RIP3 Associates with RIP1, TRIF, MAVS, and Also IRF3/7 in Host Innate Immune Signaling in Large Yellow Croaker Larimichthys crocea
Abstract
:1. Introduction
2. Results
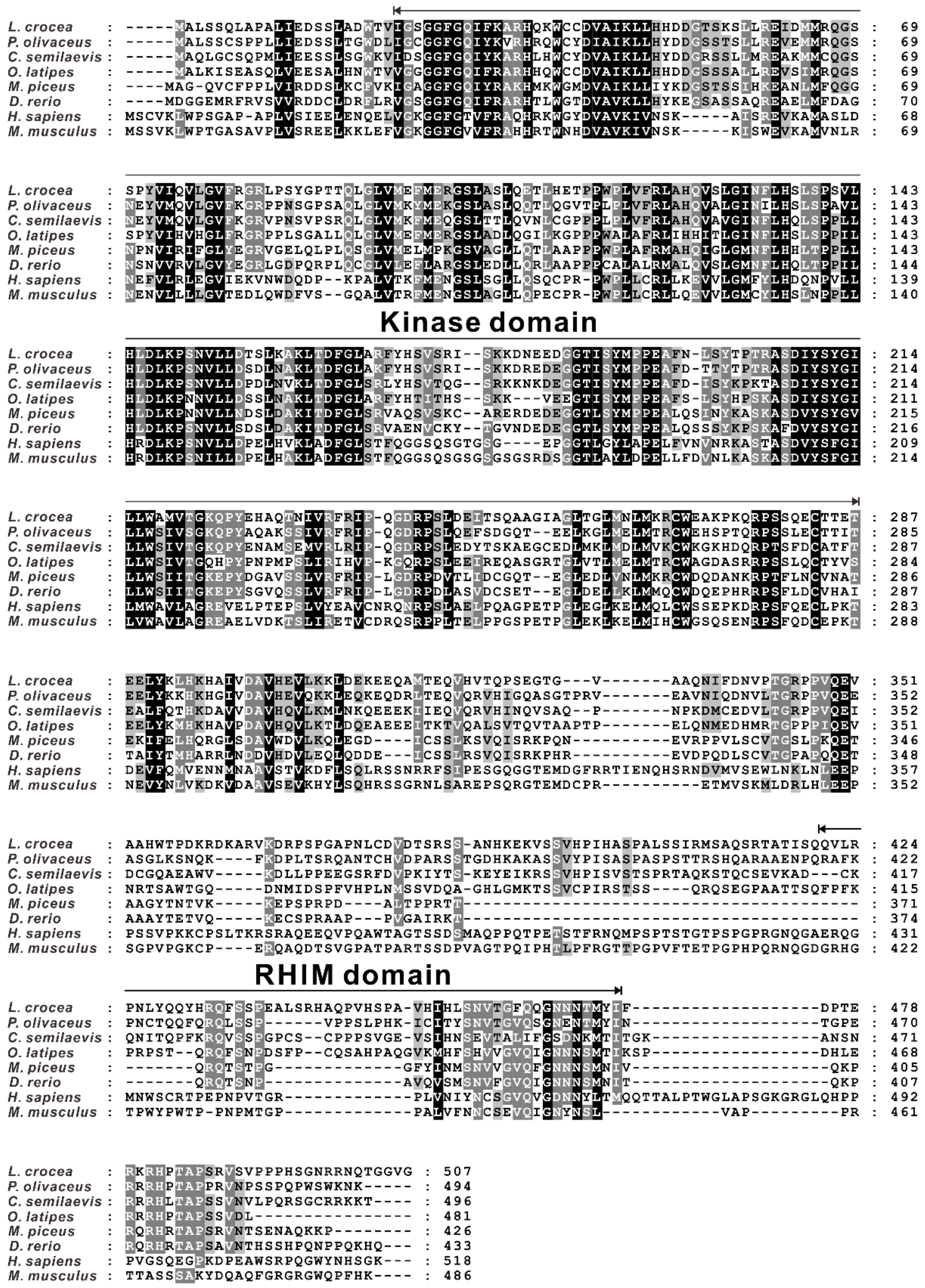
2.1. Identification and Sequence Analysis of RIP3 in Large Yellow Croakers
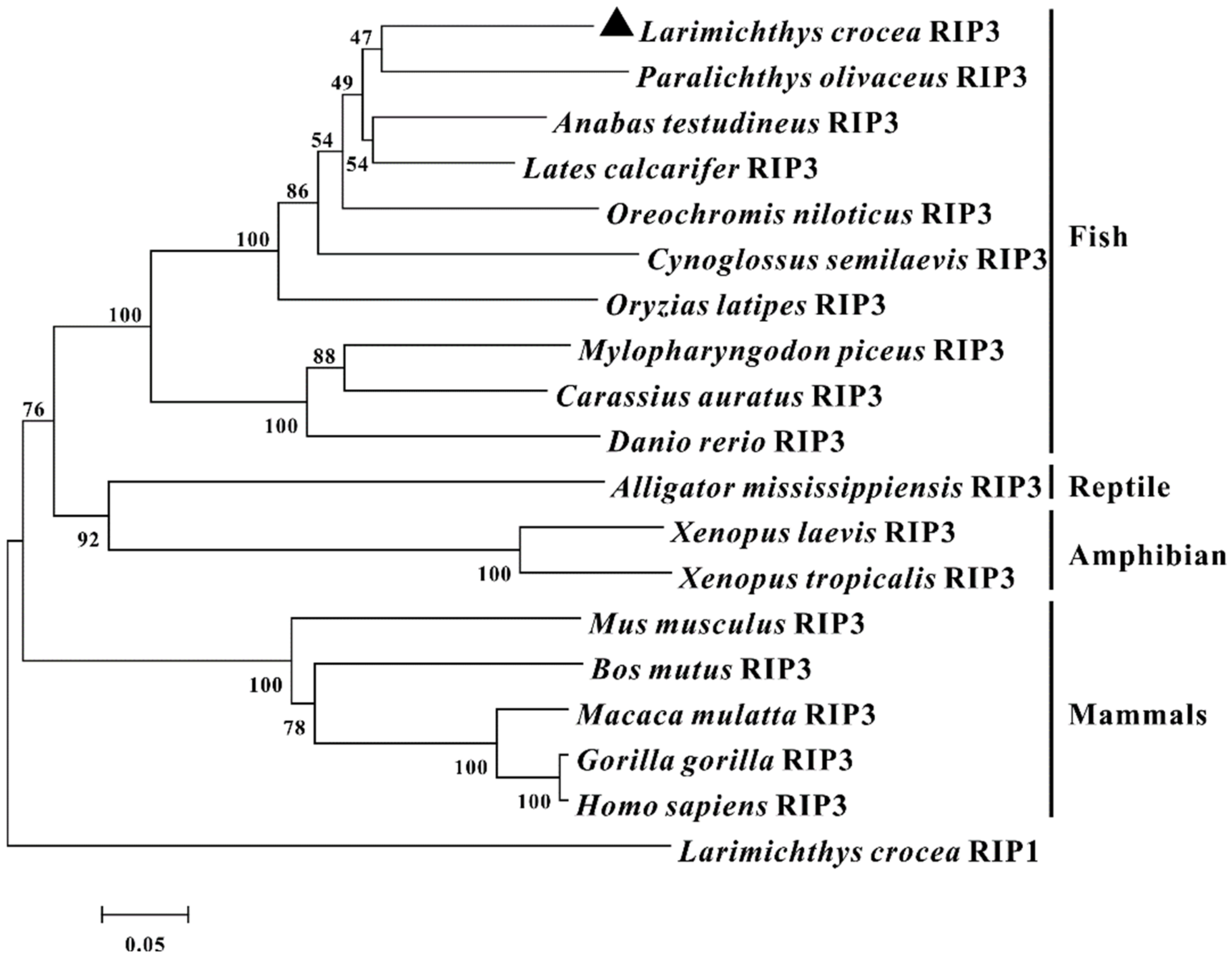
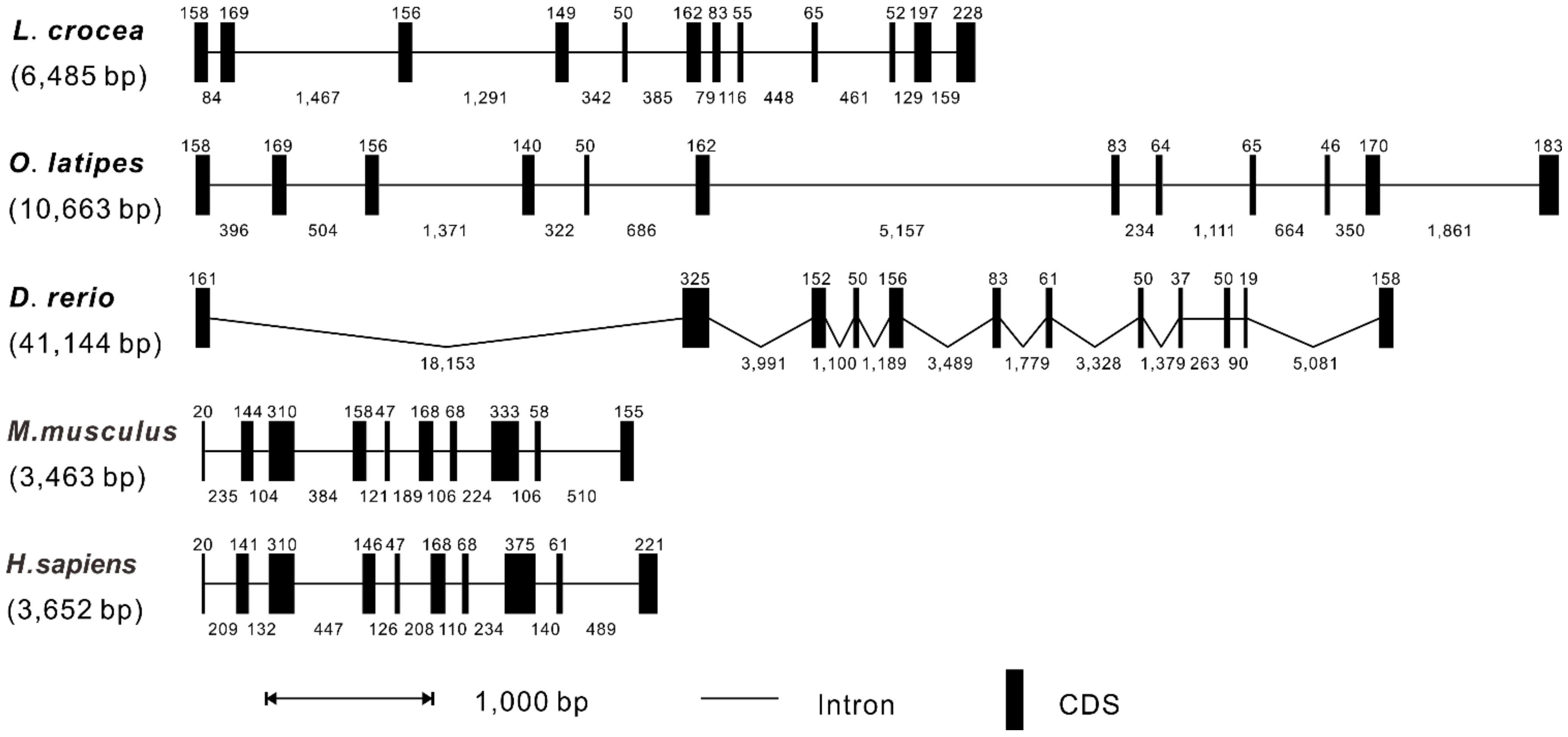
2.2. Genomic Organization of RIP3 Genes in Vertebrates
2.3. Subcellular Localization of Lc-RIP3
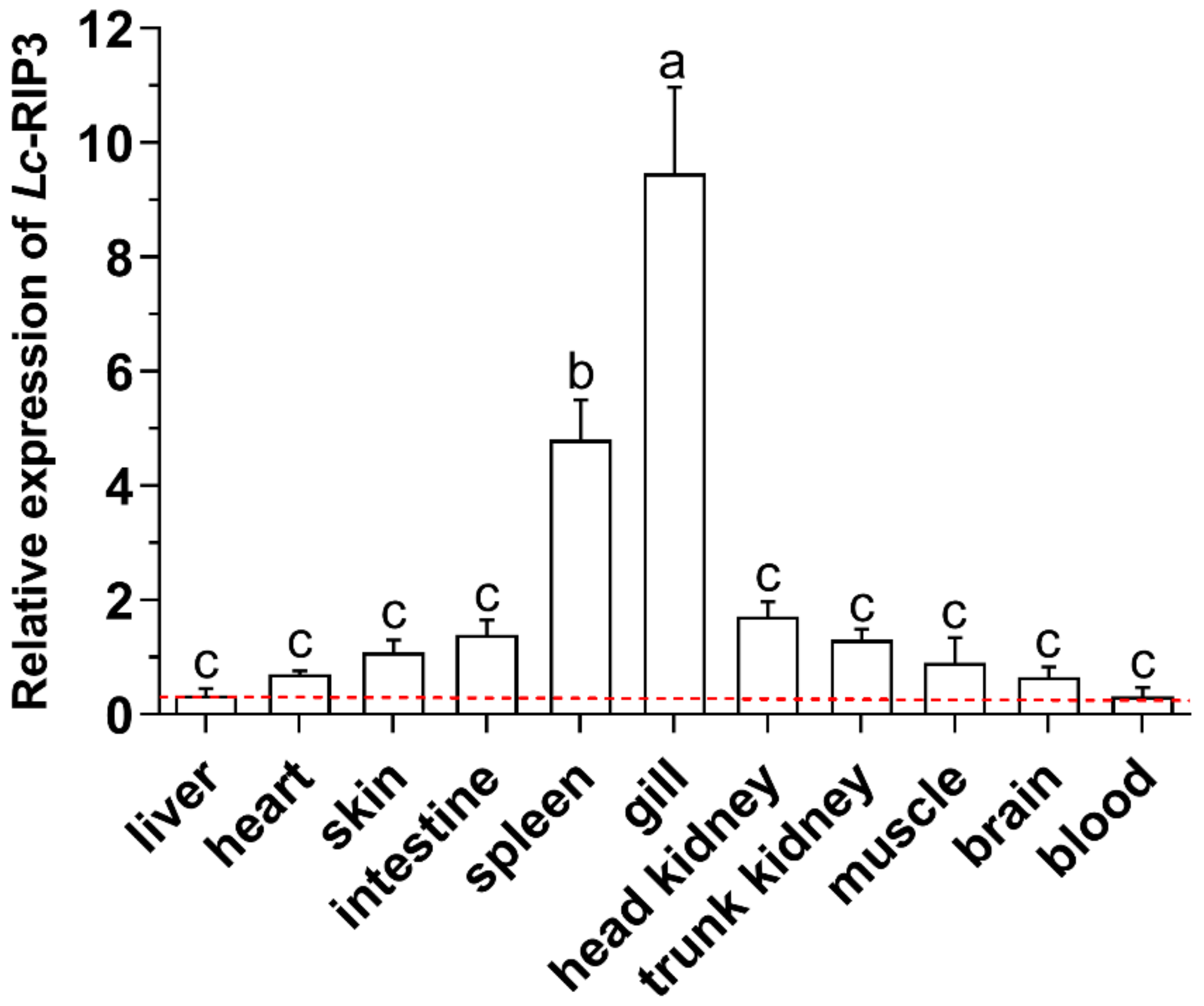
2.4. Expression Pattern Analysis of Lc-RIP3 in Organs/Tissues
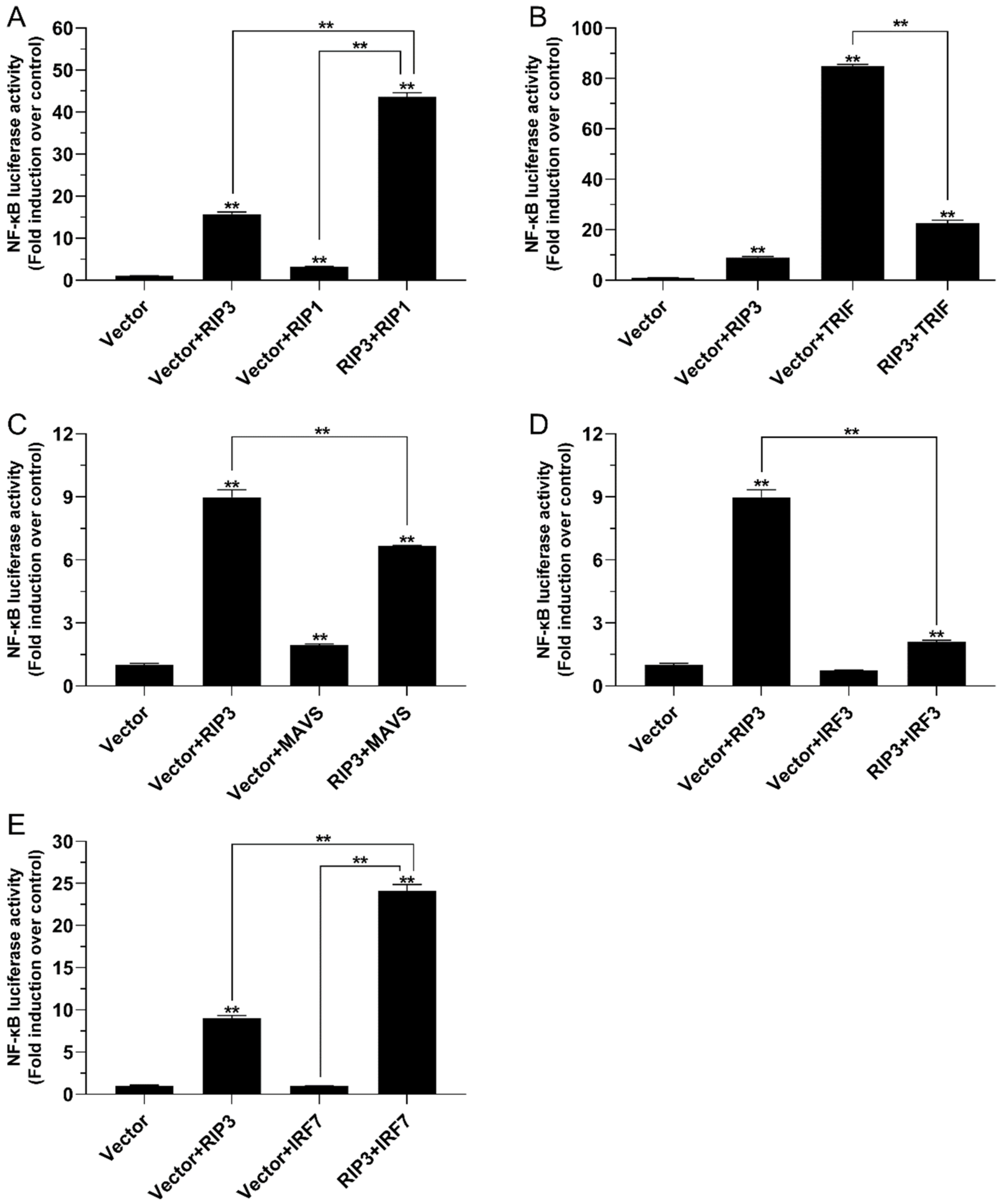
2.5. The Role of Lc-RIP3 in the Regulation of NF-κB Signaling
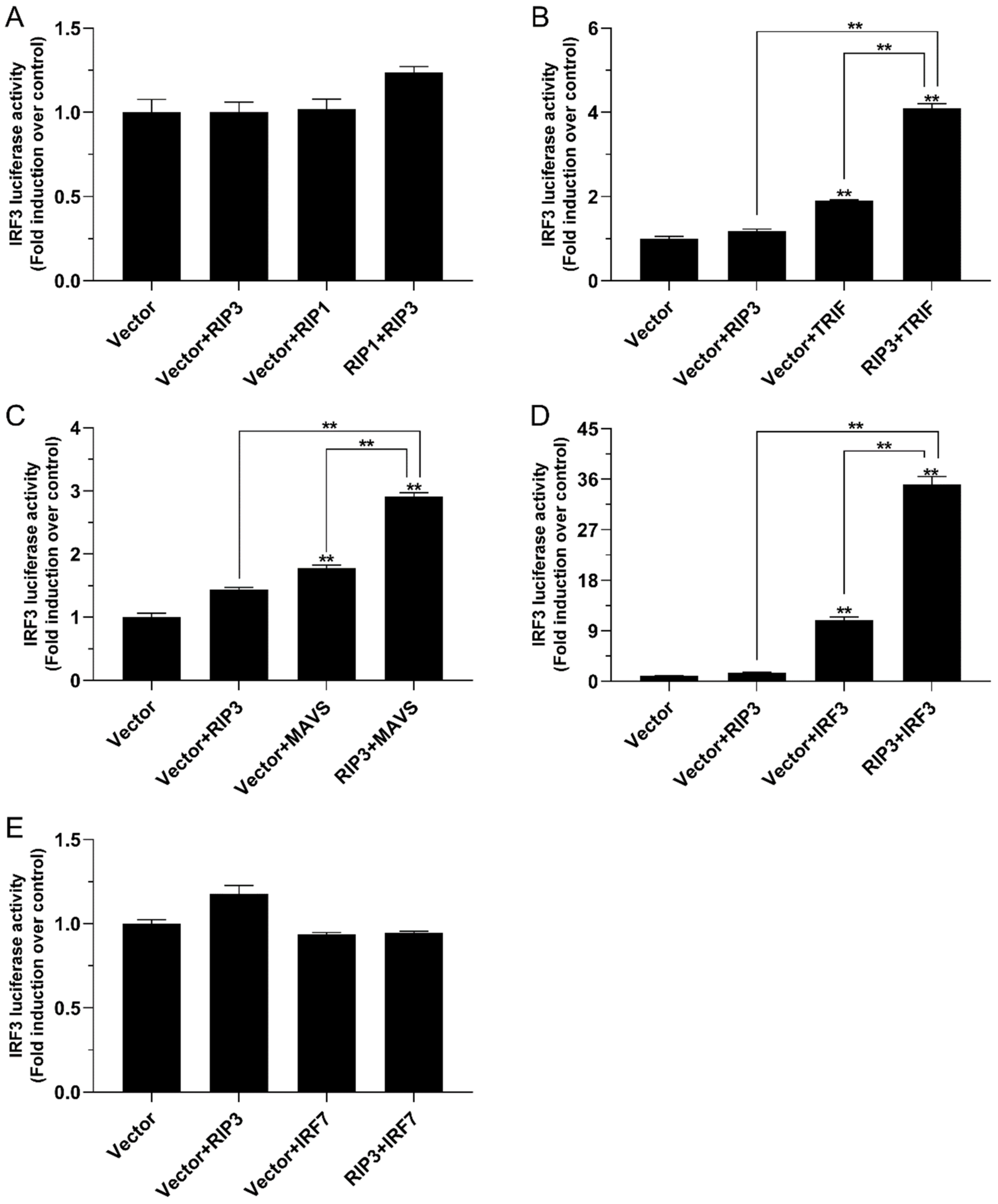
2.6. The Role of Lc-RIP3 in the Regulation of IRF3 Signaling
3. Discussion
4. Materials and Methods
4.1. Fish, Cell Lines, and Transfection
4.2. Gene Cloning and Construction of Plasmids
4.3. Immune Stimulation and qRT-PCR
4.4. Bioinformatics Analysis
4.5. Confocal Microscopy
4.6. Luciferase Activity Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Gan, Z.; Chen, S.N.; Huang, B.; Zou, J.; Nie, P. Fish type I and type II interferons: Composition, receptor usage, production and function. Rev. Aquac. 2020, 12, 773–804. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.Y.; Ding, L.G.; Huang, Z.Y.; Xu, H.Y.; Xu, Z. Commensal bacteria-immunity crosstalk shapes mucosal homeostasis in teleost fish. Rev. Aquac. 