Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Organoid Formation, Viability, and Encapsulation in Hydrogel
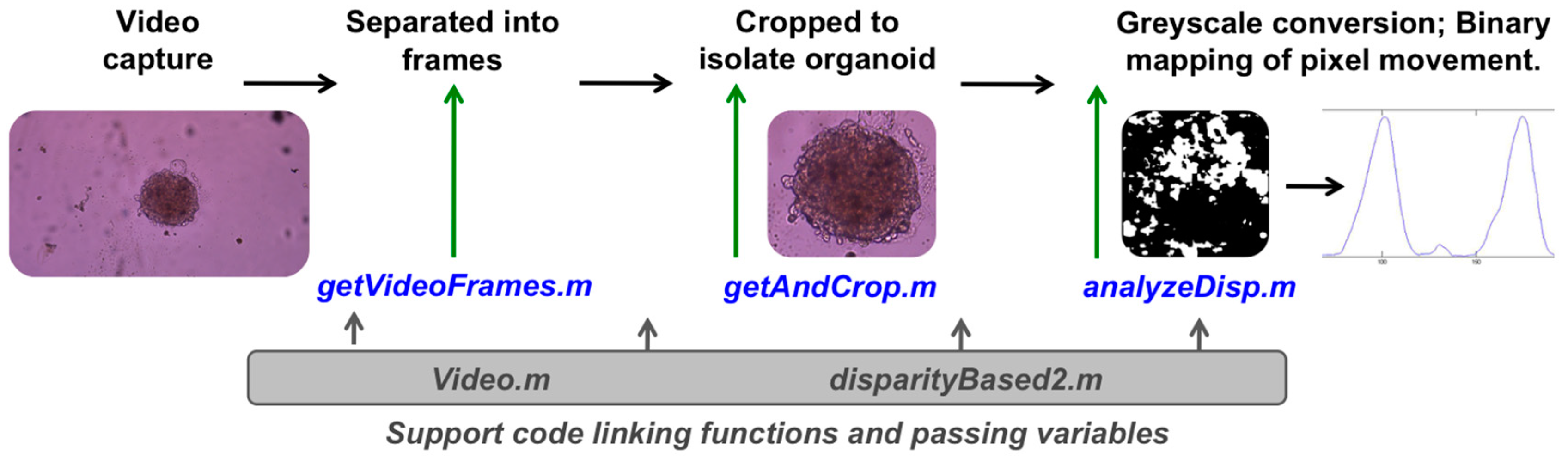
2.2. Software Code
2.3. Beating Kinetics Evaluation and Drug Response Studies
2.4. Statistical Methods
3. Results
3.1. Cardiac Organoid Formation and Initial Assessment
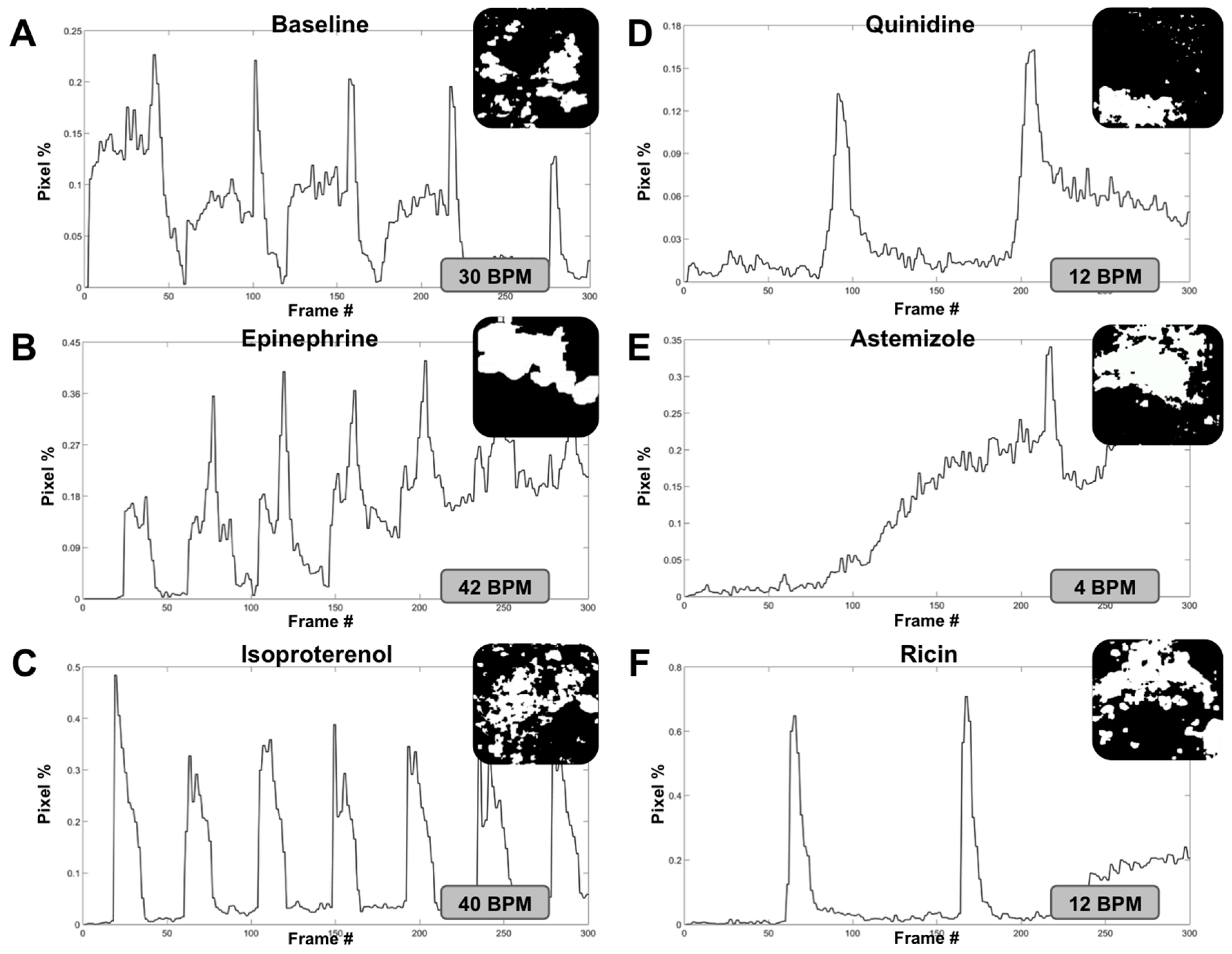
3.2. Optical Tracking and Organoid Heartbeat Quantification
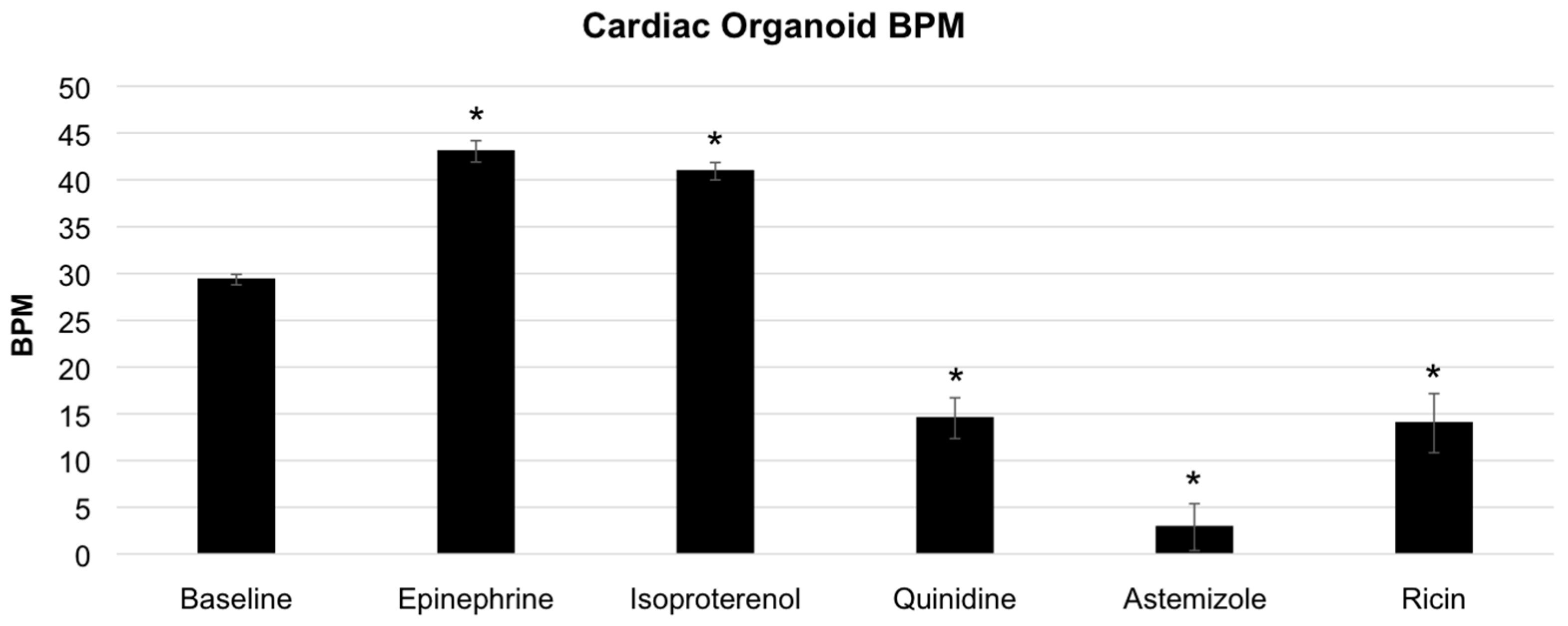
3.3. Modulation of Organoid Beat Rate through Drug and Toxin Exposure
3.4. Accuracy Comparison—Digital Quantification Versus Manual Counting
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Bhise, N.S.; Ribas, J.; Manoharan, V.; Zhang, Y.S.; Polini, A.; Massa, S.; Dokmeci, M.R.; Khademhosseini, A. