Current and Prospective Methods for Plant Disease Detection
Abstract
:1. Introduction
2. Current Methods for Crop Disease Detection
2.1. Direct Detection Methods
2.1.1. Polymerase Chain Reaction
| Techniques | Limit of Detection (CFU/mL) [12] | Advantages | Limitations |
|---|---|---|---|
| PCR | 103–104 | Mature and common technology, portable, easy to operate. | Effectiveness is subjected to DNA extraction, inhibitors, polymerase activity, concentration of PCR buffer and deoxynucleoside triphosphate. |
| FISH | 103 | High sensitivity. | Autofluorescence, photobleaching. |
| ELISA | 105–106 | Low cost, visual color change can be used for detection. | Low sensitivity for bacteria. |
| IF | 103 | High sensitivity, target distribution can be visualized. | Photobleaching. |
| FCM | 104 | Simultaneous measurement of several parameters, rapid detection. | High cost, overwhelming unnecessary information. |
2.1.2. Fluorescence in-situ Hybridization
2.1.3. Enzyme-Linked Immunosorbent Assay
2.1.4. Immunofluorescence
2.1.5. Flow Cytometry
2.2. Indirect Detection Methods
2.2.1. Thermography
2.2.2. Fluorescence Imaging
2.2.3. Hyperspectral Techniques
2.2.4. Gas Chromatography
3. Detection of Plant Diseases Using Portable Sensors
3.1. Biosensor Platforms Based on Nanomaterials
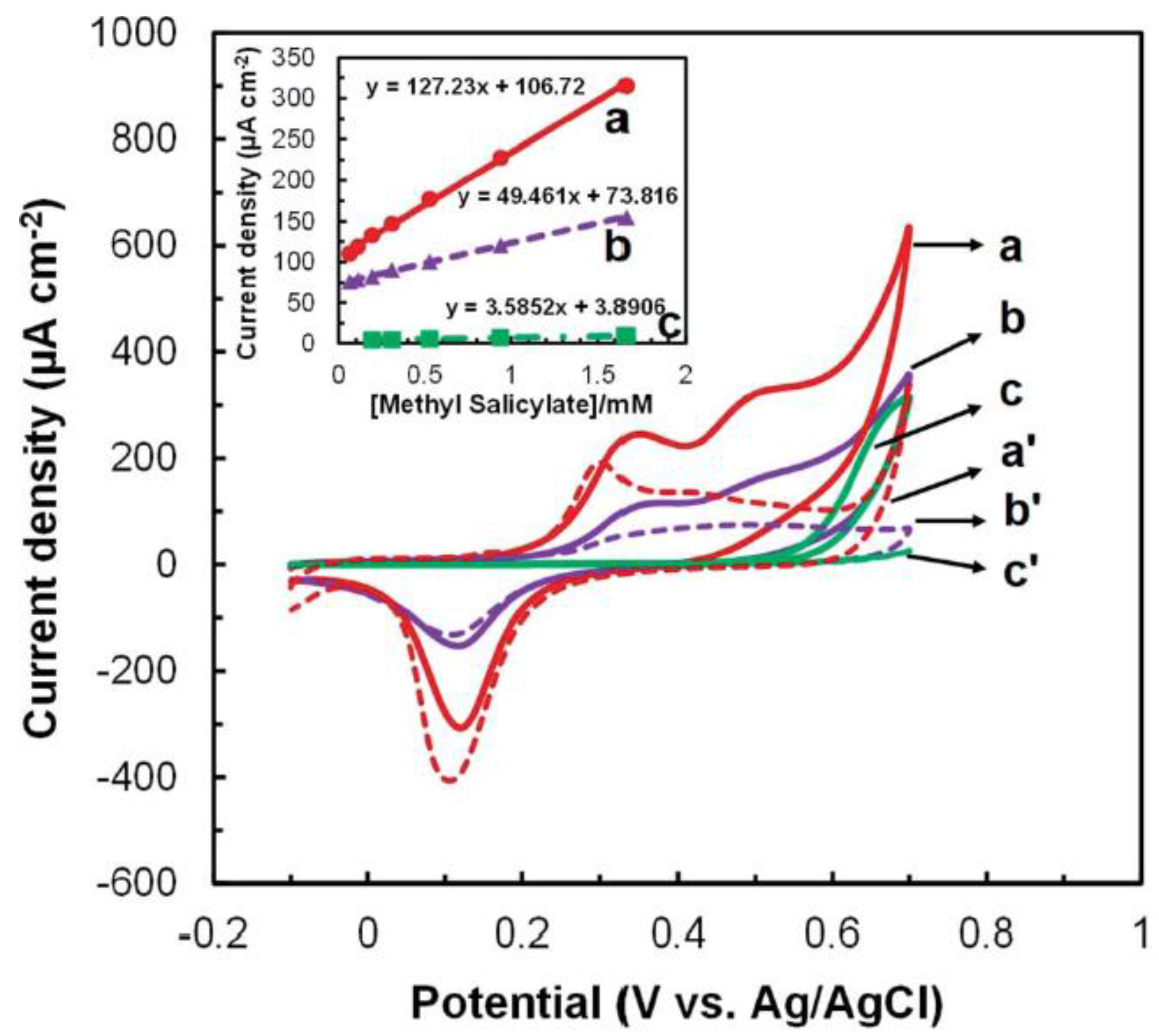
3.2. Affinity Biosensors
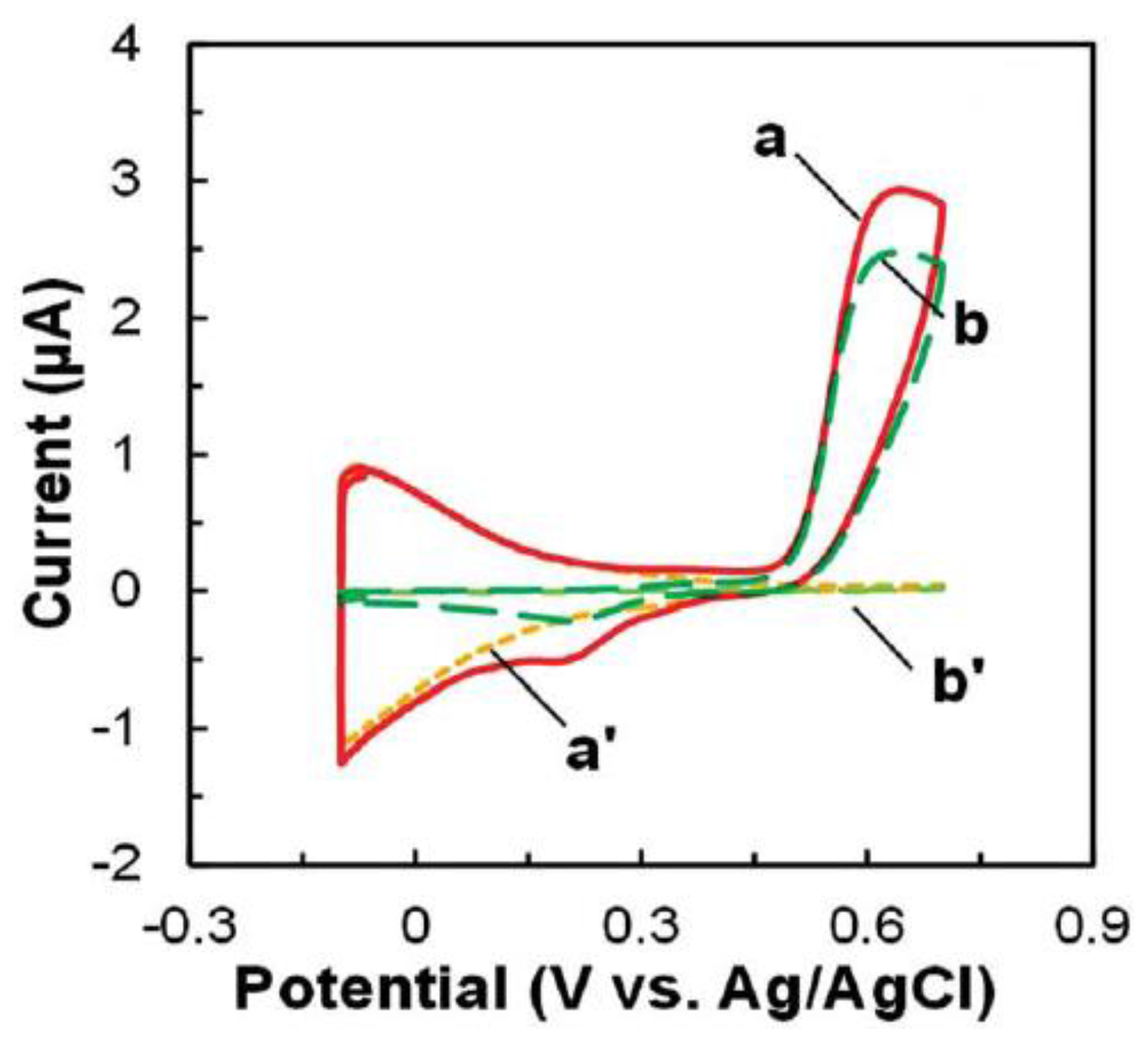
3.2.1. Antibody-Based Biosensors
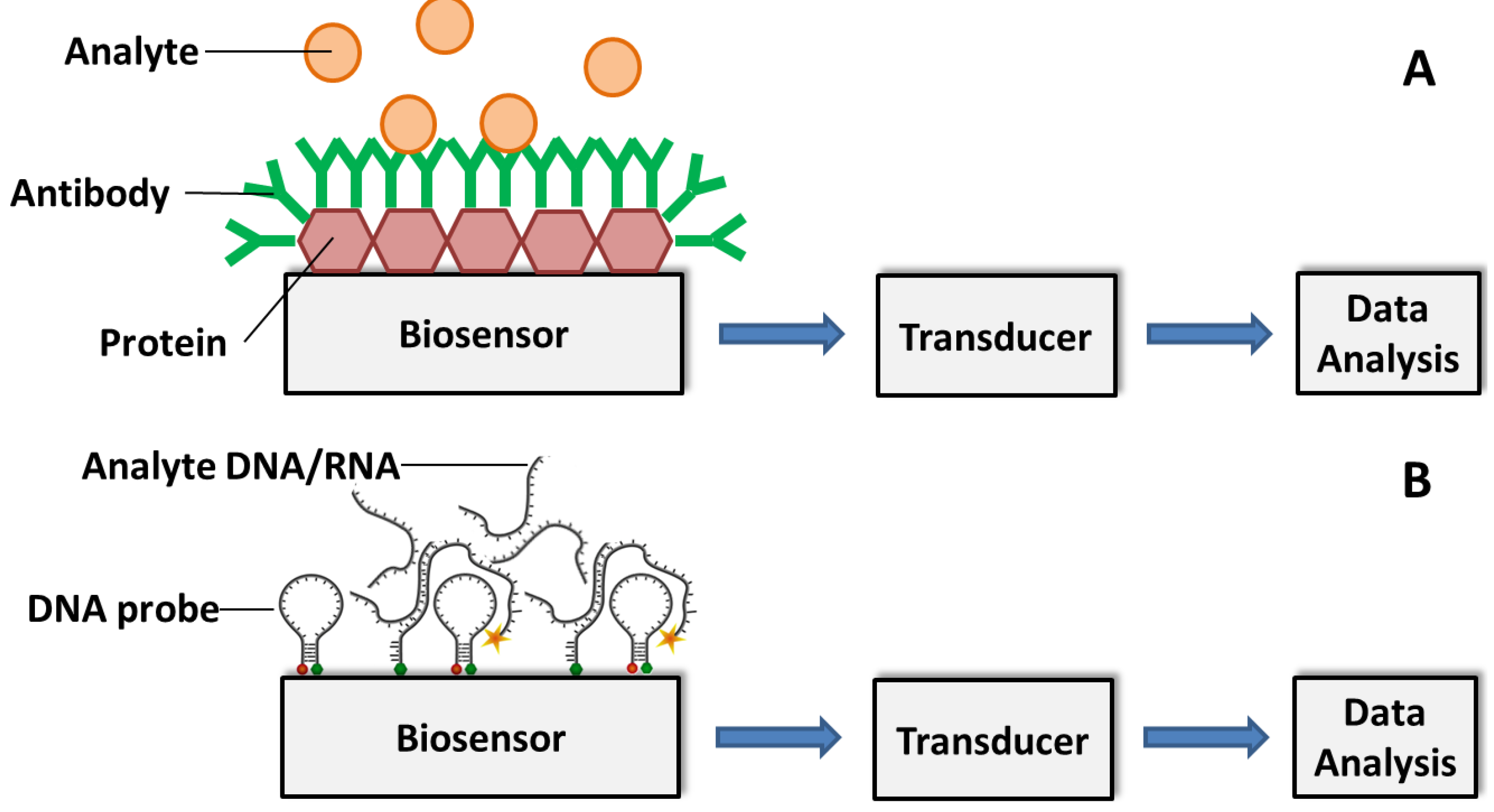
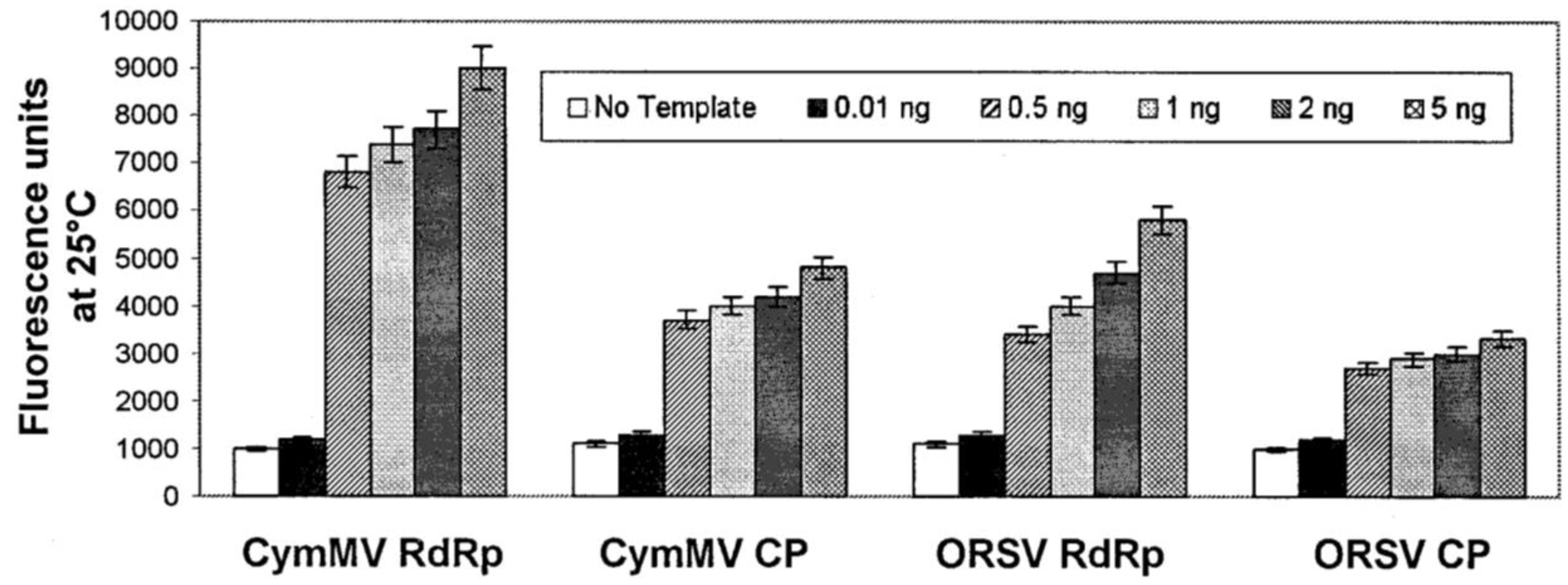
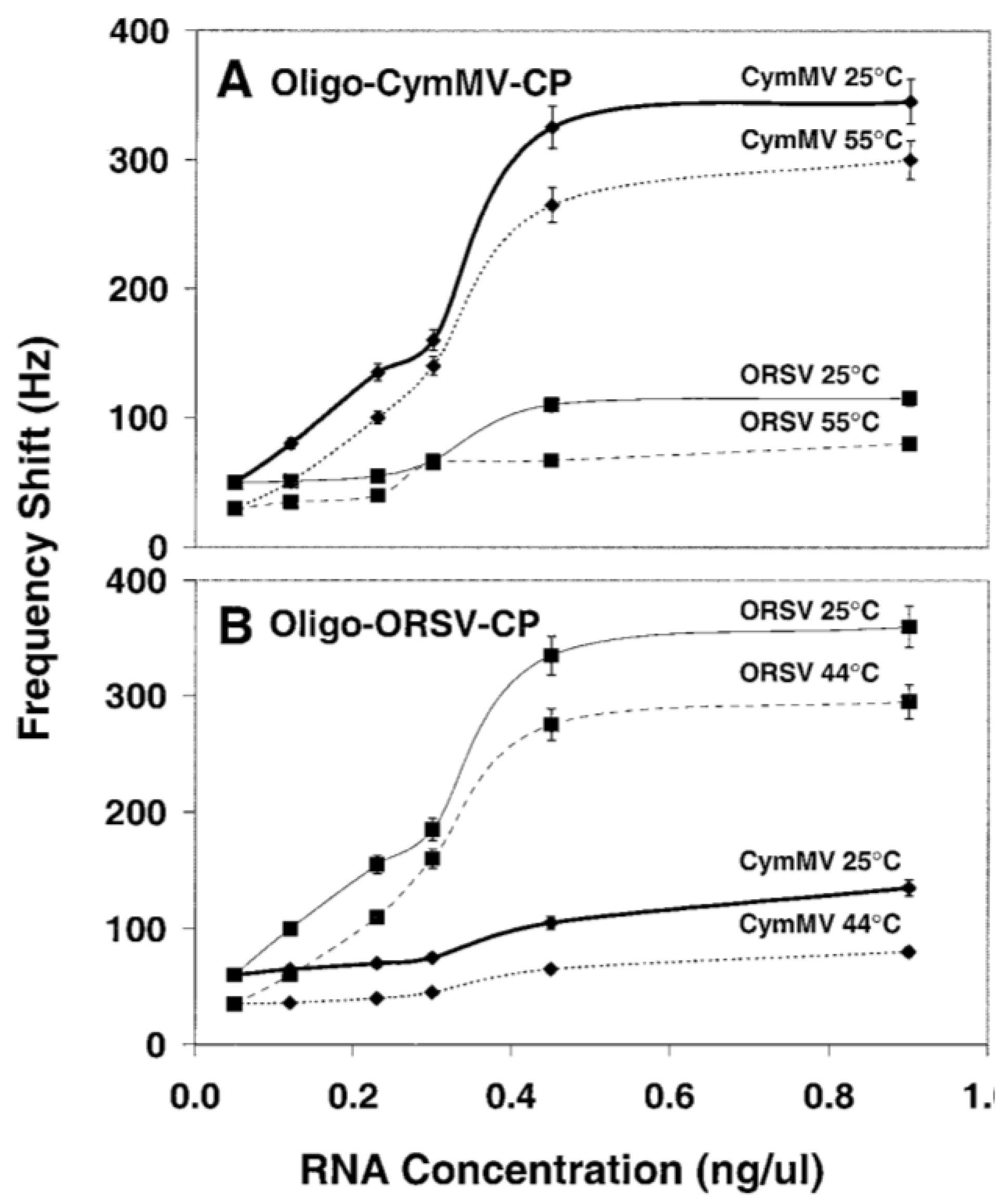
3.2.2. DNA/RNA-Based Affinity Biosensor
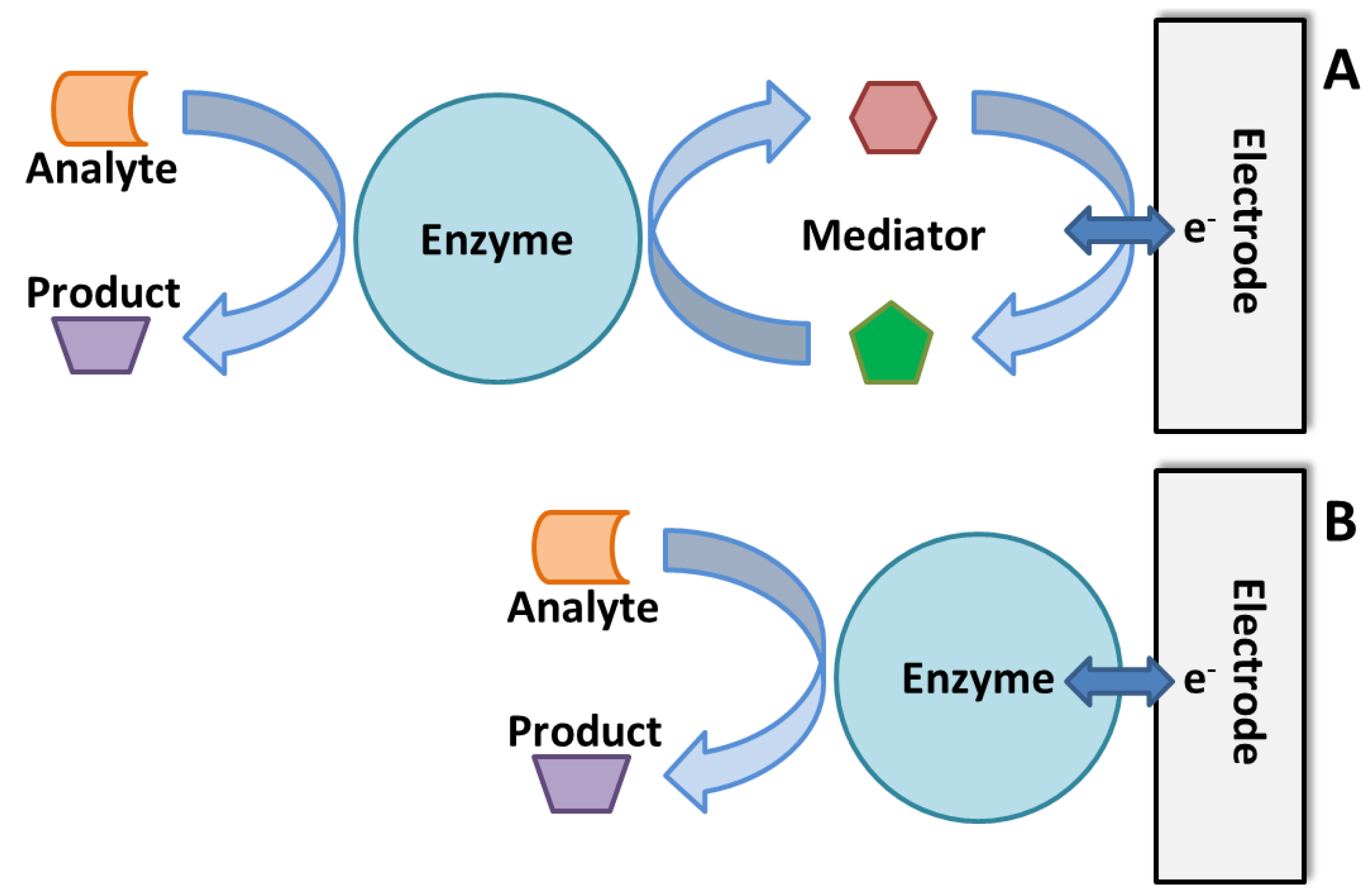
3.3. Enzymatic Electrochemical Biosensors
| Volatiles | Biotic Stress Causing Agents that Increase VOC Emissions |
|---|---|
| cis-3-hexen-1-ol | Botrytis cinerea, Spodoptera littoralis, Lirimyza huidobrensis, Spodoptera exigua, Manduca sexta, Macrosiphum euphorbiae, Helicoverpa armigera |