2021, 13, 2322–2343. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Dolasia, K.; Bisht, M.K.; Pradhan, G.; Udgata, A.; Mukhopadhyay, S. TLRs/NLRs: Shaping the landscape of host immunity. Int. Rev. Immunol. 2018, 37, 3–19. [Google Scholar] [CrossRef]
- He, S.; Wang, X. RIP kinases as modulators of inflammation and immunity. Nat. Immunol. 2018, 19, 912–922. [Google Scholar] [CrossRef]
- Silke, J.; Rickard, J.A.; Gerlic, M. The diverse role of RIP kinases in necroptosis and inflammation. Nat. Immunol. 2015, 16, 689–697. [Google Scholar] [CrossRef]
- Newton, K. RIPK1 and RIPK3: Critical regulators of inflammation and cell death. Trends Cell Biol. 2015, 25, 347–353. [Google Scholar] [CrossRef]
- Sun, X.; Lee, J.; Navas, T.; Baldwin, D.T.; Stewart, T.A.; Dixit, V.M. RIP3, a novel apoptosis-inducing kinase. J. Biol. Chem. 1999, 274, 16871–16875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, P.W.; Huang, B.C.; Shen, M.; Quast, J.; Chan, E.; Xu, X.; Nolan, G.P.; Payan, D.G.; Luo, Y. Identification of RIP3, a RIP-like kinase that activates apoptosis and NFkappaB. Curr. Biol. 1999, 9, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, D.; Najjar, M.; Zelic, M.; Shah, S.; Nogusa, S.; Polykratis, A.; Paczosa, M.K.; Gough, P.J.; Bertin, J.; Whalen, M.; et al. Kinase activities of RIPK1 and RIPK3 can direct IFN-beta synthesis induced by lipopolysaccharide. J. Immunol. 