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 2014, 190, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3d cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, A.M.; Hancock, M.J.; Harrington, H.; Kaji, H.; Khademhosseini, A. Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 2011, 17, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Devarasetty, M.; Soker, S.; Hall, A.R. In situ patterned micro 3d liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 2015, 7, 031001. [Google Scholar] [CrossRef] [PubMed]
- Benam, K.H.; Dauth, S.; Hassell, B.; Herland, A.; Jain, A.; Jang, K.J.; Karalis, K.; Kim, H.J.; MacQueen, L.; Mahmoodian, R.; et al. Engineered in vitro disease models. Annu. Rev. Pathol. 2015, 10, 195–262. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Devarasetty, M.; Forsythe, S.; Atala, A.; Soker, S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. 2016, 113, 2020–2032. [Google Scholar] [CrossRef] [PubMed]
- Vunjak-Novakovic, G.; Bhatia, S.; Chen, C.; Hirschi, K. Heliva platform: Integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res. Ther. 2013, 4 (Suppl. 1), S8. [Google Scholar] [CrossRef] [PubMed]
- Piccini, J.P.; Whellan, D.J.; Berridge, B.R.; Finkle, J.K.; Pettit, S.D.; Stockbridge, N.; Valentin, J.P.; Vargas, H.M.; Krucoff, M.W.; Group, C.H.W. Current challenges in the evaluation of cardiac safety during drug development: Translational medicine meets the critical path initiative. Am. Heart J. 2009, 158, 317–326. [Google Scholar] [CrossRef] [PubMed]
- FDA. Recalls, Market Withdrawals, & Safety Alerts; in archives; Administration UFaD, Ed.; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2012.