| trans-2-hexanal | Botrytis cinerea, Spodoptera littoralis, Lirimyza huidobrensis, Spodoptera exigua, Manduca sexta, Helicoverpa armigera |
| Methyl salicylate | Botrytis cinerea, Spodoptera littoralis, Tetranychus urticae, Manduca sexta, Macrosiphum euphoria, Tobacco mosaic virus |
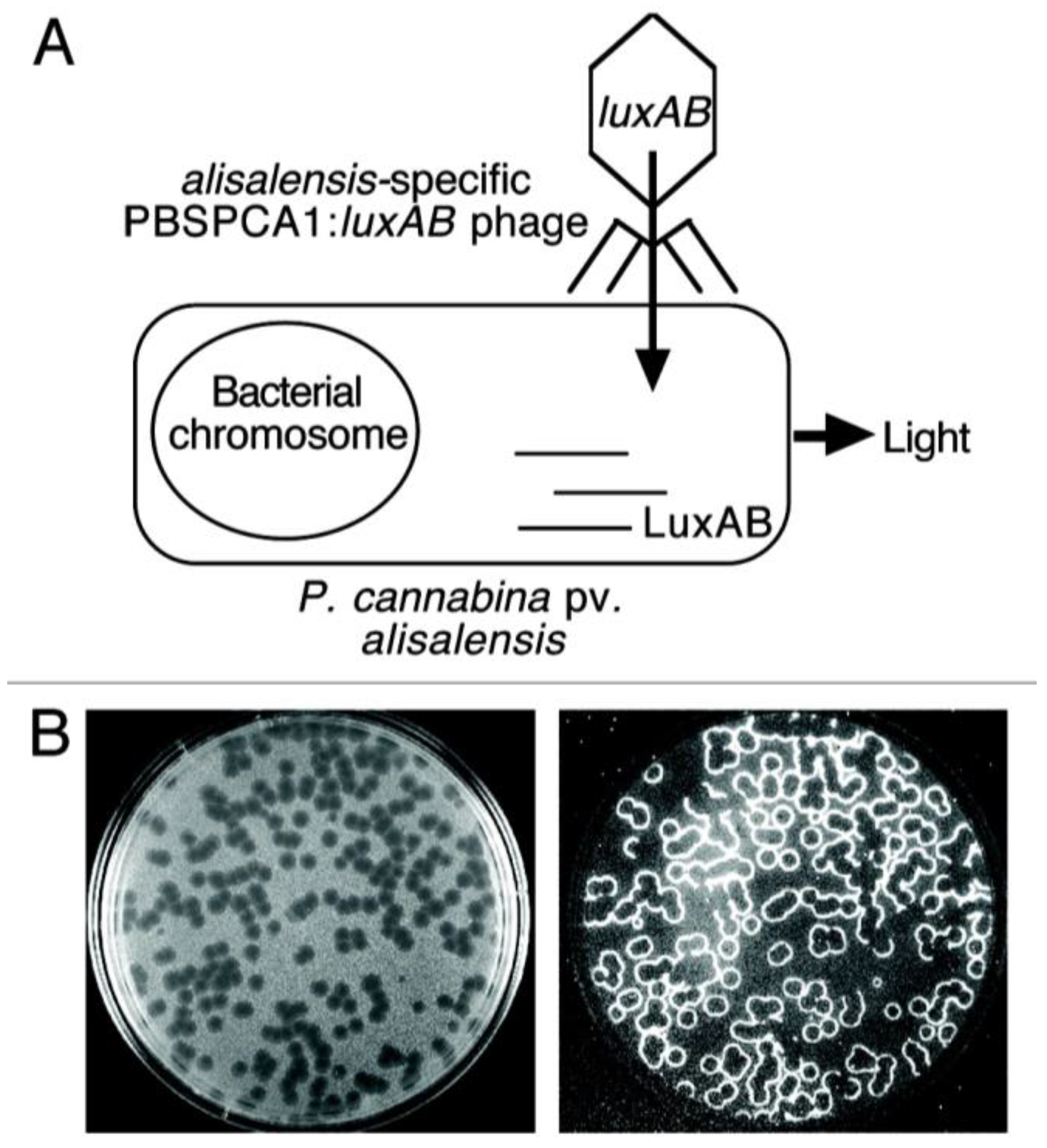
3.4. Bacteriophage-Based Biosensors
4. Challenges and Future Directions
| Techniques | Limit of Detection | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Nanomaterial sensor | 35 nM (p-ethylguaiacol) | Real-time, easy to fabricate, stable | Low specificity compared to enzyme, DNA, antibody based biosensors | [57] |
| Enzymatic biosensor | 0.98 µM (methyl salicylate) | High specificity, real-time | Interference from pH change | [105] |
| DNA-based piezoelectric biosensor | 10 ng (virus) | Low cost | Not real-time, DNA may need to be amplified | [100] |
| DNA-based molecular beacon biosensor | 0.5 ng (virus) | Real-time, low limit of detection | High cost, DNA may need to be amplified | [118] |
| Antibody-based quartz crystal biosensor | 10 ng (virus) | Low cost | Not real-time | [86] |
| ELISA | 2.5 ng (virus) | Low cost, low limit of detection for virus | Not real-time for bacteria, poor sensitivity for bacteria | [119] |
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ingram, J. A food systems approach to researching food security and its interactions with global environmental change. Food Secur. 2011, 3, 417–431. [Google Scholar] [CrossRef]
- Keinan, A.; Clark, A.G. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 2012, 336, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Senauer, B.; Vaclav, S. Feeding the World: A Challenge for the Twenty-First Century. Available online: http://onlinelibrary.wiley.com/doi/10.1111/j.1728-4457.2000.00827.x/pdf (accessed on 5 January 2015).