2017, 198, 4435–4447. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, K.E.; Khan, N.; Mildenhall, A.; Gerlic, M.; Croker, B.A.; D’Cruz, A.A.; Hall, C.; Kaur Spall, S.; Anderton, H.; Masters, S.L.; et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015, 6, 6282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viringipurampeer, I.A.; Shan, X.; Gregory-Evans, K.; Zhang, J.P.; Mohammadi, Z.; Gregory-Evans, C.Y. Rip3 knockdown rescues photoreceptor cell death in blind pde6c zebrafish. Cell Death Differ. 2014, 21, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Yang, H.; Zhao, L.; Luo, S.; Zhang, H.; Chen, S. Structural and functional conservation of half-smooth tongue sole Cynoglossus semilaevis RIP3 in cell death signalling. Fish Shellfish Immunol. 2018, 82, 573–578. [Google Scholar] [CrossRef]
- Dai, Y.; Cao, Y.; Chen, Z.; Huang, J.; Xiao, J.; Zou, J.; Feng, H. RIPK3 collaborates with RIPK1 to inhibit MAVS-mediated signaling during black carp antiviral innate immunity. Fish Shellfish Immunol. 2021, 115, 142–149. [Google Scholar] [CrossRef]
- Chen, X.H.; Lin, K.B.; Wang, X.W. Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J. Fish Dis. 2003, 26, 615–619. [Google Scholar] [CrossRef]
- Zhang, J.T.; Zhou, S.M.; An, S.W.; Chen, L.; Wang, G.L. Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, Pseudomonas plecoglossicida. J. Fish Dis. 2014, 37, 113–121. [Google Scholar] [CrossRef]
