762. [Google Scholar] [CrossRef]
- Dai, X.; Zhou, D.; Xiong, W.; Feng, J.; Luo, W.; Luo, G.; Wang, H.; Sun, F.; Zhou, X. The IncP-6 Plasmid p10265-KPC from Pseudomonas aeruginosa Carries a Novel DeltaISEc33-Associated blaKPC-2 Gene Cluster. Front. Microbiol. 2016, 7, 310. [Google Scholar] [CrossRef]
- Galetti, R.; Andrade, L.N.; Varani, A.M.; Darini, A.L.C. A Phage-Like Plasmid Carrying bla (KPC-2) Gene in Carbapenem-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 572. [Google Scholar] [CrossRef]
- Hu, Y.Y.; Wang, Q.; Sun, Q.L.; Chen, G.X.; Zhang, R. A novel plasmid carrying carbapenem-resistant gene bla(KPC-2) in Pseudomonas aeruginosa. Infect. Drug Resist. 2019, 12, 1285–1288. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liang, Q.; Feng, J.; Zhan, Z.; Zhao, Y.; Yang, W.; Yang, H.; Chen, Y.; Huang, M.; Tong, Y.; et al. Coexistence of two novel resistance plasmids, bla(KPC-2)-arrying p14057A and tetA(A)-carrying p14057B, in Pseudomonas aeruginosa. Virulence 2018, 9, 306–311. [Google Scholar] [CrossRef]
- Sun, F.; Zhou, D.; Wang, Q.; Feng, J.; Feng, W.; Luo, W.; Liu, Y.; Qiu, X.; Yin, Z.; Xia, P. Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J. Antimicrob. Chemother. 2016, 71, 909–912. [Google Scholar] [CrossRef]
- Xiong, J.; Alexander, D.C.; Ma, J.H.; Deraspe, M.; Low, D.E.; Jamieson, F.B.; Roy, P.H. Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob. Agents Chemother. 2013, 57, 3775–3782. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, P.; Merlino, J.; Labbate, M.; Cheong, E.Y.; Gottlieb, T.; Stokes, H.W. Tn6060, a transposon from a genomic island in a Pseudomonas aeruginosa clinical isolate that includes two class 1 integrons. Antimicrob. Agents Chemother. 2009, 53, 5294–5296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Pilato, V.; Pollini, S.; Rossolini, G.M. Tn6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaVIM-1 metallo-beta-lactamase gene in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 1583–1587. [Google Scholar] [CrossRef]
- Coyne, S.; Courvalin, P.; Galimand, M. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 2010, 156, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Toleman, M.A.; Walsh, T.R. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 2011, 35, 912–935. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot., the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Varani, A.; Perochon, J.; Chandler, M. Exploring bacterial insertion sequences with ISfinder: Objectives, uses, and future developments. Methods Mol. Biol. 2012, 859, 91–103. [Google Scholar]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Moura, A.; Soares, M.; Pereira, C.; Leitao, N.; Henriques, I.; Correia, A. INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 2009, 25, 1096–1098. [Google Scholar] [CrossRef]
- Tansirichaiya, S.; Rahman, M.A.; Roberts, A.P. The Transposon Registry. Mob. DNA 2019, 10, 40. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Gardner, S.N.; Slezak, T.; Hall, B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015, 31, 2877–2878. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
| MIC Values | MIC Breakpoints µg/mL | ||||
|---|---|---|---|---|---|
| µg/mL | R or S | S | I | R | |
| Piperacillin | >1024 | R | ≤16 | 32–64 | ≥128 |
| Piperacillin/tazobactam | >1024/4 | R | ≤16/4 | 32/4–64/4 | ≥128/4 |
| Ceftazidime | 128 | R | ≤8 | 16 | ≥32 |
| Cefepime | >512 | R | ≤8 | 16 | ≥32 |
| Imipenem | >128 | R | ≤2 | 4 | ≥8 |
| Meropenem | >128 | R | ≤2 | 4 | ≥8 |
| Aztreonam | >512 | R | ≤8 | 16 | ≥32 |
| Gentamicin | 4 | S | ≤4 | 8 | ≥16 |
| Amikacin | <8 | S | ≤16 | 32 | ≥64 |
| Tobramycin | <2 | S | ≤4 | 8 | ≥16 |
| Ciprofloxacin | 8 | R | ≤0.5 | 1 | ≥2 |
| Levofloxacin | 32 | R | ≤1 | 2 | ≥4 |
| Colistin | 1 | S | ≤2 | - | ≥4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, M.; Guan, H.; Sha, D.; Cao, W.; Song, X.; Che, J.; Kan, B.; Li, J. Characterization of blaKPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China. Antibiotics 2021, 10, 1234. https://doi.org/10.3390/antibiotics10101234
Yuan M, Guan H, Sha D, Cao W, Song X, Che J, Kan B, Li J. Characterization of blaKPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China. Antibiotics. 2021; 10(10):1234. https://doi.org/10.3390/antibiotics10101234
Chicago/Turabian StyleYuan, Min, Hongxia Guan, Dan Sha, Wenting Cao, Xiaofeng Song, Jie Che, Biao Kan, and Juan Li. 2021. "Characterization of blaKPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China" Antibiotics 10, no. 10: 1234. https://doi.org/10.3390/antibiotics10101234
APA StyleYuan, M., Guan, H., Sha, D., Cao, W., Song, X., Che, J., Kan, B., & Li, J. (2021). Characterization of blaKPC-2-Carrying Plasmid pR31-KPC from a Pseudomonas aeruginosa Strain Isolated in China. Antibiotics, 10(10), 1234. https://doi.org/10.3390/antibiotics10101234