- Rosset, P. Food sovereignty and the contemporary food crisis. Development 2008, 51, 460–463. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Conway, G. One Billion Hungry: Can We Feed the World? Cornell University Press: Ithaca, NY, USA, 2012. [Google Scholar]
- Savary, S.; Ficke, A.; Aubertot, J.; Hollier, C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Pimentel, D.; Zuniga, R.; Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 2005, 52, 273–288. [Google Scholar] [CrossRef]
- Roberts, M.J.; Schimmelpfennig, D.E.; Ashley, E.; Livingston, M.J.; Ash, M.S.; Vasavada, U. The Value of Plant Disease Early-Warning Systems: A Case Study of USDA’s Soybean Rust Coordinated Framework; United States Department of Agriculture, Economic Research Service: Washington, DC, USA, 2006.
- Cai, H.; Caswell, J.; Prescott, J. Nonculture molecular techniques for diagnosis of bacterial disease in animals a diagnostic laboratory perspective. Vet. Pathol. Online 2014, 51, 341–350. [Google Scholar] [CrossRef] [PubMed]
- López, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Corris, M.T.; Llop, P.; Renyalver, R.; Cambra, M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef] [PubMed]
- James, D. A simple and reliable protocol for the detection of apple stem grooving virus by RT-PCR and in a multiplex PCR assay. J. Virol. Methods 1999, 83, 1–9. [Google Scholar] [CrossRef]
- Nassuth, A.; Pollari, E.; Helmeczy, K.; Stewart, S.; Kofalvi, S.A. Improved RNA extraction and one-tube RT-PCR assay for simultaneous detection of control plant RNA plus several viruses in plant extracts. J. Virol. Methods 2000, 90, 37–49. [Google Scholar] [CrossRef]
- Osiowy, C. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J. Clin. Microbiol. 1998, 36, 3149–3154. [Google Scholar] [PubMed]
- Pallisgaard, N.; Hokland, P.; Riishøj, D.C.; Pedersen, B.; Jørgensen, P. Multiplex reverse transcription-polymerase chain reaction for simultaneous screening of 29 translocations and chromosomal aberrations in acute leukemia. Blood 1998, 92, 574–588. [Google Scholar] [PubMed]
- Williams, K.; Blake, S.; Sweeney, A.; Singer, J.T.; Nicholson, B.L. Multiplex reverse transcriptase PCR assay for simultaneous detection of three fish viruses. J. Clin. Microbiol. 1999, 37, 4139–4141. [Google Scholar] [PubMed]
- Lievens, B.; Brouwer, M.; Vanachter, A.C.R.C.; Cammue, B.P.A.; Thomma, B.P.H.J. Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 2006, 171, 155–165. [Google Scholar] [CrossRef]
- Schaad, N.W.; Frederick, R.D. Real-time PCR and its application for rapid plant disease diagnostics. Can. J. Plant Pathol. 2002, 24, 250–258. [Google Scholar] [CrossRef]
- Van der Wolf, J.; van Bechhoven, J.R.C.M.; Bonants, P.J.M.; Schoen, C.D. New technologies for sensitive and specific routine detection of plant pathogenic bacteria. In Plant Pathogenic Bacteria; Springer: Berlin, Germany, 2001; pp. 75–77. [Google Scholar]
- Kempf, V.A.; Trebesius, K.; Autenrieth, I.B. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J. Clin. Microbiol. 2000, 38, 830–838. [Google Scholar] [PubMed]
- Hijri, M. The use of Fluorescent in situ hybridisation in plant fungal identification and genotyping. In Plant Pathology; Springer: Berlin, Germany, 2009; pp. 131–145. [Google Scholar]
- Kliot, A.; Kontsedalov, S.; Lebedev, G.; Brumin, M.; Cathrin, P.B.; Marubayashi, J.M.; Skaljac, M.; Belausov, E.; Czosnek, H.; Ghanim, M. Fluorescence in situ hybridizations (FISH) for the localization of viruses and endosymbiotic bacteria in plant and insect tissues. J. Vis. Exp. 2014, 84, e51030. [Google Scholar] [CrossRef] [PubMed]
- Moter, A.; Göbel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- DeLong, E.F.; Wickham, G.S.; Pace, N.R. Phylogenetic stains: Ribosomal RNA-based probes for the identification of single cells. Science 1989, 243, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993, 14, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.F.; Adams, A. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.T.; Alarcon, B.; Lopez, M.; Cambra, M. Characterization of monoclonal antibodies specific for Erwinia carotovora subsp. atroseptica and comparison of serological methods for its sensitive detection on potato tubers. Appl. Environ. Microbiol. 1994, 60, 2076–2085. [Google Scholar] [PubMed]
- López, M.M.; Llop, P.; Cubero, J.; Penyalver, R.; Caruso, P.; Bertolini, E.; Penalver, J.; Gorris, M.T.; Cambra, M. Strategies for improving serological and molecular detection of plant pathogenic bacteria. In Plant Pathogenic Bacteria; Springer: Berlin, Germany, 2001; pp. 83–86. [Google Scholar]
- Ward, E.; Foster, S.J.; Fraaije, B.A.; Mccartney, H.A. Plant pathogen diagnostics: Immunological and nucleic acid-based approaches. Ann. Appl. Biol. 2004, 145, 1–16. [Google Scholar] [CrossRef]
- Dewey, F.; Marshall, G. Production and use of monoclonal antibodies for the detection of fungi. In Proceeding of British Crop Protection Council Symposium, Farnham, UK, 18–21 November 1996.