- Yin, F.; Gong, H.; Ke, Q.; Li, A. Stress, antioxidant defence and mucosal immune responses of the large yellow croaker Pseudosciaena crocea challenged with Cryptocaryon irritans. Fish Shellfish Immunol. 2015, 47, 344–351. [Google Scholar] [CrossRef]
- Zou, P.F.; Shen, J.J.; Li, Y.; Yan, Q.; Zou, Z.H.; Zhang, Z.P.; Wang, Y.L. Molecular cloning and functional characterization of TRIF in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2019, 91, 108–121. [Google Scholar] [CrossRef]
- Zou, P.F.; Tang, J.C.; Li, Y.; Feng, J.J.; Zhang, Z.P.; Wang, Y.L. MAVS splicing variants associated with TRAF3 and TRAF6 in NF-kappaB and IRF3 signaling pathway in large yellow croaker Larimichthys crocea. Dev. Comp. Immunol. 2021, 121, 104076. [Google Scholar] [CrossRef]
- Upton, J.W.; Shubina, M.; Balachandran, S. RIPK3-driven cell death during virus infections. Immunol. Rev. 2017, 277, 90–101. [Google Scholar] [CrossRef]
- Orozco, S.; Oberst, A. RIPK3 in cell death and inflammation: The good, the bad, and the ugly. Immunol. Rev. 2017, 277, 102–112. [Google Scholar] [CrossRef] [Green Version]
- Zou, P.F.; Shen, J.J.; Li, Y.; Zhang, Z.P.; Wang, Y.L. TRAF3 enhances TRIF-mediated signaling via NF-kappaB and IRF3 activation in large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2020, 97, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ao, J.; Chen, X. Comparative study of interleukin-17C (IL-17C) and IL-17D in large yellow croaker Larimichthys crocea reveals their similar but differential functional activity. Dev. Comp. Immunol. 