- DiMasi, J.A.; Hansen, R.W.; Grabowski, H.G. The price of innovation: New estimates of drug development costs. J. Health Econ. 2003, 22, 151–185. [Google Scholar] [CrossRef]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Aleman, J.; Arneri, A.; Bersini, S.; Piraino, F.; Shin, S.R.; Dokmeci, M.R.; Khademhosseini, A. From cardiac tissue engineering to heart-on-a-chip: Beating challenges. Biomed. Mater. 2015, 10, 034006. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Klos, M.; Bollensdorff, C.; Hou, L.; Ewart, P.; Kamp, T.J.; Zhang, J.; Bizy, A.; Guerrero-Serna, G.; Kohl, P.; et al. Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell-derived cardiac myocyte monolayers. Circ. Res. 2012, 110, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hubner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Polini, A.; Prodanov, L.; Bhise, N.S.; Manoharan, V.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-chip: A new tool for drug discovery. Expert Opin. Drug Discov. 2014, 9, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Materne, E.M.; Brincker, S.; Sussbier, U.; Fradrich, C.; Busek, M.; Sonntag, F.; Sakharov, D.A.; Trushkin, E.V.; Tonevitsky, A.G.; et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3d human liver and skin tissue co-culture. Lab Chip 2013, 13, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Mousavi Shaegh, S.A.; De Ferrari, F.; Zhang, Y.S.; Nabavinia, M.; Binth Mohammad, N.; Ryan, J.; Pourmand, A.; Laukaitis, E.; Banan Sadeghian, R.; Nadhman, A.; et al. A microfluidic optical platform for real-time monitoring of ph and oxygen in microfluidic bioreactors and organ-on-chip devices. Biomicrofluidics 2016, 10, 044111. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.R.; Zhang, Y.S.; Kim, D.J.; Manbohi, A.; Avci, H.; Silvestri, A.; Aleman, J.; Hu, N.; Kilic, T.; Keung, W.; et al. Aptamer-based microfluidic electrochemical biosensor for monitoring cell-secreted trace cardiac biomarkers. Anal. Chem. 2016, 88, 10019–10027. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Smith, A.S.; Prot, J.M.; Oleaga, C.; Hickman, J.J.; Shuler, M.L. How multi-organ microdevices can help foster drug development. Adv. Drug Deliv. Rev. 2014, 69–70, 158–169. [Google Scholar] [CrossRef] [PubMed]

| Condition | Drug Concentration | Exposure Time (min) | Average Video Time (s) | Average Observed Beats | Average Observed Effective Heart Rate (BPM) | Average Digitized Heart Rate (BPM) | Accuracy |
|---|---|---|---|---|---|---|---|
| Baseline | n/a | n/a | 30 | 15 | 30 | 30 | 100% |
| Isoproterenol | 100 uM | 30 | 21 | 14 | 40 | 40 | 100% |
| Epinephrine | 500 nM | 15 | 33 | 23 | 41.8 | 42 | 99.6% |
| Quinidine | 1 uM | 10 | 15 | 4 | 16 | 12 | 75% |
| Astemizole | 100 nM | 30 | 30 | 2 | 4 | 4 | 100% |
| Ricin A Chain | 100 nM | 30 | 30 | 6 | 12 | 12 | 100% |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devarasetty, M.; Forsythe, S.; Shupe, T.D.; Soker, S.; Bishop, C.E.; Atala, A.; Skardal, A. Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors 2017, 7, 24. https://doi.org/10.3390/bios7030024
Devarasetty M, Forsythe S, Shupe TD, Soker S, Bishop CE, Atala A, Skardal A. Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors. 2017; 7(3):24. https://doi.org/10.3390/bios7030024
Chicago/Turabian StyleDevarasetty, Mahesh, Steven Forsythe, Thomas D. Shupe, Shay Soker, Colin E. Bishop, Anthony Atala, and Aleksander Skardal. 2017. "Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids" Biosensors 7, no. 3: 24. https://doi.org/10.3390/bios7030024
APA StyleDevarasetty, M., Forsythe, S., Shupe, T. D., Soker, S., Bishop, C. E., Atala, A., & Skardal, A. (2017). Optical Tracking and Digital Quantification of Beating Behavior in Bioengineered Human Cardiac Organoids. Biosensors, 7(3), 24. https://doi.org/10.3390/bios7030024