- Wullings, B.A.; VanBeuningen, A.R.; Janse, J.D.; Akkermans, A.D.L. Detection of Ralstonia solanacearum, which causes brown rot of potato, by fluorescent in situ hybridization with 23S rRNA-targeted probes. Appl. Environ. Microbiol. 1998, 64, 4546–4554. [Google Scholar] [PubMed]
- Chitarra, L.G.; van den Bulk, R.W. The application of flow cytometry and fluorescent probe technology for detection and assessment of viability of plant pathogenic bacteria. Eur. J. Plant Pathol. 2003, 109, 407–417. [Google Scholar] [CrossRef]
- Diaper, J.; Edwards, C. Flow cytometric detection of viable bacteria in compost. FEMS Microbiol. Ecol. 1994, 14, 213–220. [Google Scholar] [CrossRef]
- Porter, J.; Pickup, R.; Edwards, C. Evaluation of flow cytometric methods for the detection and viability assessment of bacteria from soil. Soil Biol. Biochem. 1997, 29, 91–100. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; Oberti, R.; West, J.; McCartney, A.; Bodria, L.; Ramon, H. Foliar Disease Detection in the Field Using Optical Sensor Fusion. Available online: http://ecommons.cornell.edu/bitstream/handle/1813/10394/FP%2004%20008%20Bravo-Moshou%20Final%2022Dec2004.pdf?sequence=1&isAllowed=y (accessed on 2 March 2015).
- Moshou, D.; Bravo, C.; Oberti, R.; West, J.; Bodria, A.; McCartney, A.; Ramon, H. Plant disease detection based on data fusion of hyper-spectral and multi-spectral fluorescence imaging using Kohonen maps. Real Time Imaging 2005, 11, 75–83. [Google Scholar] [CrossRef]
- Nilsson, H.-E. Remote sensing and image analysis in plant pathology. Can. J. Plant Pathol. 1995, 17, 154–166. [Google Scholar] [CrossRef]
- West, J.S.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, H.A. The potential of optical canopy measurement for targeted control of field crop diseases. Annu. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.K.; Oerke, E.; Steiner, U.; Dehne, H. Recent advances in sensing plant diseases for precision crop protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- Chaerle, L.; van der Straeten, D. Imaging techniques and the early detection of plant stress. Trends Plant Sci. 2000, 5, 495–501. [Google Scholar] [CrossRef]
- Chaerle, L.; Leinonen, I.; Jones, H.G.; Van Der Straeten, D. Monitoring and screening plant populations with combined thermal and chlorophyll fluorescence imaging. J. Exp. Bot. 2007, 58, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Lindenthal, M.; Steiner, U.; Dehne, H.W.; Oerke, E.C. Effect of downy mildew development on transpiration of cucumber leaves visualized by digital infrared thermography. Phytopathology 2005, 95, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.; Steiner, U.; Dehne, H.W.; Lindenthal, M. Thermal imaging of cucumber leaves affected by downy mildew and environmental conditions. J. Exp. Bot. 2006, 57, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.-C.; Fröhling, P.; Steiner, U. Thermographic assessment of scab disease on apple leaves. Precis. Agric. 2011, 12, 699–715. [Google Scholar] [CrossRef]
- Stoll, M.; Schultz, H.R.; Baecker, G.; Berkelmann-Loehnertz, B. Early pathogen detection under different water status and the assessment of spray application in vineyards through the use of thermal imagery. Precis. Agric. 2008, 9, 407–417. [Google Scholar] [CrossRef]
- Lindenthal, M. Visualisierung der Krankheitsentwicklung von Falschem Mehltau an Gurken durch Pseudoperonospora cubensis mittels Thermografie; Universitäts-und Landesbibliothek Bonn: Bonn, Germany, 2005. [Google Scholar]
- Hillnhütter, C.; Mahlein, A.K.; Sikora, R.A.; Oerke, E.-C. Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. Field Crops Res. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Bürling, K.; Hunsche, M.; Noga, G. Use of blue-green and chlorophyll fluorescence measurements for differentiation between nitrogen deficiency and pathogen infection in winter wheat. J. Plant Physiol. 2011, 168, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Kuckenberg, J.; Tartachnyk, I.; Noga, G. Temporal and spatial changes of chlorophyll fluorescence as a basis for early and precise detection of leaf rust and powdery mildew infections in wheat leaves. Precis. Agric. 2009, 10, 34–44. [Google Scholar] [CrossRef]
- Chaerle, L.; Lenk, S.; Leinonen, I.; Jones, H.G.; Van Der Straeten, D.; Dr., Buschmann, C. Multi-sensor plant imaging: Towards the development of a stress-catalogue. Biotechnol. J. 2009, 4, 1152–1167. [Google Scholar] [CrossRef] [PubMed]
- Cséfalvay, L.; Gaspero, G.D.; Matous, K.; Bellin, D.; Ruperti, B.; Olejnickova, J. Pre-symptomatic detection of Plasmopara viticola infection in grapevine leaves using chlorophyll fluorescence imaging. Eur. J. Plant Pathol. 2009, 125, 291–302. [Google Scholar] [CrossRef]
- Scholes, J.D.; Rolfe, S.A. Chlorophyll fluorescence imaging as tool for understanding the impact of fungal diseases on plant performance: A phenomics perspective. Funct. Plant Biol. 2009, 36, 880–892. [Google Scholar] [CrossRef]
- Delalieux, S.; van Aardt, J.; Keulemans, W.; Schrevens, E.; Coppin, P. Detection of biotic stress (Venturia inaequalis) in apple trees using hyperspectral data: Non-parametric statistical approaches and physiological implications. Eur. J. Agron. 2007, 27, 130–143. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kanda, E.; Kitada, K.; Ishiguro, K.; Torigoe, Y. Detection of rice panicle blast with multispectral radiometer and the potential of using airborne multispectral scanners. Phytopathology 2001, 91, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qin, Z.; Liu, X.; Ustin, S.L. Detection of stress in tomatoes induced by late blight disease in California, USA, using hyperspectral remote sensing. Int. J. Appl. Earth Observ. Geoinf. 2003, 4, 295–310. [Google Scholar] [CrossRef]
- Fang, Y.; Umasankar, Y.; Ramasamy, R.P. Electrochemical detection of p-ethylguaiacol, a fungi infected fruit volatile using metal oxide nanoparticles. Analyst 2014, 139, 3804–3810. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Grove, G. Fruit rots cause losses in Ohio strawberries. Ohio Rep. Res. Dev. 1982, 67, 3–4. [Google Scholar]
- Umasankar, Y.; Rains, G.C.; Ramasamy, R.P. Electroanalytical studies on green leaf volatiles for potential sensor development. Analyst 2012, 137, 3138–3145. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.M.; Hofstee, J.W.; Wildt, J.; Verstappen, F.W.; Bouwmeester, H.; van Henten, E.J. Induced plant volatiles allow sensitive monitoring of plant health status in greenhouses. Plant Signal. Behav. 2009, 4, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Isidorov, V.; Zenkevich, I.; Ioffe, B. Volatile organic compounds in the atmosphere of forests. Atmos. Environ. 1985, 19, 1–8. [Google Scholar] [CrossRef]