2017, 76, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Mu, Y.; Ding, Y.; Ao, J.; Chen, X. Molecular and functional characterization of Toll-like receptor 21 in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2016, 59, 179–188. [Google Scholar] [CrossRef]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pazdernik, N.J.; Donner, D.B.; Goebl, M.G.; Harrington, M.A. Mouse receptor interacting protein 3 does not contain a caspase-recruiting or a death domain but induces apoptosis and activates NF-kappaB. Mol. Cell. Biol. 1999, 19, 6500–6508. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.W.; Lin, J.A.; Han, J.H. Receptor-interacting protein (RIP) kinase family. Cell. Mol. Immunol. 2010, 7, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Ting, A.T.; Pimentel-Muiños, F.X.; Seed, B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappa B but not Fas/APO-1-initiated apoptosis. EMBO J. 1996, 15, 6189–6196. [Google Scholar] [CrossRef]
- He, S.; Liang, Y.; Shao, F.; Wang, X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 20054–20059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gay, N.J.; Symmons, M.F.; Gangloff, M.; Bryant, C.E. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014, 14, 546–558. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Bowie, A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef]
- Ji, J.; Rao, Y.; Wan, Q.; Liao, Z.; Su, J. Teleost-specific TLR19 localizes to endosome, recognizes dsRNA, recruits TRIF, triggers both IFN and NF-kappaB pathways, and protects cells from grass carp reovirus infection. J. Immunol. 2018, 200, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, A.; Oshiumi, H.; Tsujita, T.; Mitani, H.; Kasai, H.; Yoshimizu, M.; Matsumoto, M.; Seya, T. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 2008, 181, 3474–3485. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.N.; Zou, P.F.; Nie, P. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) in fish: Current knowledge and future perspectives. Immunology 2017, 151, 16–25. [Google Scholar] [CrossRef]
- Mogensen, T.H. IRF and STAT transcription factors—From basic biology to roles in infection, protective immunity, and primary immunodeficiencies. Front. Immunol. 2018, 9, 3047. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef] [Green Version]
- Zou, P.F.; Chang, M.X.; Li, Y.; Xue, N.N.; Li, J.H.; Chen, S.N.; Nie, P. NOD2 in zebrafish functions in antibacterial and also antiviral responses via NF-kappaB, and also MDA5, RIG-I and MAVS. Fish Shellfish Immunol. 2016, 55, 173–185. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Common Name | Scientific Name | Accession No. | Length (aa) | Identity | Similarity |