- Kesselmeier, J.; Staudt, M. Biogenic volatile organic compounds (VOC): An overview on emission, physiology and ecology. J. Atmos. Chem. 1999, 33, 23–88. [Google Scholar] [CrossRef]
- Perera, R.M.; Marriott, P.J.; Galbally, I.E. Headspace solid-phase microextraction—Comprehensive two-dimensional gas chromatography of wound induced plant volatile organic compound emissions. Analyst 2002, 127, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Shipway, A.N.; Katz, E.; Willner, I. Nanoparticle arrays on surfaces for electronic, optical, and sensor applications. Chem. Phys. Chem. 2000, 1, 18–52. [Google Scholar] [PubMed]
- Yao, K.S.; Li, S.J.; Tzeng, K.C.; Cheng, T.C.; Chang, C.Y.; Chiu, C.Y.; Liao, C.Y.; Hsu, J.J.; Lin, Z.P. Fluorescence silica nanoprobe as a biomarker for rapid detection of plant pathogens. Adv. Mater. Res. 2009, 79, 513–516. [Google Scholar] [CrossRef]
- Singh, S.; Singh, M.; Agrawal, V.V.; Kumar, A. An attempt to develop surface plasmon resonance based immunosensor for Karnal bunt (Tilletia indica) diagnosis based on the experience of nano-gold based lateral flow immuno-dipstick test. Thin Solid Films 2010, 519, 1156–1159. [Google Scholar] [CrossRef]
- López, M.M.; Llop, P.; Olmos, A.; Marco-Noales, E.; Cambra, M.; Bertolini, E. Are molecular tools solving the challenges posed by detection of plant pathogenic bacteria and viruses? Curr. Issues Mol. Biol. 2009, 11, 13–46. [Google Scholar] [PubMed]
- Frasco, M.F.; Chaniotakis, N. Semiconductor quantum dots in chemical sensors and biosensors. Sensors 2009, 9, 7266–7286. [Google Scholar] [CrossRef] [PubMed]
- Algar, W.R.; Krull, U.J. Quantum dots as donors in fluorescence resonance energy transfer for the bioanalysis of nucleic acids, proteins, and other biological molecules. Anal. Bioanal. Chem. 2008, 391, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Rad, F.; Mohsenifar, A.; Tabatabaei, M.; Safarnejad, M.R.; Shahryari, F.; Safarpour, H.; Foroutan, A.; Mardi, M.; Davoudi, D.; Fotokian, M. Detection of Candidatus Phytoplasma aurantifolia with a quantum dots fret-based biosensor. J. Plant Pathol. 2012, 94, 525–534. [Google Scholar]
- Safarpour, H.; Safarnejad, M.R.; Tabatabaei, M.; Mohsenifar, A.; Rad, F.; Basirat, M.; Shahryari, F.; Hasanzadeh, F. Development of a quantum dots FRET-based biosensor for efficient detection of Polymyxa betae. Can. J. Plant Pathol. 2012, 34, 507–515. [Google Scholar]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. Recent advances in graphene-based biosensors. Biosens. Bioelectron. 2011, 26, 4637–4648. [Google Scholar] [CrossRef] [PubMed]
- Pérez-López, B.; Merkoçi, A. Nanoparticles for the development of improved (bio) sensing systems. Anal. Bioanal. Chem. 2011, 399, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Shiddiky, M.J.; Torriero, A.A. Application of ionic liquids in electrochemical sensing systems. Biosens. Bioelectron. 2011, 26, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ye, Y.; Liu, S. Gold nanoparticle-based signal amplification for biosensing. Anal. Biochem. 2011, 417, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Mandler, D.; Kraus-Ophir, S. Self-assembled monolayers (SAMs) for electrochemical sensing. J. Solid State Electrochem. 2011, 15, 1535–1558. [Google Scholar] [CrossRef]
- Umasankar, Y.; Ramasamy, R.P. Highly sensitive electrochemical detection of methyl salicylate using electroactive gold nanoparticles. Analyst 2013, 138, 6623–6631. [Google Scholar] [CrossRef] [PubMed]
- Boonham, N.; Glover, R.; Tomlinson, J.; Mumford, R. Exploiting generic platform technologies for the detection and identification of plant pathogens. In Sustainable Disease Management in a European Context; Springer: Berlin, Germany, 2008; pp. 355–363. [Google Scholar]
- Chartuprayoon, N.; Rheem, Y.; Chen, W.; Myung, N. Detection of plant pathogen using LPNE grown single conducting polymer Nanoribbon. In Meeting Abstracts; The Electrochemical Society: Pennington, NJ, USA, 2010. [Google Scholar]
- Sadanandom, A.; Napier, R.M. Biosensors in plants. Curr. Opin. Plant Biol. 2010, 13, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O’Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzyme Microb. Technol. 2003, 32, 3–13. [Google Scholar] [CrossRef]
- Palchetti, I.; Mascini, M. Electroanalytical biosensors and their potential for food pathogen and toxin detection. Anal. Bioanal. Chem. 2008, 391, 455–471. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.; Stack, E.; Gilmartin, N.; O’Kennedy, R. Antibody-based sensors: Principles, problems and potential for detection of pathogens and associated toxins. Sensors 2009, 9, 4407–4445. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Huang, X.; Xu, J.; Li, G.; Ma, J.; Ji, H.F.; Zhu, S.; Chen, H. Rapid and sensitive detection of maize chlorotic mottle virus using surface plasmon resonance-based biosensor. Anal. Biochem. 2013, 440, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Eun, A.J.-C.; Huang, L.; Chew, F.-T.; Li, S.F.-Y.; Wong, S.-M. Detection of two orchid viruses using quartz crystal microbalance (QCM) immunosensors. J. Virol. Methods 2002, 99, 71–79. [Google Scholar] [CrossRef]
- Campbell, G.A.; Mutharasan, R. Detection of Bacillus anthracis spores and a model protein using PEMC sensors in a flow cell at 1 mL/min. Biosens. Bioelectron. 2006, 22, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Lot, H.; German-Retana, S.; Krause-Sakate, R.; Thomas, J.; Souche, S.; Delaunay, T.; Lanneau, M.; Le Gall, O. Analysis of the serological variability of Lettuce mosaic virus using monoclonal antibodies and surface plasmon resonance technology. J. Gen. Virol. 2007, 88, 2605–2610. [Google Scholar] [CrossRef] [PubMed]
- Dickert, F.L.; Hayden, O.; Bindeus, R.; Mann, K.-J.; Blaas, D.; Waigmann, E. Bioimprinted QCM sensors for virus detection—Screening of plant sap. Anal. Bioanal. Chem. 2004, 378, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Nugaeva, N.; Gfeller, K.Y.; Backmann, N.; Duggelin, M.; Lang, H.P.; Guntherodt, H.-J.; Hegner, M. An antibody-sensitized microfabricated cantilever for the growth detection of Aspergillus niger spores. Microsc. Microanal. 2007, 13, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Skottrup, P.; Frokiaer, H.; Hearty, S.; O’Kennedy, R.; Hejgaard, J.; Nicolaisen, M.; Justesen, A.F. Monoclonal antibodies for the detection of Puccinia striiformis urediniospores. Mycol. Res. 2007, 111, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Skottrup, P.; Hearty, S.; Frokiaer, H.; Leonard, P.; Hejgaard, J.; O’Kennedy, R.; Nicolaisen, M.; Justesen, A.F. Detection of fungal spores using a generic surface plasmon resonance immunoassay. Biosens. Bioelectron. 2007, 22, 2724–2729. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Ziegler, A.; Pittman, H.; Paterson, M.; Toth, R.; Eggleston, I. Oriented immobilisation of engineered single-chain antibodies to develop biosensors for virus detection. J. Virol. Methods 2006, 134, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Zezza, F.; Pascale, M.; Mule, G.; Visconti, A. Detection of Fusarium culmorum in wheat by a surface plasmon resonance-based DNA sensor. J. Microbiol. Methods 2006, 66, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-Y.; Huang, C.-H.; Lu, S.-H.; Kuo, I.-T.; Chau, L.-K. Direct detection of orchid viruses using nanorod-based fiber optic particle plasmon resonance immunosensor. Biosens. Bioelectron. 2014, 51, 371–378. [Google Scholar] [CrossRef] [PubMed]
- James, C. Polypyrrole nanoribbon based chemiresistive immunosensors for viral plant pathogen detection. Anal. Methods 2013, 5, 3497–3502. [Google Scholar]
- Perdikaris, A.; Vassilakos, N.; Yiakoumettis, I.; Kektsidou, O.; Kintzios, S. Development of a portable, high throughput biosensor system for rapid plant virus detection. J. Virol. Methods 2011, 177, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dash, S.K.; Suman, D.P.S. DNA based biosensors for detection of pathogens. In Plant Fungal Disease Management, 1st ed.; Westville: New York, NY, USA, 2015; pp. 31–35. [Google Scholar]
- Eun, A.J.-C.; Wong, S.-M. Molecular beacons: A new approach to plant virus detection. Phytopathology 2000, 90, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Eun, A.J.-C.; Huang, L.; Chew, F.-T.; Li, S.F.-Y.; Wong, S.-M. Detection of two orchid viruses using quartz crystal microbalance-based DNA biosensors. Phytopathology 2002, 92, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E.; Stricker, S. Application of electrochemical biosensors for detection of food pathogenic bacteria. Electroanalysis 2000, 12, 317–325. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Haccoun, J.; Piro, B.; Tran, L.D.; Dang, L.A.; Pham, M.C. Reagentless amperometric detection of L-lactate on an enzyme-modified conducting copolymer poly (5-hydroxy-1,4-naphthoquinone-co-5-hydroxy-3-thioacetic acid-1,4-naphthoquinone). Biosens. Bioelectron. 2004, 19, 1325–1329. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Umasankar, Y.; Ramasamy, R.P. Plant Volatile Sensor: Enzymatic Transducer for Selective and Sensitive Determination of Methyl Salicyalte. In Meeting Abstracts; The Electrochemical Society: Pennington, NJ, USA, 2014. [Google Scholar]
- Kulagina, N.V.; Shankar, L.; Michael, A.C. Monitoring glutamate and ascorbate in the extracellular space of brain tissue with electrochemical microsensors. Anal. Chem. 1999, 71, 5093–5100. [Google Scholar] [CrossRef] [PubMed]
- Mc Grath, S.; van Sinderen, D. Bacteriophage: Genetics and Molecular Biology; Horizon Scientific Press: Norfolk, UK, 2007. [Google Scholar]
- Brigati, J.R.; Petrenko, V.A. Thermostability of landscape phage probes. Anal. Bioanal. Chem. 2005, 382, 1346–1350. [Google Scholar] [CrossRef] [PubMed]
- Kretzer, J.W.; Lehmann, R.; Schmelcher, M.; Banz, M.; Kim, K.-P.; Korn, C.; Loessner, M.J. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 2007, 73, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, T.; Schwartz-Mittelmann, A.; Biran, D.; Ron, E.Z.; Rishpon, J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal. Chem. 2003, 75, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; van Vaerenbergh, J.; Vandenheuvel, D.; Dunon, V.; Ceyssens, P.-J.; de Proft, M.; Kropinski, A.M.; Noben, J.-P.; Maes, M.; Lavigne, R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by “Dickeya solani”. PLoS ONE 2012, 7, e33227. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl. Environ. Microbiol. 2011, 77, 4155–4162. [Google Scholar] [CrossRef] [PubMed]
- Tlili, C.; Sokullu, E.; Safavieh, M.; Tolba, M.; Ahmed, M.U.; Zourob, M. Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal. Chem. 2013, 85, 4893–4901. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Bull, C.T.; Rubio, I.; Wechter, W.P.; Westwater, C.; Molineux, I.J. “Light-tagged” bacteriophage as a diagnostic tool for the detection of phytopathogens. Bioengineered 2013, 4, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Askora, A.; Kawasaki, T.; Usami, S.; Fujie, M.; Yamada, T. Host recognition and integration of filamentous phage ϕRSM in the phytopathogen, Ralstonia solanacearum. Virology 2009, 384, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Frampton, R.A.; Taylor, C.; Moreno, A.V.H.; Visnovsky, S.B.; Petty, N.K.; Pitman, A.R.; Fineran, P.C. Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Appl. Environ. Microbiol. 2014, 80, 2216–2228. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Kawasaki, T.; Nagata, S.; Fujiwara, A.; Usami, S.; Fujie, M. New bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 2007, 153, 2630–2639. [Google Scholar] [CrossRef] [PubMed]
- Eun, A.J.-C.; Wong, S.-M. Detection of cymbidium mosaic potexvirus and odontoglossum ringspot tobamovirus using immuno-capillary zone electrophoresis. Phytopathology 1999, 89, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Bryant, G.; Durand, D. Detection of plant virus by using purified IgG in ELISA. J. Virol. Methods 1981, 3, 27–35. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.; Ramasamy, R.P. Current and Prospective Methods for Plant Disease Detection. Biosensors 2015, 5, 537-561. https://doi.org/10.3390/bios5030537
Fang Y, Ramasamy RP. Current and Prospective Methods for Plant Disease Detection. Biosensors. 2015; 5(3):537-561. https://doi.org/10.3390/bios5030537
Chicago/Turabian StyleFang, Yi, and Ramaraja P. Ramasamy. 2015. "Current and Prospective Methods for Plant Disease Detection" Biosensors 5, no. 3: 537-561. https://doi.org/10.3390/bios5030537
APA StyleFang, Y., & Ramasamy, R. P. (2015). Current and Prospective Methods for Plant Disease Detection. Biosensors, 5(3), 537-561. https://doi.org/10.3390/bios5030537