|---|---|---|---|---|---|
| Japanese flounder | Paralichthys olivaceus | XP_019967771.1 | 494 | 61% | 74% |
| Half-smooth tongue sole | Cynoglossus semilaevis | XP_008316289.1 | 496 | 52% | 67% |
| Medaka | Oryzias latipes | XP_023821938.1 | 481 | 50% | 66% |
| Black carp | Mylopharyngodon piceus | QXJ87326.1 | 426 | 37% | 55% |
| Zebrafish | Danio rerio | XP_001343827.1 | 433 | 36% | 52% |
| Human | Homo sapiens | NP_006862.2 | 518 | 23% | 40% |
| Mouse | Mus musculus | NP_064339.2 | 486 | 24% | 39% |
| Primer Name | Sequence (5′–3′) | Application |
|---|---|---|
| Lc-RIP3-F | ATGGCACTGTCCAGCCAGCT | Lc-RIP3 ORF cloning |
| Lc-RIP3-R | GCCAACTCCTCCTGTTTGGTTC | |
| pcDNA3.1-RIP3-F | CGGGATCCATGGCACTGTCCAGCCAGCT | pcDNA3.1-RIP3 |
| pcDNA3.1-RIP3-R | GGGGTACCGCCAACTCCTCCTGTTTGGTTC | |
| pcDNA3.1-TRIF-F | CCGCTCGAGCGATGGCTAGCCGCGAGGGAGAAGA | pcDNA3.1-TRIF |
| pcDNA3.1-TRIF-R | CGGGGTACCTTGCTCATCTAAATCATCT | |
| pcDNA3.1-MAVS-F | CGCGGATCCATGGCTTCGTTTGCCAGAGACAG | pcDNA3.1-MAVS |
| pcDNA3.1-MAVS-R | CCCAAGCTTGTTCTTAAACTTCCACG | |
| pcDNA3.1-IRF3-F | CCGGAATTCCAATGGCTTCTCATTCTAAACCTC | pcDNA3.1-IRF3 |
| pcDNA3.1-IRF3-R | CCCAAGCTTGTACAGCTCCATCATCT | |
| pcDNA3.1-IRF7-F | CGCGGATCCATGGCTCAAAGCCCTCCCAAG | pcDNA3.1-IRF7 |
| pcDNA3.1-IRF7-R | CGGGGTACCATAAAGCTCAGCAGCCAG | |
| pTurbo-RIP3-F | GGGGTACCGATGGCACTGTCCAGCCAGCT | pTurbo-RIP3-GFP |
| pTurbo-RIP3-R | CGGGATCCCGGCCAACTCCTCCTGTTTGGT | |
| qRIP3-F | TCAAGGAAGCAGCCCGTATG | qRT-PCR |
| qRIP3-R | GAGCCAGTCTGAACACCAACG | |
| qβ-actin-F | TTATGAAGGCTATGCCCTGCC | qRT-PCR |
| qβ-actin-R | TGAAGGAGTAGCCACGCTCTGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, P.; Li, K.; Li, Y.; Shen, Y.; Zhang, Z.; Wang, Y. RIP3 Associates with RIP1, TRIF, MAVS, and Also IRF3/7 in Host Innate Immune Signaling in Large Yellow Croaker Larimichthys crocea. Antibiotics 2021, 10, 1199. https://doi.org/10.3390/antibiotics10101199
Zou P, Li K, Li Y, Shen Y, Zhang Z, Wang Y. RIP3 Associates with RIP1, TRIF, MAVS, and Also IRF3/7 in Host Innate Immune Signaling in Large Yellow Croaker Larimichthys crocea. Antibiotics. 2021; 10(10):1199. https://doi.org/10.3390/antibiotics10101199
Chicago/Turabian StyleZou, Pengfei, Kaiqing Li, Ying Li, Yingjia Shen, Ziping Zhang, and Yilei Wang. 2021. "RIP3 Associates with RIP1, TRIF, MAVS, and Also IRF3/7 in Host Innate Immune Signaling in Large Yellow Croaker Larimichthys crocea" Antibiotics 10, no. 10: 1199. https://doi.org/10.3390/antibiotics10101199
APA StyleZou, P., Li, K., Li, Y., Shen, Y., Zhang, Z., & Wang, Y. (2021). RIP3 Associates with RIP1, TRIF, MAVS, and Also IRF3/7 in Host Innate Immune Signaling in Large Yellow Croaker Larimichthys crocea. Antibiotics, 10(10), 1199. https://doi.org/10.3390/antibiotics10101199







